Abstract
Background
Human respiratory syncytial virus (hRSV) detection in nasopharyngeal aspirates (NPAs) from infants with acute respiratory tract infection (ARTI) does not prove the hRSV etiology of the current ARTI episode. HRSV RNA quantification may help in affording this issue.
Objectives
hRSV was detected by quantitative reverse transcription-PCR in NPAs taken upon admission to hospital and, whenever possible, at discharge and subsequent medical visits.
Study design
Prospective study, including 63 infants affected by either hRSV upper or lower ARTI.
Results
Based on the kinetics of viral load, hRSV etiology was identified in 25 infants in whom hRSV load dropped from 2.5 × 106 upon admission (presence of respiratory symptoms) to 7.5 × 102 RNA copies/ml NPA upon discharge (absence of symptoms) after a median time of 5 days, and in 19 infants, in whom hRSV load was determined at admission only, in association with clinical symptoms (2.4 × 106 copies/ml). Furthermore, low levels of hRSV RNA (<1 × 105 copies/ml NPA) identified 14 patients with non-hRSV ARTI. Finally, in 14 infants with hRSV coinfections or sequential infections, hRSV quantification defined the hRSV role in the current ARTI episode.
Conclusions
hRSV RNA quantification is critical in defining the hRSV role in respiratory infections.
Abbreviations: hRSV, human respiratory syncytial virus; NPA, nasopharyngeal aspirate; ARTI, acute respiratory tract infection; CC, cell culture; DFA, direct fluorescent antibody; RT-PCR, reverse transcription-PCR
Keywords: Respiratory syncytial virus, Viral respiratory infections, RT-PCR, hRSV RNA quantification
1. Introduction
Human respiratory syncytial virus (hRSV) is the major cause of acute respiratory tract infections (ARTI) in infants during their first year of life and has become the single most common cause of infant hospitalization (Boyce et al., 2000, Leader and Kohlhase, 2002, Shay et al., 1999). Diagnosis of hRSV infection in nasopharyngeal aspirate (NPA) secretions is currently achieved by cell culture (CC) (Falsey et al., 2002, Johnston and Siegel, 1990, Malley et al., 1998), by the direct fluorescent antibody (DFA) assay (Fong et al., 2000, Halstead et al., 1990, Landry and Ferguson, 2000, Rovida et al., 2005) or by reverse transcription-polymerase chain reaction (RT-PCR) (Falsey et al., 2003, Freymuth et al., 1995, Paton et al., 1992, Perkins et al., 2005). However, the sole presence of hRSV in NPA is not per se indicative of the etiologic involvement of the virus in the current episode of ARTI.
Comparable viral loads have been found in nasal secretions from the upper respiratory tract and tracheal secretions from the lower respiratory tract of children hospitalized for an ARTI (Buckingham et al., 2002, Malley et al., 1998, Perkins et al., 2005). In addition, hRSV subgroups A and B have been identified. Although this finding is not universally confirmed (Brouard et al., 1993, De Vincenzo, 2004), hRSV-A strains have been repeatedly shown to be shed at a higher titer than hRSV-B strains (Perkins et al., 2005) and to cause more severe diseases than hRSV-B infections (Mufson et al., 1991, Walsh et al., 1997).
In the present study, we quantified the hRSV RNA load (both subgroups A and B) in NPAs from hRSV-infected infants by quantitative RT-PCR to investigate the possibility of defining the etiologic role of hRSV in the current ARTI episode from a series of infants admitted to the hospital either with a single hRSV infection or with two or three simultaneous or sequential viral infections including hRSV.
2. Patients and methods
2.1. Patients
NPAs from a series of 253 infants aged less than 1 year and admitted to the hospital in the period November 2005–May 2006 with a diagnosis of ARTI were examined for hRSV as well as other respiratory viruses by DFA, CC, and qualitative RT-PCR, as reported (Rovida et al., 2005). NPAs were collected by trained personnel using a standardized procedure, as reported (De Vincenzo, 2004). In 63 hRSV-infected infants, hRSV RNA was quantified upon admission and, whenever possible, upon discharge, and at subsequent medical visits, as reported below. Overall, 49 infants had a single hRSV infection. In addition, 14 infants with multiple simultaneous (coinfections) or sequential (within 30 days) infections in the respiratory tract (including hRSV) were examined prospectively in order to identify the virus responsible for the current ARTI episode.
2.2. Quantitative RT-PCR assay
hRSV RNA was quantified by a single-step RT-PCR, using the SuperScript™ III Platinum® One-Step quantitative RT-PCR System (Invitrogen). Nucleic acids from NPAs were extracted using MINI MAG and NucliSens® Magnetic Extraction Reagents (BioMérieux, Lyon, France). Degenerated primers AB1 and AB2 targeting the gene F, as reported elsewhere (Coiras et al., 2003) were used to amplify both hRSV subgroups A and B. Using the same primer set, a plasmid containing the PCR product of the reference hRSV Long strain (subgroup A) cloned in TA-Cloning (Invitrogen) was serially diluted in the range of 105 to 100 input copies to construct external reference standards. In addition, an internal RNA control was used, consisting of a predetermined amount of a synthetic heterologous RNA sequence (Armored RNA®, Ebola Virus, subtype Zaire, Ambion RNA Diagnostic, Ambion, Inc., Austin, TX). Armored RNA® was extracted in parallel with clinical samples and plasmid serial dilutions, and a fixed amount of the extracted RNA (100 copies) was added to both RT-PCR reaction mixtures (samples and plasmids) containing, besides the primer pair hRSV AB1 (10 μM) and hRSV AB2 (10 μM), Ebola-specific primers ArmEBOFor (10 μM) and ArmEBORev (10 μM) (Sanchez et al., 1999). The thermal profile was: 10 cycles at 95 °C for 30 s, 55 °C for 2 min and 68 °C for 1 min + 3 s/cycle; then, 40 cycles at 95 °C for 30 s, 50 °C for 2 min, and 68 °C for 1 min + 3 s/cycle. Amplicons were visualized in 3% agarose gel, and quantified densitometrically. By using the Quantity One Programme software (Bio-Rad, Hercules, CA), external standards normalized with the internal control allowed construction of a standard curve, from which the RNA copy number of test samples was derived. The sensitivity of the assay was 10 DNA copies/10 μl for both subgroups A and B. Quantitative hRSV results were expressed as RNA copy number/ml NPA following data multiplication by 100.
2.3. hRSV subgrouping
The hRSV subgroup was initially determined on a series of 10 subgroup A and 10 subgroup B hRSV strains by two-step RT-PCR targeting the gene F (nt 1–705) for the first step, and the gene F fragments nt 347–682 for subgroup A, and nt 30–614 for subgroup B strains for the second step, as previously reported (Rovida et al., 2005), in agreement with Coiras et al. (2003). Subgrouping results were totally overlapping with those obtained by DFA staining of NPA cells by monoclonal antibodies MAB8581 and MAB8582 (Chemicon, Temecula, CA).
2.4. Statistics
Comparison between medians was done by using the Mann–Whitney U-test for unpaired data, while the Wilcoxon test was used for paired data. Distribution of frequencies was examined by the chi-square test.
3. Results
3.1. Comparative sensitivity of RT-PCR targeting the gene F in detecting hRSV plasmid DNA and RNA of both subgroups A and B
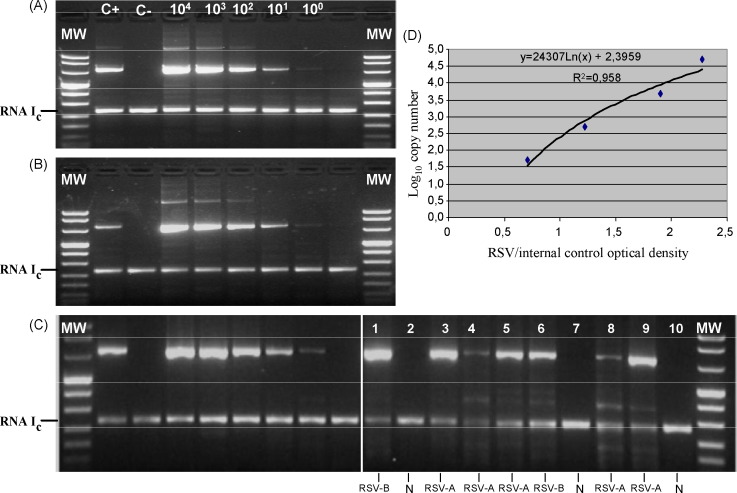
DNA plasmids containing the amplified region were constructed for both subgroups A and B, showing that the PCR method has a sensitivity of at least 10 DNA copies for each of the two subgroups (Fig. 1 ). In repeated experiments, at least 10 Ebola Armored RNA copies could be consistently retrotranscribed and amplified (Fig. 1A–C). Potential variation in RT-PCR hRSV efficiency amplification of clinical samples was compensated through the normalization of target (hRSV) densitometric signals by internal control (Ebola) signals (Fig. 1C).
Fig. 1.
Sensitivity of the quantitative RT-PCR method in detecting: (A) hRSV subgroup A plasmid; (B) hRSV subgroup B plasmid dilutions; and (C): C+, C− and plasmid subgroup A dilutions used for quantification; lanes 1–10: clinical subgroup A (lanes 3, 4, 5, 8, and 9) and B (lanes 1 and 6) strains. (D) Quantification curve. MW, molecular weight markers; N (lanes 2, 7, 10), negative samples; RNA Ic, armored RNA internal control; C+, positive control; C−, negative control.
3.2. Epidemiology and diagnosis of hRSV infections
Overall, of the 253 infants (399 NPAs) examined, 186 (73.5%) were found positive for a respiratory virus (278 NPAs, 69.7%). In detail, hRSV was the predominant infection, involving a total of 63 infants, i.e. 49 infants with a single infection, and 14 infants with a coinfection/sequential coinfection. The incidence of the other viral infections (for both infants and NPAs examined) is reported in Table 1 .
Table 1.
Incidence of respiratory viral infections of different etiology in the infant population admitted to hospital in the winter–spring season 2005–2006
| Parameters | No. (%) |
|
|---|---|---|
| Infants | NPAs | |
| No. examined | 253 | 399 |
| No. positive | 186 (74) | 322 (81) |
| hRSV | 49 (26) | 91 (29) |
| Rhino | 42 (23) | 60 (20) |
| hCoV | 13 (7) | 19 (6) |
| hMPV | 9 (5) | 17 (5) |
| hPIV | 9 (5) | 11 (3) |
| Influenza | 7 (4) | 11 (3) |
| AdV | 7 (4) | 16 (4) |
| Sequential infectionsa | 12 (6) | 18 (6) |
| Coinfectionsb | 19 (10) | 53 (17) |
| Sequential coinfectionsc | 19 (10) | 26 (7) |
hRSV, human respiratory syncytial virus; rhino, rhinoviruses; hCoV, human coronaviruses; hMPV, human metapneumoviruses; hPIV, human parainfluenzaviruses; influenza, influenzaviruses A and B; AdV, adenoviruses; NPA, nasopharyngeal aspirate.
Four involving hRSV. Sequential infections indicate viral respiratory infections caused by single viruses within 30 days of follow-up after hospital admission.
Five involving hRSV. Coinfections indicate a single simultaneous detection of two or more viruses in the respiratory tract of the same infant during follow-up.
Five involving hRSV. Sequential coinfections indicate simultaneous detection of two or more viruses at least twice during follow-up.
Of a total of 134 NPAs examined for hRSV by CC, DFA and RT-PCR, 19 were found to be negative by each assay (Table 2 ). Of the 115 samples found positive, 78 were positive by CC, 98 by DFA, and 102 by RT-PCR. In addition, the circulation rate of hRSV-A strains (n = 96) was found to be about five times higher than that of hRSV-B strains (n = 19) among children admitted to hospital in the winter-spring season 2005–2006 in our area.
Table 2.
Comparison of hRSV detection rates by CC, DFA and RT-PCR in 134 NPAs
| No. of NPAs |
Result combinationa |
|||||
|---|---|---|---|---|---|---|
| hRSV-A | hRSV-B | Total | CC | DFA | RT-PCR | |
| 58 | 9 | 67 | + | + | + | |
| 13 | 6 | 19 | − | + | + | |
| 2 | 1 | 3 | + | − | + | |
| 7 | 0 | 7 | + | + | − | |
| 5 | 0 | 5 | − | + | − | |
| 11 | 2 | 13 | − | − | + | |
| 0 | 1 | 1 | + | − | − | |
| 0 | 0 | 19 | − | − | − | |
| Total pos. | 96 | 19 | 134 | 78 | 98 | 102 |
+, positive; −, negative; CC, cell culture; DFA, direct fluorescent antibody staining; RT-PCR, reverse transcription-PCR.
3.3. Identification of hRSV as the causative agent of ARTI
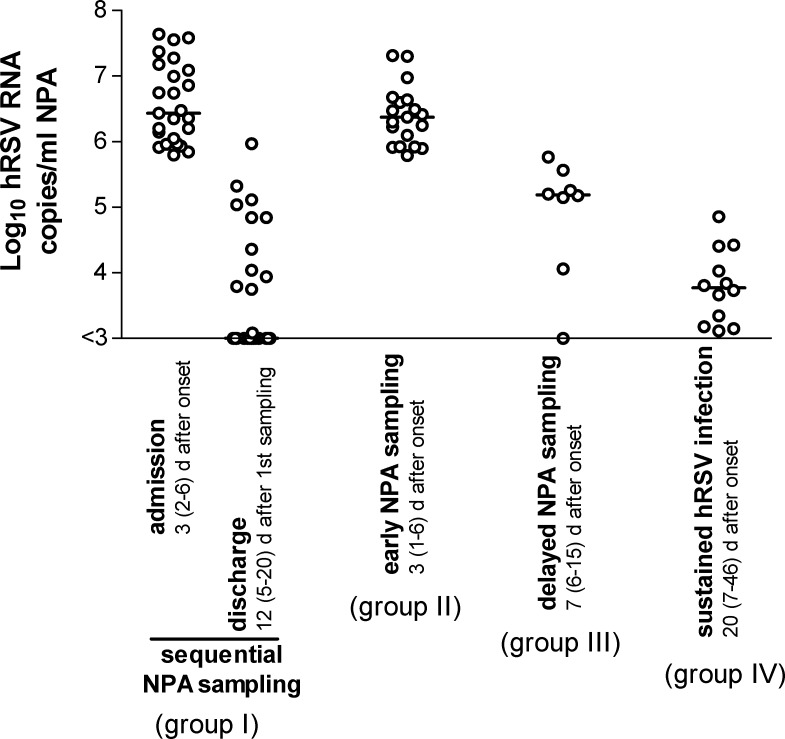
We were able to identify, among the 63 infants admitted to hospital with an acute episode of ARTI, four different clinical situations involving hRSV, which are reported in summary in Fig. 2 .
Fig. 2.
Individual and median levels of viral load in the four groups of infants examined upon admission to hospital and during follow-up. For group IV, both minimal and maximal viral loads observed in NPAs during the follow-up period of six infants are reported.
The first involved 25 infants, in whom the sharp drop in hRSV RNA load from a high level (2.5 × 106 RNA copies/ml NPA, range 6.30 × 105 to 4.35 × 107) a median time of 3 (range 1–5) days after onset of symptoms, to a low level (7.5 × 102 RNA copies: range 3.0 × 102 to 9.3 × 105) a median time of 8 (range 5–20) days after admission, unequivocally documented the resolution or attenuation of clinical symptoms, in association with the hRSV etiology of the ARTI episode (group I).
The second interested 19 additional infants, in whom the high hRSV load at admission (2.4 × 106 RNA copies/ml NPA; range 6.2 × 105 to 2.1 × 107) at a median time of 3 (1–6) days after onset of symptoms, in association with clinical symptoms, indicated per se the hRSV etiology of ARTI (group II).
The third included eight infants admitted late (with respect to onset of hRSV infection) to hospital at a median time of 7 (range 7–15) days after onset of symptoms, with an intermediate viral load (1.4 × 105, range 3.0 × 102 to 3.7 × 105) which rapidly became negative, thus representing the late stage of an hRSV ARTI (group III).
Finally, six infants showed a sustained hRSV infection, with a low viral load in NPA (5.9 × 103, range 1.3 × 103 to 7.2 × 104 RNA copies/ml NPA) persistent at a median time of 20 (7–46) days after onset. Thus, in these patients hRSV was presumably not directly involved in the current episode of ARTI (group IV).
With the exception of median viral loads of groups I and II at admission, and groups IV and I at discharge (p > 0.05), all the other medians were significantly different from each other (p < 0.05).
3.4. Determination of the hRSV role in respiratory viral coinfections/sequential infections
Overall, 9/58 infants belonging to the four groups reported above, as well as five additional infants (within the total number of 63 infants examined), were found to be affected by hRSV coinfections or sequential infections.
Based on previously acquired criteria, the hRSV etiology could be either confirmed by high hRSV load (>105 RNA copies/ml NPA) at admission or a sharp drop in hRSV load between admission (>105 RNA copies/ml NPA) and discharge (<102 RNA copies/ml NPA), or excluded by detection of a stable low (<102 copies/ml NPA) or medium (103 to 104 copies/ml NPA) level of viral load at follow-up visits (Table 3 ). Respiratory viruses other than hRSV detected in hRSV coinfections and sequential infections are reported in Table 3.
Table 3.
Importance of hRSV RNA quantification at time of admission to hospital in defining the etiologic role of hRSV in 14 infants with hRSV coinfections or sequential infections during episodes of ARTI
| Pt # | hRSV VL (copies/ml NPA) | hRSV VL kinetics | Assessment of hRSV etiology | Other respiratory viruses detected and potentially responsible |
|---|---|---|---|---|
| 2 | High (>106) | Sharp drop | hRSV-A | AdV |
| 9 | Low (102) | ND | Ruled out | hCoV 229E |
| 25 | Low (102) | ND | Ruled out | AdV, rhino |
| 28 | High (>105) | ND | hRSV-A | Rhino |
| 29 | Medium (104) | Stable | Ruled out | hMPV-A, rhino |
| 30 | High (>105) | Sharp drop | hRSV-A | hMPV-B |
| 31 | Low (102) | Stable | Ruled out | hMPV-A |
| 33 | High (>107) | Sharp drop | hRSV-A | Rhino, influenza B |
| 34 | High (>106) | Sharp drop | hRSV-A | hCoV NL63 |
| 36 | High (>106) | ND | hRSV-A | Rhino |
| 39 | High (>105) | Sharp drop | hRSV-B | hCoV NL63 |
| 58 | High (>107) | Sharp drop | hRSV-A | Rhino |
| 61 | Low (102) | Stable | Ruled out | Parainfluenza 3 |
| 63 | Low (102) | ND | Ruled out | hCoV NL63 |
Pt#, patient number; VL, viral load; ND, not done; AdV, adenoviruses; hCoV, human coronaviruses; hMPV, human metapneumoviruses; rhino, rhinoviruses.
3.5. hRSV subgrouping
Overall, hRSV strains from 56 infants could be typed, and found to be either subgroup A (n = 45 strains) or subgroup B (n = 11 strains). Among the 44 infants in whom a high viral load in NPA was detected upon admission to hospital (group I + group II), the 33 infants infected by a subgroup A strain showed a median viral load of 1.77 × 106 RNA copies/ml NPA (range 6.30 × 105 to 4.35 × 107), while the nine infants infected by a subgroup B strain had a median viral load of 5.50 × 106 RNA copies/ml NPA (range 7.00 × 105 to 3.84 × 107). The difference was significant (p = 0.05). However, infants with subgroup B infections did not show a significantly greater severity of clinical symptoms (upper versus lower ARTIs) nor a greater duration of the stay in the hospital (8 days versus 9 days) as compared to infants with subgroup A infections.
4. Discussion
Two were the main objectives of this study investigating the impact of hRSV RNA quantification in NPAs on the diagnosis of hRSV ARTIs: (1) the definition or exclusion of hRSV etiology in episodes of ARTI at the time of infant admission to hospital; (2) the discrimination of the role of hRSV infection among infections by other respiratory viruses occurring simultaneously or sequentially in the same infant.
As for the definition of hRSV etiology, results of our study indicate that infants admitted with a respiratory syndrome caused by hRSV displayed hRSV RNA load greater than 105 RNA copies/ml NPA with a rapidly decreasing trend in the following days. In the past, experimental infection of adult healthy volunteers with hRSV had already shown that viral RNA does not persist for a prolonged period (Falsey et al., 2003). In addition, it was shown that high viral load peaks at admission rapidly decline during the following days (Buckingham et al., 2000). In this study, the kinetics observed in infants examined upon admission and at discharge from the hospital allowed the correct interpretation of findings observed both in infants examined only upon admission (high viral load indicated per se hRSV etiology even in the absence of a decreasing trend) and in those admitted late (with respect to hRSV infection) to hospital with a low hRSV load rapidly declining.
Virus persistence at a low level was often associated with detection of coinfections, i.e. multiple infections simultaneously occurring in the same patient. In some infants reported in this study with a sustained low level of hRSV RNA load, quantification of hMPV (a recently discovered virus often causing respiratory syndromes similar to those caused by hRSV) was sufficient to identify the virus responsible for the current episode of ARTI (data not reported). In case of coinfection, quantification of all viruses present in the same NPA sample may be the sole approach available for the identification of the virus actually causing the acute respiratory syndrome.
As for the different pathogenicity of hRSV subgroups A and B, it was reported that subgroup A strains may yield significantly higher viral loads than subgroup B strains (Perkins et al., 2005) and may cause clinical syndromes of greater severity (Hall et al., 1990, Mufson et al., 1991, Walsh et al., 1997). In this study, we had a predominance of subgroup A infections. Within these limits, we observed a significantly higher median viral load for subgroup B infections, which was not associated with greater clinical severity or a longer duration of the hospital stay.
In conclusion, results of our study emphasize the need for the quantification of hRSV RNA load in order to establish the etiologic role of the virus in the current ARTI episode. Due to the high number of respiratory viruses causing similar syndromes in the infantile patient population, the discrimination among the different agents is a necessary requirement for the performance of both accurate epidemiological and clinical surveys and may become critical for the correct administration of specific antiviral drugs.
Acknowledgements
We thank all the technical staff of the Servizio di Virologia for performing all the assays. We also are grateful to Daniela Sartori for preparing the manuscript, and to Laurene Kelly for revision of the English. This work was partially supported by Ministero della Salute, Ricerca Finalizzata Fondazione IRCCS Policlinico San Matteo, grants 89282(05)/A and 89288(05).
References
- Boyce T.G., Mellen B.G., Mitchel E.F., Jr, Wright P.F., Griffin M.R. Rates of hospitalization for respiratory syncytial virus infection among children in Medicaid. J Pediatr. 2000;137:865–870. doi: 10.1067/mpd.2000.110531. [DOI] [PubMed] [Google Scholar]
- Brouard J., Freymuth F., Constantini S., Petitjean J., de Schrevel G., Duhamel J.F. Prevalence and clinical aspects of A and B subgroups of respiratory syncytial virus infection. Observation of 8 consecutive epidemics between 1982 and 1990. Arch Fr Pediatr. 1993;50:639–643. [PubMed] [Google Scholar]
- Buckingham S., Bush A., De Vincenzo J. Nasal quantity of respiratory syncytial virus correlates with disease severity in hospitalized infants. Ped Infect Dis J. 2000;19:113–117. doi: 10.1097/00006454-200002000-00006. [DOI] [PubMed] [Google Scholar]
- Buckingham S.C., Jafri H.S., Bush A.J., Carubelli C.M., Sheeran P., Hardy R.D. A randomised, double-blind, placebo-controlled trial of dexamethasone in severe respiratory syncytial virus (RSV) infections: effects on RSV quantity and clinical outcome. J Infect Dis. 2002;185:1222–1228. doi: 10.1086/340024. [DOI] [PubMed] [Google Scholar]
- Coiras M.T., Perez-Breña P., García M.L., Casas I. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription-nested PCR assay. J Med Virol. 2003;69:132–144. doi: 10.1002/jmv.10255. [DOI] [PubMed] [Google Scholar]
- De Vincenzo J.P. Natural infection of infants with respiratory syncytial virus subgroups A and B: a study of frequency, disease severity and viral load. Pediatr Res. 2004;56:914–917. doi: 10.1203/01.PDR.0000145255.86117.6A. [DOI] [PubMed] [Google Scholar]
- Falsey A.R., Formica M.A., Walsh E.E. Diagnosis of respiratory syncytial virus infection: comparison of reverse transcription-PCR to viral culture and serology in adults with respiratory illness. J Clin Microbiol. 2002;40:817–820. doi: 10.1128/JCM.40.3.817-820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Formica M.A., Treanor J.J., Walsh E.E. Comparison of quantitative reverse transcription-PCR to viral culture for assessment of respiratory syncytial virus shedding. J Clin Microbiol. 2003;41:4160–4165. doi: 10.1128/JCM.41.9.4160-4165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong C.K.Y., Lee M.K., Griffith B.P. Evaluation of R-Mix fresh cells in shell vials for detection of respiratory viruses. J Clin Microbiol. 2000;38:4660–4662. doi: 10.1128/jcm.38.12.4660-4662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F., Eugene G., Vabret A., Petitjean J., Gennetay E., Brouard J. Detection of respiratory syncytial virus by reverse-transcription-PCR and hybridization with a DNA enzyme immunoassay. J Clin Microbiol. 1995;33:3352–3355. doi: 10.1128/jcm.33.12.3352-3355.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C.B., Walsh E.E., Schnabel K.C., Long C.E., McConnochie K.M., Hildreth S.W. Occurrence of groups A and B respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162:1283–1290. doi: 10.1093/infdis/162.6.1283. [DOI] [PubMed] [Google Scholar]
- Halstead D.C., Todd S., Fritch G. Evaluation of five methods for respiratory syncytial virus detection. J Clin Microbiol. 1990;28:1021–1025. doi: 10.1128/jcm.28.5.1021-1025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.L., Siegel C.S. Evaluation of direct immunofluorescence, enzyme immunoassay, centrifugation culture, and conventional culture for the detection of respiratory syncytial virus. J Clin Microbiol. 1990;28:2394–2397. doi: 10.1128/jcm.28.11.2394-2397.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry M.L., Ferguson D. SimulFluor respiratory screen for rapid detection of multiple respiratory viruses in clinical specimens by immunofluorescence staining. J Clin Microbiol. 2000;38:708–711. doi: 10.1128/jcm.38.2.708-711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader S., Kohlhase K. Respiratory syncytial virus-coded pediatric hospitalisation, 1997–1999. Pediatr Infect Dis J. 2002;21:629–632. doi: 10.1097/00006454-200207000-00005. [DOI] [PubMed] [Google Scholar]
- Malley R., De Vincenzo J., Ramilo O., Dennehy P.H., Meissner H.C., Gruber W.C. Reduction of respiratory syncytial virus (RSV) in tracheal aspirates in intubated infants using humanized monoclonal antibody to RSV F protein. J Infect Dis. 1998;178:1555–1561. doi: 10.1086/314523. [DOI] [PubMed] [Google Scholar]
- Mufson M.A., Akerlind-Stopner B., Orvell C., Belshe R.B., Norrby E. A single-season epidemic with respiratory syncytial virus subgroup B2 during 10 epidemic years, 1978 to 1988. J Clin Microbiol. 1991;29:162–165. doi: 10.1128/jcm.29.1.162-165.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton A.W., Paton J.C., Lawrence A.J., Goldwater P.N., Harris R.J. Rapid detection of respiratory syncytial virus in nasopharyngeal aspirates by reverse transcription and polymerase chain reaction amplification. J Clin Microbiol. 1992;30:901–904. doi: 10.1128/jcm.30.4.901-904.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S.M., Webb D.L., Torrance S.A., El Saleeby C., Harrison L.M., Aitken J.A. Comparison of a real-time reverse transcription PCR assay and a culture technique for quantitative assessment of viral load in children naturally infected with respiratory syncytial virus. J Clin Microbiol. 2005;43:2356–2362. doi: 10.1128/JCM.43.5.2356-2362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida F., Percivalle E., Zavattoni M., Torsellini M., Sarasini A., Campanini G. Monoclonal antibodies versus reverse transcription-PCR for detection of respiratory viruses in a patient population with respiratory tract infections admitted to hospital. J Med Virol. 2005;75:336–347. doi: 10.1002/jmv.20276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Ksiazed T.G., Rollin P.E., Miranda M.E., Trappier S.G., Khan A.S. Detection and molecular characterization of Ebola viruses causing disease in human and nonhuman primates. J Infect Dis. 1999;179(Suppl 1):S164–S169. doi: 10.1086/514282. [DOI] [PubMed] [Google Scholar]
- Shay D.K., Holman R.C., Newman R.D., Liu L.L., Stout J.W., Anderson L.J. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA. 1999;282:1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- Walsh E.E., McConnochie K.M., Long C.E., Hall C.B. Severity of respiratory virus infection is related to virus strain. J Infect Dis. 1997;175:814–820. doi: 10.1086/513976. [DOI] [PubMed] [Google Scholar]