Summary
Human enterovirus (HEV) 105 was first reported in 2012 in children from Peru and Congo. We report on the identification of a novel HEV-C105 strain in a pediatric patient in Cyprus with an upper respiratory tract infection. Sequence alignment and phylogenetic analysis of 5′-UTRs of all known HEVs revealed that our isolate belongs to a group of recently identified HEV-C viruses exhibiting a 5′-UTR distinct from all other previously known enteroviruses. This has important implications for diagnosis, as this region is the primary target for diagnostic assays. Increased awareness in laboratories may thus increase the rate of detection of enteroviruses belonging to this subspecies, or lead to the discovery of further genotypes.
Keywords: Enteroviruses, Epidemiology, Real-Time RT-PCR, Emerging viruses, Cyprus
1. Introduction
Human enteroviruses (HEVs) are small non-enveloped positive-strand RNA viruses that belong to the genus Enterovirus of the Picornaviridae family. They are associated with a wide range of clinical manifestations, ranging from mild respiratory illness to aseptic meningitis or even flaccid paralysis. Sequence analysis of the VP1 coding gene, which encodes important serotype-specific neutralization epitopes, has revealed four major genetic clusters of human enteroviruses. Phylogenetic analysis of VP1 sequences today forms the basis of a new classification scheme, in which enteroviruses are assigned to one of four species (enterovirus A to enterovirus D), with a continuously growing number of genotypes (http://www.picornaviridae.com).
The typing of enteroviruses is important for studying associations between clinical manifestations and specific types, as well as for guiding the development of new diagnostic tests and therapies.
2. Case investigation
In February 2012 a sample from a pediatric patient who had presented to the Archbishop Makarios Hospital Nicosia, Cyprus with an acute respiratory infection, was routinely analyzed for a panel of respiratory viruses (influenza viruses A and B, parainfluenza viruses, metapneumovirus, respiratory syncytial virus (RSV), adenovirus, coronaviruses E229, NL-63, and OC-43, bocavirus, rhinovirus, and enterovirus). The sample was found to be positive for rhinovirus, while being negative for all the other viruses analyzed.
For research purposes the sample was further subjected to typing, which in our laboratory is based on sequencing of two amplicons spanning part of the 5′-untranslated region (UTR) plus the VP4 and part of the VP2 region, using published protocols.1, 2 BLAST results for the amplicon spanning the VP4/VP2 region (nucleotide (nt) position 562–1061, relative to rhinovirus A1 strain ATCC VR-1559) showed highest homology to enterovirus isolates recently classified as HEV-C105, while sequencing of the amplicon in the 5′-UTR region (nt position 188–544, relative to rhinovirus A1 strain ATCC VR-1559) revealed close matches with a rhinovirus HRV-A species, thereby indicating a mixed infection.
For this reason the sample was further investigated using degenerate primers amplifying part of the enterovirus VP1 region.3 Sequencing confirmed that this enterovirus indeed belongs to the newly designated enterovirus HEV-C105 genotype, for which so far only five GenBank entries exist. All these isolates were collected in November 2010, one each from Peru, Romania, and Congo, and two from Burundi.4, 5 However, the fact that the sample had not been detected with our broad specificity enterovirus real-time reverse transcriptase (RT)-PCR assay, which targets highly conserved loci in the 5′-UTR region, demanded further investigation.
Primers were designed on the basis of EV-C105 strain Per153, which had shown highest homology to our isolate, in order to determine the complete 5′-UTR region as well as the complete VP1 region. Both sequences were deposited at GenBank (accession numbers KF322115 and KF322116). Subsequently, the determined sequences of the complete 5′-UTR region and VP1 coding region were aligned with the respective regions of all human enteroviruses classified so far, which were available in GenBank, as well as representatives of rhinovirus species A, B, and C, using ClustalW. The alignments, which comprise 124 sequences, are available in Fasta-Format on request.
3. Results
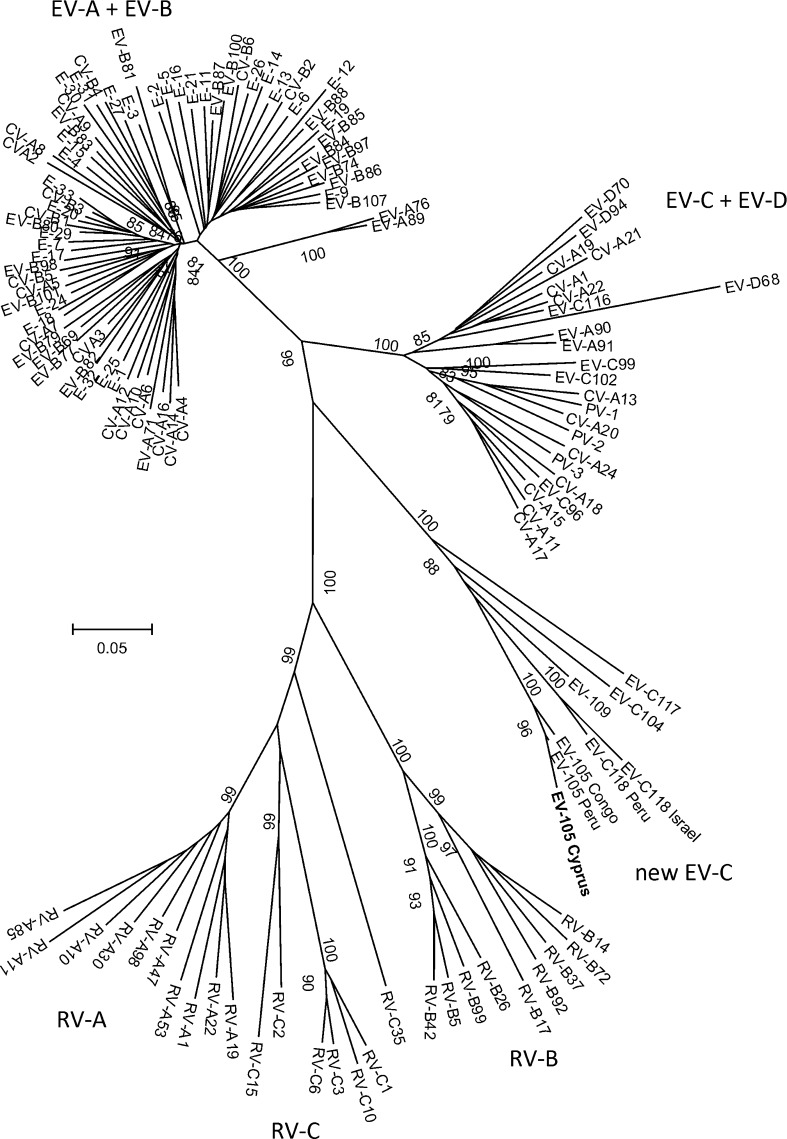
The phylogenetic tree obtained for the 5′-UTR region is shown in Figure 1 . As can be seen, the Cyprus isolate belongs to a novel monophyletic clade comprising the five recently discovered genotypes EV-C104, EV-C105, EV-C109, EV-C117, and EV-C118.5, 6, 7 The 5′-UTR of enteroviruses belonging to EV-A and EV-B species form a mixed clade, just as the enteroviruses belonging to the ‘classical’ EV-C and EV-D species. All three rhinovirus species are clearly distinguishable.
Figure 1.
Phylogenetic tree based on the 5′-UTR region of all known enteroviruses and representatives of the three rhinovirus species. The tree was inferred using the neighbor-joining method. Distances were computed using the maximum composite likelihood method and are in units of the number of base substitutions per site. The Cyprus isolate is shown in bold.
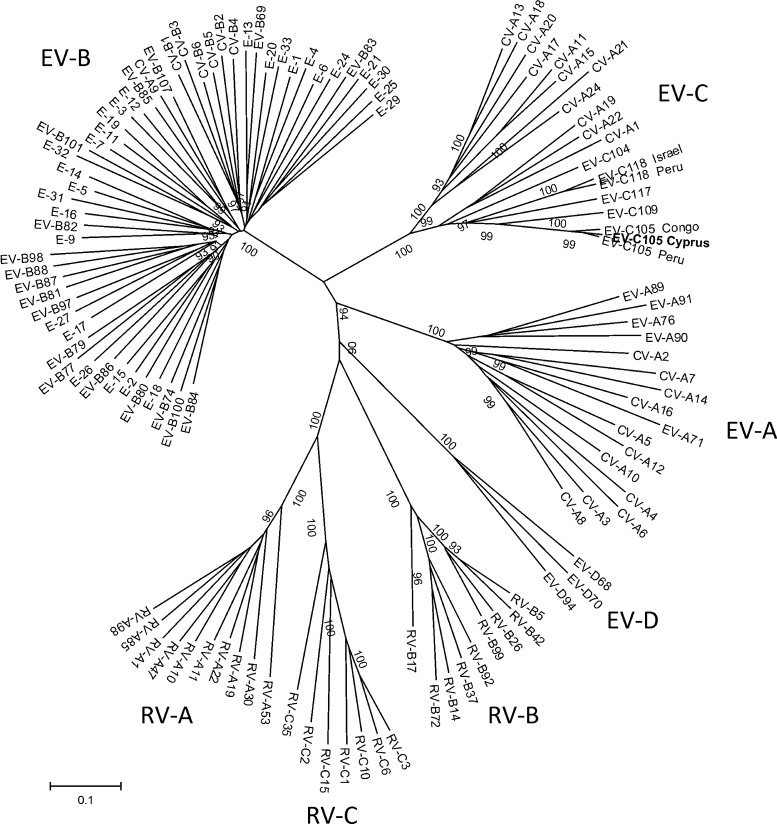
The phylogenetic tree based on the complete VP1 coding region is displayed in Figure 2 . All seven species are clearly distinguished, and EV-C105 again clusters most closely with the other recently discovered genotypes (EV-C104, EV-C105, EV-C109, EV-C117, and EV-C118) in an EV-C sub-branch that, in contrast to the 5′-UTR tree, also contains the ‘classical’ serotypes CV-A1, CV-A22, and CV-A24.
Figure 2.
Phylogenetic tree based on the VP1 coding region from available complete enteroviruses and representatives of the three rhinovirus species. The tree was inferred using the neighbor-joining method. The bootstrap consensus tree is inferred from 500 replicates; only bootstrap values >90% are shown. Distances were computed using the maximum composite likelihood method and are in units of the number of base substitutions per site. The Cyprus isolate is shown in bold.
4. Discussion
EV-C104 was first described in Switzerland in patients with acute otitis media or pneumonia and was later also found in Italy and Japan.7, 8, 9 EV-C109 was initially described in patients with influenza-like illness in Nicaragua,6 but has since also been found in Hungary10 and Italy (unpublished). EV-C117 was first isolated from a pediatric patient with pneumonia in 2010 in Lithuania, and EV-C118 from two children in Israel diagnosed with acute otitis media and community-acquired pneumonia.11, 12 The genotype EV-C105, to which the Cyprus isolate belongs, was only recently discovered in a respiratory sample in Peru and a fecal sample from a case of fatal acute flaccid paralysis in Congo.4, 5
A characteristic of all members of this new EV-C subspecies is their distinct 5′-UTR. Lukashev et al. hypothesized that this 5′-UTR may have been derived from an animal enterovirus.5 Future recombination with other EV-C or even interspecies recombination, could lead to an increasing number of enteroviruses exhibiting this new 5′-UTR.13, 14 Even though these enteroviruses have so far been isolated mostly from respiratory samples, Tapparel et al.7 noted that based on its genomic features, EV-C104 could theoretically infect the central nervous system.
Despite marked differences in the 5′-UTR of the EV-A, EV-B, EV-D, and ‘classic’ EV-C, completely conserved regions exist, which has allowed for the design of non-degenerate primers and/or probes that are able to amplify all enteroviruses with a high sensitivity, which is necessary for diagnostic applications. However, the alignment of all EV 5′-UTRs revealed that the sequences of the enteroviruses belonging to this new clade exhibit differences in several of the exact regions that are conventionally used for diagnosis. In addition to our own observation, Tapparel et al. also noted in the case of EV-C104 that their enterovirus specific real-time PCR assay did not amplify this new genotype.7
For this reason, new real-time RT-PCR primers and probes were designed based on the alignment, in order to detect all enteroviruses belonging to this new clade (see Table 1 ). A plasmid containing the 5′-UTR sequence of the EV-C105 Cyprus isolate was used as positive control. The new primer/probe set was able to amplify the new isolate as well as serial dilutions of the plasmid down to a concentration of 200 plasmids/ml. Further validation will verify the reliability of the assay to detect all members of the new HEV-C subtype.
Table 1.
Primers and probes used for detection of conventional enteroviruses as well as the genotypes belonging to the novel HEV-C subspecies
| Primer/probe | Sequence | Position (relative to PV1 Mahoney) |
|---|---|---|
| HEV F | CCCTGAATGCGGCTAATCC | 449–467 |
| HEV R | ATTGTCACCATAAGCAGCCA | 593–574 |
| HEV P | FAM-ACGGACACCCAAAGTAGTCGGTTCC-BHQ1 | 554–530 |
| HEV F_new_C | GCCCCTGAATGTGGATAATCC | 447–467 |
| HEV R_new_C | ATTGTCACCATAAACATTCAa | 593–574 |
| HEV P_new_C | FAM-CACCCAAAGTAGTTGGTTCCGCCA-BHQ1 | 558–535 |
HEV, human enterovirus.
ZNA oligo (zip nucleic acid). The modification improves affinity for the target sequence and increases the melting temperature of AT-rich oligonucleotides.
With this report, the authors hope to increase awareness in laboratories to these newly emerging enteroviruses. As the detection of EV-C105 so far in Peru, Congo, Burundi, and now Cyprus indicates a worldwide distribution, more countries are likely to report this novel enterovirus soon. Improved surveillance should improve detection rates and the discovery of further genotypes belonging to this enterovirus subspecies. Further studies are needed to clarify the actual prevalence and clinical importance of these novel enteroviruses.
Acknowledgements
This work was co-funded in part by the European Regional Development Fund and the Republic of Cyprus through the Research Promotion Foundation (Project YΓEIA/ΔYΓEIA/0609(BIE/25).
Conflict of interest: No conflict of interest to declare.
Corresponding Editor: Eskild Petersen, Aarhus, Denmark
References
- 1.Savolainen C., Mulders M.N., Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002;85:41–46. doi: 10.1016/s0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 2.Kiang D., Kalra I., Yagi S., Louie J.K., Boushey H., Boothby J. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008;46:3736–3745. doi: 10.1128/JCM.00674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nix W.A., Oberste M.S., Pallansch M.A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokarz R., Hirschberg D.L., Sameroff S., Haq S., Luna G., Bennett A.J. Genomic analysis of two novel human enterovirus C genotypes found in respiratory samples from Peru. J Gen Virol. 2013;94:120–127. doi: 10.1099/vir.0.046250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukashev A.N., Drexler J.F., Kotova V.O., Amjaga E.N., Reznik V.I., Gmyl A.P. Novel serotypes 105 and 116 are members of distinct subgroups of human enterovirus C. J Gen Virol. 2012;93:2357–2362. doi: 10.1099/vir.0.043216-0. [DOI] [PubMed] [Google Scholar]
- 6.Yozwiak N.L., Skewes-Cox P., Gordon A., Saborio S., Kuan G., Balmaseda A. Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J Virol. 2010;84:9047–9058. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tapparel C., Junier T., Gerlach D., Van-Belle S., Turin L., Cordey S. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis. 2009;15:719–726. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaida A., Kubo H., Sekiguchi J., Hase A., Iritani N. Enterovirus 104 infection in adult, Japan, 2011. Emerg Infect Dis. 2012;18:882–883. doi: 10.3201/eid1805.111890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piralla A., Rovida F., Baldanti F., Gerna G. Enterovirus genotype EV-104 in humans, Italy, 2008–2009. Emerg Infect Dis. 2010;16:1018–1021. doi: 10.3201/eid1606.091533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pankovics P., Boros A., Szabo H., Szekely G., Gyurkovits K., Reuter G. Human enterovirus 109 (EV109) in acute paediatric respiratory disease in Hungary. Acta Microbiol Immunol Hung. 2012;59:285–290. doi: 10.1556/AMicr.59.2012.2.13. [DOI] [PubMed] [Google Scholar]
- 11.Daleno C., Piralla A., Scala A., Baldanti F., Usonis V., Principi N. Complete genome sequence of a novel human enterovirus C (HEV-C117) identified in a child with community-acquired pneumonia. J Virol. 2012;86:10888–10889. doi: 10.1128/JVI.01721-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daleno C., Greenberg D., Piralla A., Scala A., Baldanti F., Principi N. A novel human enterovirus C (EV-C118) identified in two children hospitalised because of acute otitis media and community-acquired pneumonia in Israel. J Clin Virol. 2013;56:159–162. doi: 10.1016/j.jcv.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schibler M., Gerlach D., Martinez Y., Belle S.V., Turin L., Kaiser L. Experimental human rhinovirus and enterovirus interspecies recombination. J Gen Virol. 2012;93:93–101. doi: 10.1099/vir.0.035808-0. [DOI] [PubMed] [Google Scholar]
- 14.Jegouic S., Joffret M.L., Blanchard C., Riquet F.B., Perret C., Pelletier I. Recombination between polioviruses and co-circulating coxsackie A viruses: role in the emergence of pathogenic vaccine-derived polioviruses. PLoS Pathog. 2009;5:e1000412. doi: 10.1371/journal.ppat.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]