Most viruses have RNA genomes. We must understand how they generate diversity so we can anticipate and respond to the arrival of new strains or entirely new viruses. Owing to the high error rate of RNA-dependent replication, RNA viruses exist as heterogeneous populations of molecules known as quasispecies[1]. The advantage of quasispecies to the virus is that, as selection pressures change, a fit genome might already exist in the quasispecies population or can evolve rapidly. The disadvantage of the high error rate is that accumulation of too many mutations is damaging[1]. However, these genetic defects can be rescued by recombination with other molecules in the quasispecies that have functional genes[2]. Furthermore, by recombination, a virus can suddenly acquire entirely new traits (whole genes) in one step.
Recombination is common in RNA viruses2, 3and it might be more important than accumulation of point mutations for significant evolutionary change and speciation of RNA viruses. Viruses in groups as diverse as the alphaviruses[4], coronaviruses[2]and luteoviruses[5]contain genes closely related to those of other groups, indicating recent recombination events in their evolution.
Understanding viral recombination is important for the safe deployment of transgenic, virus-resistant plants. Dozens of crop species have been engineered to express viral genes. This confers resistance to the virus from which the transgene was derived[6]. However, recombination between an invading virus and the transgenic viral RNA can yield viable progeny[7]. To assess the risks of new, virulent viruses arising from such events, a better understanding of RNA recombination mechanisms is essential.
Recombination has been well studied in poliovirus[8], coronaviruses[2], brome mosaic virus (BMV)[3], turnip crinkle virus (TCV)[3]and others. In some cases, recombination has been detected by the use of different selectable markers on different portions of each parental genome. The occurrence of both markers on the same genome demonstrates that replication has occurred2, 8. This method detects rare events but generally gives a low-resolution view of the junction of the recombined genomes. For higher resolution, recombination is analyzed by sequencing across the junctions of parental and progeny RNAs.
RNA virus recombination events have been classified into homologous, aberrant homologous and nonhomologous recombination[2]. However, Nagy and Simon[9]have suggested revising the classification into sequence similarity- essential, similarity-assisted and similarity-nonessential recombination. Most evidence supports a copy-choice mechanism8, 9, in which the replicase switches from copying one RNA (donor template) to another (acceptor template) without releasing the nascent strand (Fig. 1 ).
Fig. 1.
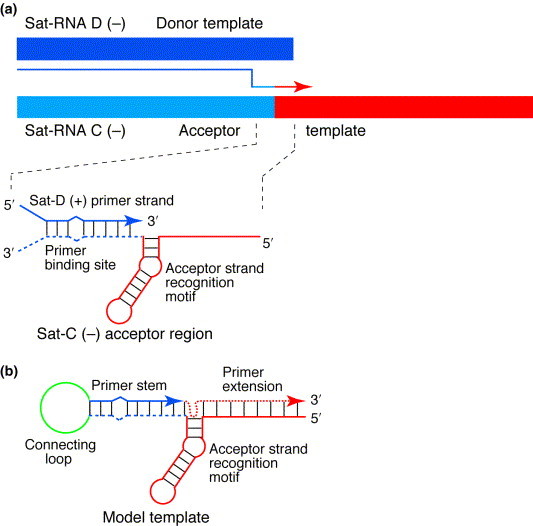
Structures involved in recombination between turnip crinkle virus (TCV) satellite RNA D (sat-RNA D) and sat-RNA C. (a) Maps of template [negative (−)] strands of 194-nt sat-RNA D and 356-nt sat-RNA C are shown. The light blue portion of sat-RNA C has 88% homology to sat-RNA D (dark blue)[11]. The portion of sat-RNA C that is derived from the TCV genome is shown in red. The light blue and red arrow indicates strand switching and the direction of movement of replicase during positive (+) strand synthesis. The dark blue arrow shows the nascent strand from the donor template; the dashed blue line indicates the portion of sat-RNA C that is homologous to sat-RNA D. (b) The structure used for primer extension assays by Nagy et al.[10]Primer and acceptor strands are connected by a large loop (green). The dashed red arrow indicates RNA synthesis, which includes the bulged stem–loop portion of the template, in the in vitro assay. Modified with permission from Ref. [10].
1. Recombination in vitro
The details of viral recombination mechanisms have remained a mystery owing to the dependence on in vivo systems (cell culture or whole organism). In vivo assays require the progeny RNA to be able to replicate and survive in the cell independently of the actual recombination event. In vivo, the recombination event cannot be separated into its individual biochemical steps: (1) generation of the nascent (primer) strand on the donor template, (2) transfer of the primer strand and replicase to the acceptor template, and (3) elongation on the acceptor template[10]. Moreover, if there is sequence identity across the recombination junction, the precise site of recombination cannot be determined by sequencing.
A recent paper from Simon's research group describes a cell-free replication system that overcomes these obstacles and opens the way for detailed biochemical analysis of RNA recombination[10]. This system examines recombination between 194-nt satellite RNA D (sat-RNA D) and 356-nt satellite RNA C of TCV (Fig. 1). Sat-RNA C is a chimera of sat-RNA D and a TCV defective interfering (DI) RNA. DI RNAs are efficient replicons comprising small portions of a viral genome from which most of the coding regions have been deleted by recombination. Sat-RNAs have no homology to their helper viruses. Their origins are unknown. Both sat- and DI RNAs contain cis-acting signals for replication by the replicase of the helper virus. These RNAs have advantages for recombination studies: there is less ambiguity regarding the effects of mutations because of the reduced gene coding and regulatory functions. Many recombination studies exploit such trans-replicated defective RNAs (Refs 2, 9).
Previously, Simon's lab identified a recombination hot spot between bases 175–179 of sat-RNA C and the 3′ end of sat-RNA D, which produces a hybrid RNA molecule[11]. They found that a bulged stem– loop structure in the acceptor template strand is required for recombination in vivo[11](Fig. 1). This is a form of similarity-assisted recombination[9]. The recent paper by Nagy et al.[10]focuses on recognition of the acceptor template by the replicase and extension of the nascent strand, which acts as a primer. The latter function is of particular interest because TCV (and BMV) replicases cannot normally extend a primer annealed to single-stranded RNA. In contrast, poliovirus replicase can copy a template consisting only of poly(A) using either oligo(U) or its genome-linked protein (VPg) as a primer[12]. However, the model for TCV recombination involves extension of the nascent strand in a way that is biochemically the same as primer-extended RNA synthesis (Fig. 1).
2. A primer and a `handle'
To test both the primer function and the requirements of the acceptor RNA to be recognized by the replicase, Nagy et al.[10]have developed an elegant assay in which they have constructed the putative recombination complex of sat-RNA C and sat-RNA D as a single RNA molecule. The primer and acceptor strands were connected by a large loop (Fig. 1), resulting in a partially duplex structure comprising this artificial stem–loop, and the upstream bulged stem–loop, which had been previously identified as being important for recombination[11]. The replicase could then extend the primer strand, copying the bulged stem–loop and the single-stranded 5′ end of the molecule. The small products were then rapidly quantified and sized. However, sizes of products were difficult to interpret because of the high degree of secondary structure, which resisted denaturation.
Using an exhaustive set of point mutations, Nagy et al.[10]went on to compare the effects of mutations on primer extension in vitro with their effects on recombination in vivo (in which case the bulged stem was located in the negative strand of sat-RNA C)[11]. Importantly, there was a good correlation between in vitro and in vivo effects. Generally, mutations that reduced efficiency of primer extension to <24% of that on the `wild-type' molecule eliminated recombination in vivo. Both stems in the bulged stem–loop upstream (in the template sense) of the acceptor strand were necessary for efficient primer extension, although their sequence was not as important. Deletion of bases in the bulge greatly reduced primer extension and eliminated recombination. Some specific bases in the terminal loop were also necessary.
The second feature shown to be necessary was the complementarity between the primer strand and the acceptor strand. The length, sequence and 3′ position of the primer were not important in determining whether extension could take place. Less stable priming stems gave less efficient extension. Thus, primer binding to the acceptor serves only as an extendible nascent RNA strand and is not involved in specific recognition of the acceptor by the replicase. In support of the role of the bulged stem–loop in specific recognition by the replicase, RNA molecules containing only this region, without the primer stem, were found to inhibit primer extension in trans. Nonfunctional mutant bulged stem–loops did not inhibit primer extension significantly. Thus, two structures are necessary to attract the nascent RNA and the replicase to the acceptor strand. The region of complementarity attracts the nascent RNA strand, and the bulged stem–loop may be the `handle' by which the replicase grabs the acceptor strand.
3. Future questions
The above model could not be supported without an in vitro assay. Although Simon's work is a promising early step in the dissection of recombination mechanisms, many questions remain. For example, what features on the donor template encourage dissociation of the replicase and nascent strand? Possibilities proposed for a variety of RNAs include a stable secondary structure, the 5′ end of the template itself or AU-rich regions that weaken the interaction between the nascent strand and the template. In coronaviruses, dissociation has been attributed to the nonprocessive nature of the replicase[2]. In BMV, the replication complex is initially highly abortive but then undergoes a modification allowing efficient elongation[13]. Perhaps, at some points of the RNA synthesis, the replication complex can revert to its loosely bound abortive state and subsequently dissociate.
What is the nature of the protein that interacts with the bulged stem–loop? Presumably, it is a component of the viral replicase, but is it the virally encoded RNA-dependent RNA polymerase itself or a host factor that participates? How does acceptor recognition by the replication machinery compare with the specific recognition of the origins of replication at the 3′ ends of positive and negative strands and internally at subgenomic RNA promoters?
More broadly speaking, how widely does this type of mechanism apply? Recognition of an internal structure on a negative strand, as described here, resembles subgenomic RNA synthesis. Subgenomic RNA synthesis may be primer dependent in coronaviruses[2]but is primer independent in plant viruses[14]. However, we propose that plant viral subgenomic RNA promoters can also be recognized by this donor-primed mechanism in the recombination events that generate variation in luteoviruses[5]. This may occur in other viruses that produce subgenomic mRNAs, including many plant viruses, coronaviruses and alphaviruses.
In contrast to the above viruses, recombination has been detected in poliovirus at hot spots for which no replication origin or subgenomic promoter would normally be predicted8, 15, 16. What are the features of the RNA and replicase that promote picornaviral recombination at specific sites? The recent demonstration of poliovirus recombination in cell-free systems15, 16bodes well for elucidation of the mechanism in the near future. Now that recombination has been demonstrated in two such very different in vitro systems, TCV and poliovirus, and numerous other cell-free replication systems are available, the next few years should yield a bounty of detailed analyses of a variety of different RNA recombination mechanisms.
References
- 1.Domingo E. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 2.Lai M.M.C. Semin. Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon A.E., Bujarski J.J. Annu. Rev. Phytopathol. 1994;32:337–362. [Google Scholar]
- 4.Hahn C.S. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5997–6001. doi: 10.1073/pnas.85.16.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller W.A., Dinesh-Kumar S.P., Paul C.P. Crit. Rev. Plant Sci. 1995;14:179–211. [Google Scholar]
- 6.Baulcombe D. Trends Microbiol. 1994;2:60–63. doi: 10.1016/0966-842x(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 7.Greene A.E., Allison R.F. Science. 1994;263:1423–1425. doi: 10.1126/science.8128222. [DOI] [PubMed] [Google Scholar]
- 8.Kirkegaard K., Baltimore D. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy P.D., Simon A.E. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 10.Nagy P.D., Zhang C., Simon A.E. EMBO J. 1998;17:2392–2403. doi: 10.1093/emboj/17.8.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascone P.J., Haydar T.F., Simon A.E. Science. 1993;260:801–805. doi: 10.1126/science.8484119. [DOI] [PubMed] [Google Scholar]
- 12.Paul A.V. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 13.Sun J-H., Kao C.C. Virology. 1997;236:348–353. doi: 10.1006/viro.1997.8742. [DOI] [PubMed] [Google Scholar]
- 14.Miller W.A., Dreher T.W., Hall T.C. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 15.Duggal R. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13786–13791. doi: 10.1073/pnas.94.25.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang R.S. RNA. 1997;3:624–633. [PMC free article] [PubMed] [Google Scholar]


