Abstract
Background
Human bocavirus (HBoV), a recently discovered virus, is prevalent among children with respiratory tract infection throughout the world. Co-infection was frequently found in HBoV-positive patients. Thus, whether HBoV is responsible for the respiratory disease is still arguable.
Objectives
A comprehensive study was carried out to integrate clinical and virological prevalence in HBoV-positive outpatient children, and to determine genetic and serologic characteristics of HBoV in Shanghai, China.
Study design
Nasal/throat swabs and sera were obtained over a 2-year period from 817 children with respiratory tract infection to examine the presence of HBoV and its co-infection. The seroepidemiology of HBoV was studied by ELISA and Western blot against the capsid protein VP2-based fragment. Persistence of HBoV was also analyzed in 12 pairs of return-visit cases.
Results
HBoV was identified in 96 samples (11.8%). The co-infection rate with other respiratory viruses was 51%. IgM was detected in 55.7% of HBoV RT-PCR-positive patients, and in 72.7% of those who had high viral genome load. In addition, persistent viral DNA positivity was detected in 10 of 12 HBoV-positive cases tested, an average of 14 days later, and one child was still HBoV-positive after 31 days.
Conclusion
HBoV was found frequently in children with respiratory tract symptoms associated with other respiratory viruses, and persisted in the respiratory tract and in serum and urine. The presence of IgM was significantly more prevalent in viremic patients and those diagnosed with high load of HBoV DNA in nasal/throat swabs.
Keywords: Bocavirus, Acute respiratory infection, Phylogeny, Humoral response, HBoV persistence, Co-infection
1. Background
In 2005, Human bocavirus (HBoV), a novel member in the genus Parvovirus, family Parvoviridae, was first identified in children with lower respiratory tract infection (LRTI).1 Since then, the detection of HBoV in acute respiratory illness was reported worldwide, with prevalence rates between 1.5% and 19%.2, 3, 4
Among parvoviruses, HBoV has the closest amino acid identity with bovine parvovirus (42%) and canine minute virus (43%). HBoV is a single-stranded DNA virus with a genome of around 5 kb, encoding 4 genes (NS1, NP1, VP1, and VP2). The majority of genetic variations occurred in the capsid VP1/VP2 genes, allowing a classification into two genotypes, ST1 and ST2.1
HBoV was detected in various samples, including nasopharyngeal aspirate,1 serum,5, 6 feces,7, 8 and urine.9 HBoV detection was significantly higher in patients with symptoms of respiratory tract or gastroenteritis than in asymptomatic individuals.3, 5, 10, 11, 12 In addition, higher HBoV prevalence was observed in children aged from 6 months to 2 years, compared to older individuals.2, 6, 13, 14, 15 Co-infection with other viruses was commonly observed in HBoV-positive patients with respiratory tract symptoms.5, 6, 15, 16, 17, 18 Thus, whether HBoV is a major cause of respiratory disease remains questionable.
Recently, several studies on the seroepidemiology of HBoV were reported by immunofluorescence assay,19 Western blot,20 or ELISA.21, 22, 23, 24, 25 Anti-HBoV IgG was detected in a large fraction of the population, especially in older children and adults, while IgM was found predominantly in HBoV-positive patients.
Only few studies have undertaken a comprehensive analysis of clinical, virological, and serological aspects in HBoV infection.24, 25 The present study aimed at better characterizing the virological and serological profiles of HBoV in children with acute respiratory infection (ARI) in samples collected during a 2-year period in one hospital in Shanghai, China. Co-infecting respiratory viruses were analyzed, and serological diagnosis using a highly antigenic fragment of VP2 was developed. In addition, persistent HBoV shedding was assessed in nasal, throat, serum and urine specimens.
2. Materials and methods
2.1. Patients and specimens
From October 2006 to September 2008, 817 samples were collected from children aged between 6 months and 9 years, who were diagnosed with an ARI with accompanying fever. All specimens were collected at the Shanghai Nanxiang Hospital, with informed consent from the children's parents. Standardized protocols accepted by the ethical committee of the hospital and reviewed by the medical committee of Institut Pasteur in Paris were used to record the patients’ clinical symptoms and medical history, and for specimen collection. Nasal and throat swab samples were collected and stored in viral transport medium, and blood samples were collected in tubes containing 3.2% sodium citrate. The doctor who was involved in the study and did the sampling remained the same throughout the study. All samples were aliquoted at arrival in the laboratory and stored at −80 °C. Twelve patients who were diagnosed positive for HBoV were recalled between 8 and 31 days after the initial visit, and nasal swabs, throat swabs, blood, as well as urine samples were collected.
2.2. Quantitative PCR for detection of HBoV in human sample extracts
Total RNA and DNA were extracted from 140 μl of each specimen by QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer's instructions. Nucleic acids were then eluted in 50 μl RNase-free water and stored at −80 °C until use. The primers and the probe used to detect HBoV were targeted to the NP1 gene, as published previously (Table 1 ).26 The real-time PCR assays were performed using Takara's Premix Ex Taq™ Perfect Real Time (Takara Biotechnology, Dalian, China) in 20 μl reaction mixture, containing 1 μl of extracted sample, 0.2 μM each of the forward and reverse primers, and 0.2 μM of probe. PCR amplification included a step at 95 °C for 10 s followed by 40 cycles of 5 s at 95 °C and 30 s at 60 °C. The reactions were run and analyzed in an ABI PRISM 7900HT (Applied Biosystems, Foster City, CA, USA). The standard curve was obtained using serial dilutions of pskHBoV containing the whole genome of HBoV ST2 genotype (a kind gift from Dr Jianming Qiu, University of Missouri-Columbia, USA). The housekeeping gene GAPDH was detected as the internal control to monitor the quality of the specimens and to normalize HBoV DNA in clinical samples for comparative analysis.
Table 1.
HBoV primers and probe used in viral genome amplification.
| Gene | Amplicon | Forward primer | Reverse primer | Size (bp) | Reference |
|---|---|---|---|---|---|
| NP1 for real-time PCR | NP-1 | 5′-AGAGGCTCGGGCTCATATCA-3′ | 5′-CACTTGGTCTGAGGTCTTCGAA-3′ | 81 | 26 |
| Probe | 5′ FAM-AGGACCACCCAATCARCCACCTATCGTCT-BHQ1 3′ | ||||
| NS1 for conventional RT-PCR | NS-1 | 5′-TATGGCCAAGGCAATCGTCCAAG-3′ | 5′-GCCGCGTGAACATGAGAAACAG-3′ | 245 | 13 |
| VP1 for sequence | VP1 | 5′-CCACTAGTATGCCTCCAATTAAGAGACAGC-3′ | 5′-CCTCTAGATTACAACACTTTATTGATGTTTGT-3′ | 2016 | |
| VP2 | VP2-P1 | 5′-CCCAAGCTTGCATGTCTGACACTGACATTCAA-3′ | 5′-CCGCTCGAGTTAATCACAAAAGATGTGAACGC-3′ | 480 | |
| VP2 | VP2-P2 | 5′-CCCAAGCTTGCATG CAAATACTTTCAAATGGTGCT-3′ | 5′-CCGCTCGAGTTATGTGCTTCCGTTTTGTCTTAT-3′ | 435 | |
| VP2 | VP2-P3 | 5′-CCCAAGCTTGCATGCTAATGTTTAATCCAAAAGTC-3′ | 5′-CCGCTCGAGTTA TCTGGTGATAGGAAATCTGTC-3′ | 465 | |
| VP2 | VP2-P4 | 5′-CCCAAGCTTGCATGCTATACAAAGGAAACCAAACC-3′ | 5′-CCGCTCGAGTTACAACACTTTATTGATGTTTG-3′ | 474 | |
2.3. Conventional RT-PCR for HBoV and other respiratory viruses
HBoV-positive specimens were also examined with conventional RT-PCR, using the One-Step RT-PCR kit (QIAGEN) that targets the NS1 gene, as described previously.13 Multiplex RT-PCR, described previously, was used to diagnose other respiratory viruses in HBoV-positive samples,27 including respiratory syncytial virus (RSV), human rhinovirus (HRV), influenza A virus (IAV), influenza B virus (IBV), human metapneumovirus (HMPV), adenovirus (ADV), human coronaviruses (HCoV) 229E, OC43, HKU-1, and NL63, and parainfluenza viruses (PIV)1–4. Briefly, 2.5 μl of the sample extract and 0.2 μM of primers were added to the 25 μl reaction mixture. The RT reaction was carried out at 50 °C for 30 min, followed by PCR, including a step at 95 °C for 15 min and 40 cycles of 30 s at 94 °C, 30 s at 55 °C, 30 s at 72 °C and the final extension at 72 °C for 10 min. The products were run in a 1% agarose gel and visualized by UV light.
2.4. Sequencing of VP1 and phylogenetic analysis
The entire VP1 gene was amplified with primer pairs (Table 1) using QIAGEN's One-Step RT-PCR kit (QIAGEN). The reaction mixture included 5 μl RNA, 0.4 μM of each primer, 400 μM of each dNTP, and 2 μl of enzyme mix in a final volume of 50 μl. Amplification included 45 min at 50 °C, 15 min at 94 °C, 8 cycles of 1st round amplification (30 s at 94 °C, 45 s at 55 °C, 2.5 min at 72 °C), followed by 35 cycles of 2nd round amplification (30 s at 94 °C, 45 s at 50 °C, 2.5 min at 72 °C), and final extension of 10 min at 72 °C. Products of 2016 bp were visualized in an agarose gel under UV light. The fragments were extracted by QIAquick Gel Extraction Kit (QIAGEN) and cloned into pMD18T (Takara Biotechnology). At least four colonies were picked from each sample and sent for sequencing to Biosune Sequence Company and Life Biotechnology (Shanghai, China). The results were analyzed by Vector NTI Advance™ 9.0 (Invitrogen, Carlsbad, CA, USA). The VP1 gene of HBoV strains collected during 2005–2009 available in Genbank were also downloaded and phylogenetic analysis was conducted with MEGA version 3.1 software. The phylogenetic tree was drawn by the Neighbor-joining method with bootstraps of 1000 replications.
2.5. Expression of VP2 in the baculovirus–insect cell system
The recombinant bacmid pFastBac1 containing HBoV VP2 gene (kindly provided by Dr Jianming Qiu) was transfected into Spodoptera frugiperda Sf21 cells to recover the recombinant baculovirus. High Five cells were infected with the virus at a multiplicity of infection of five plaque-forming units/cell and cells were harvested 3 days later by centrifugation. The cell pellet was resuspended in 25 mM NaHCO3 at a density of 2 × 107 cells/ml and left on ice for 30 min until the cells were completely lysed. Cell debris was removed by centrifugation at 13,000 rpm. The proteins were further precipitated in 20% ammonium sulfate, and resuspended in PBS.
2.6. Recombinant protein production
Four regions of VP2 were amplified by PCR from the pskHBoV clone. The forward and reverse primers (Table 1) contained Hind III and Xho I restriction sites, respectively. After digestion with the restriction enzymes, the amplicons were cloned into pET30a containing 6xHis tag at the 5′ terminal. The recombinant proteins were expressed in E. coli BL21(DE3) host strains by induction with IPTG for 3 h. Cells were collected by centrifugation and the pellets were resuspended in lysis buffer and lysed by sonication on ice. The recombinant proteins were further purified by Ni-NTA agarose (Invitrogen) under denaturing conditions, following the manufacturer's instructions. The protein needed for ELISA was further dialyzed into PBS.
2.7. Immunoblotting
Fifteen micrograms of each protein were loaded and separated on a 12.5% SDS-PAGE gel, and then transferred onto nitrocellulose membranes. After blocking for 2 h in 5% nonfat dry milk in TBS-T (0.5% Tween-20 in TBS buffer) blocking buffer, the membranes were incubated for 1 h in patient serum samples diluted in blocking buffer (dilution 1:50 for IgM test, 1:100 for IgG test). After incubation, the membranes were washed four times in TBS-T, and incubated in anti-human IgG or IgM conjugated with horseradish peroxidase (HRP) (Bethy Laboratories, Montgomery, TX, USA). After 1 h, the membranes were washed four times in TBS-T buffer and 3,3′-diaminobenzidine and H2O2 were added to reveal the peroxidase activity.
2.8. ELISA
ELISA 96-well plates (Nunc™, Roskilde, Denmark) were coated with 200 ng/well of purified proteins in coating buffer (0.05 M carbonate/bicarbonate, pH 9.6) overnight at 4 °C. After washing with PBS-T (0.05% Tween-20), the wells were saturated in 5% dry milk in PBS-T at 37 °C for 2 h. Serum samples diluted in 1% dry milk in PBS-T were added and incubated for 1 h at 37 °C. After washing, HRP-conjugated anti-human IgG (1:10,000 diluted in dry milk in PBS-T) was added for 1 h at 37 °C. Plates were washed five times with PBS-T, and OPD (o-Phenylenediamine Dihydrochloride) (Sigma, St. Louis, MO, USA) was used as a substrate. After 20 min of incubation, the absorbance at OD450 was read. All samples were measured in triplicate, and the average value was calculated for each serum sample. A purified heptapeptide fragment of SARS-HCoV spike protein produced in E. coli was used as a negative antigen (the corresponding expression plasmid pet30a-SARS spike was a kind gift of Dr. YongJin Wang, East Normal University, Shanghai, China). The threshold for positivity was calculated as twice the mean OD450 value obtained when sera were tested against the SARS spike protein fragment.
2.9. Statistical analysis
The statistical analysis was performed via SAS6.12 software (SAS institute Inc., Cary, NC, USA). The p value was measured by two-sided analysis, and a p value less than 0.05 was considered to be statistically significant. Comparisons between different variations were evaluated by chi-squared or Fisher's test.
3. Results
3.1. Viral screening from pediatric patients
During a 2-year period, a total of 817 pairs of nose and throat swabs were obtained from pediatric patients (male: 531, female: 286) with an age range from 6 months to 9 years. To detect HBoV, we first used conventional RT-PCR and then real-time PCR, targeting conserved NS1 and NP1 gene sequences,13, 25 respectively. Samples with discrepant results between negative RT-PCR and positive real-time PCR were retested by increasing the amount of nucleic acid extract in conventional RT-PCR and by duplicating real-time PCR. Results finally gave 100% concordance between the two techniques. Ninety-six children (11.8%) were positive for HBoV detected in 96 throat swabs and 78 in nose swabs. Among 103 patients with virus co-infection, 49 (47.6%) were found infected with HBoV (data not shown). Forty two samples contained another virus, six samples contained 3 viruses, and one contained 4 viruses (Table 2 ).
Table 2.
Multiple viral infections in HBoV-positive samples.
| Two virus infection (n) | Three virus infection (6 cases) | Four virus infection (1 case) |
|---|---|---|
| RSV (10) | PIV1/HRV | RSV/HRV/IBV |
| HRV (4) | RSV/HRV | |
| IAV/IBV (5/7) | RSV/HMPV | |
| HMPV (3) | RSV/229E | |
| ADV (5) | PIV4/HRV | |
| HCoV-NL63 (1) | RSV/ADV | |
| PIV1/3 (4/3) |
RSV: Respiratory syncytial virus; HRV: human rhinovirus; IAV: influenza A virus; IBV: influenza B virus; HMPV: human metapneumovirus; ADV: adenovirus; HCoV: human coronaviruses (including 229E, NL63); PIV1–4: parainfluenza virus 1–4.
The HBoV loads detected in nose and throat swabs ranged from <500 to 108 DNA copies/ml, and those in serum ranged from <500 to 105 copies/ml. Thirteen (59.1%) of 22 patients showing high copies (>104 copies/ml) of HBoV in throat swabs were also positive in their serum, whereas only 8 (14.0%) out of 57 patients showing low copies of HBoV were positive in their serum (p = 0.00012). Forty patients (62.5%) among 64 with a low level of HBoV copies in their throat were co-infected with another virus, whereas only nine (28.1%) from 32 with high copies of HBoV were co-infected (p = 0.0022).
3.2. Clinical characteristics for HBoV-positive patients
HBoV was detected in 17.1% of children younger than 1 year, 14.3% in 1–2-year-old children, 8.2% in 2–4-year-olds, and 11.2% in older children. The gender ratio of HBoV-positive patients was 55% male and 45% female.
The main clinical symptoms in HBoV-positive patients involved fever (100%), cough (74.7%), and wheezing (20.9%). No specific symptom was prevalent in the HBoV-positive patients (Table 3 ). The number of HBoV RT-PCR-positive patients with bronchitis, however, was low (5.4%) when compared to the high prevalence of this disease (Table 3).
Table 3.
Clinical diagnosis of HBoV-positive children.
| Clinical diagnosis | Total samples | HBoV-positive | Percentage of HBoV-positive/total samples |
|---|---|---|---|
| Acute laryngitis | 58 | 11 | 19.0% |
| Bronchitis | 479 | 26 | 5.4% |
| Bronchiolitis | 51 | 11 | 21.6% |
| Bronchopneumonia | 187 | 39 | 20.9% |
| Asthma | 42 | 9 | 21.4% |
| Total | 817 | 96 | 11.8% |
3.3. Phylogenetic analysis of VP1 genes of HBoV
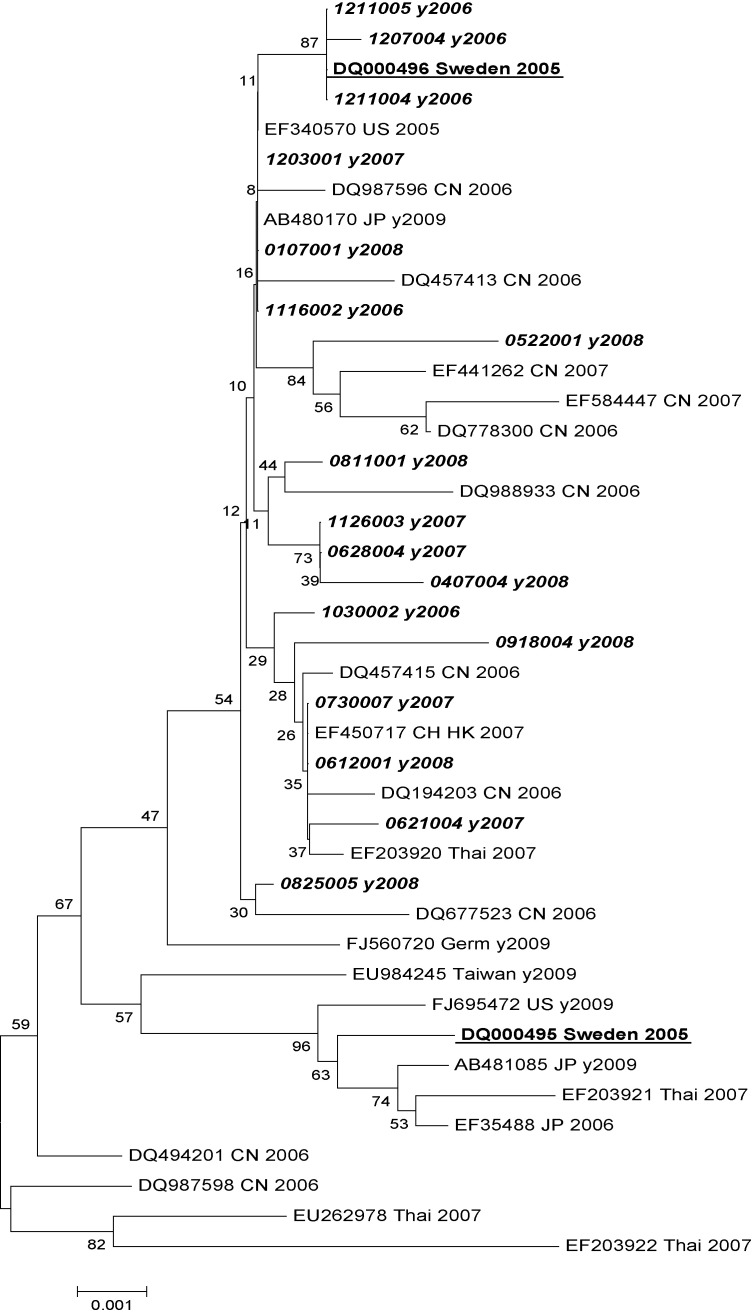
The VP1 gene of HBoV from 17 samples containing enough DNA/RNA material was amplified and sequenced. The nucleotide identity among different strains was over 99.3%. All strains tested in this study clustered with European, Asian and American strains into genotype ST2 (Fig. 1 ).
Fig. 1.
Phylogenetic tree of HBoV based on the VP1 gene sequence. Sample numbers in italic were sequenced in this study, the first two numbers represent the month and the next two numbers the date of sample collection. Samples underlined were the two first original samples identified in Sweden,1 each corresponding to one genotype. All the field strains we found were more similar to ST2 (DQ000496). CN: China, JP: Japan, Thai: Thailand, US: USA, Germ: Germany, HK: Hongkong. VP1 gene sequences from HBoV field samples of this study are given nos. GQ403967–GQ403983 in Genbank.
3.4. Analysis of antigenic regions in VP2 of HBoV immunoreactive with patient sera
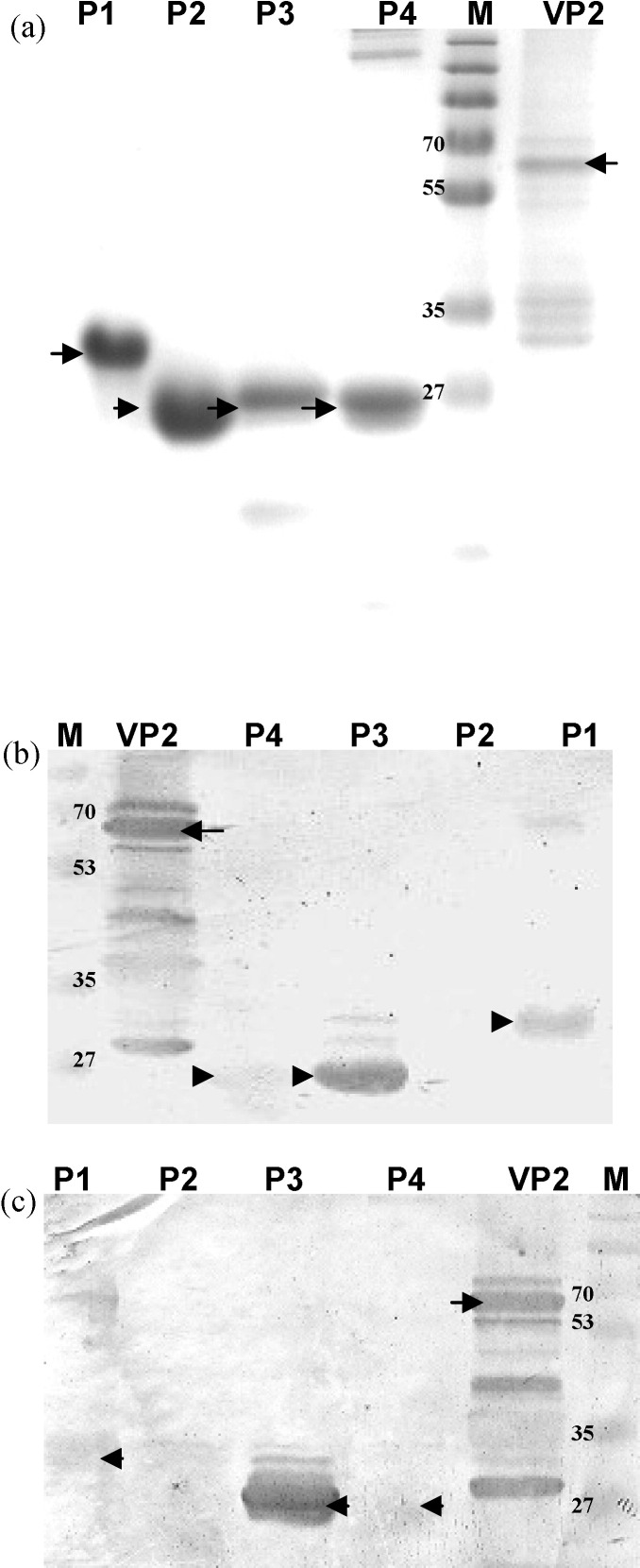
The VP2 capsid protein and four fragments of VP2 (designated P1 to P4) were first tested in a panel of serum samples by Western blots to detect HBoV-specific IgG and IgM antibodies. Eight of the samples contained high HBoV load, six contained low viral load, and six contained no HBoV. VP2 and its P3 fragment were the most immunoreactive with both IgG and IgM, and the P1 fragment was reactive mostly with IgM (Fig. 2 ). Therefore, the 79 available serum samples from HBoV RT-PCR-positive patients were tested in Western blots against VP2 protein and P1 and P3 polypeptides to detect IgM and IgG. In total, 40 (50.6%) sera were IgG-positive, and 44 (55.7%) were IgM-positive for HBoV antigens. Nineteen of 21 HBoV viremic patients had antibodies, with 15 (71.4%) IgM-positive and 11 (52.3%) IgG-positive. Among 22 children infected with high HBoV DNA levels (>104 copies/ml) in nasal or throat swabs, 16 (72.7%) were IgM-positive and 13 (59.1%) were IgG-positive. In 36 single-infected specimens, 26 (69.4%) were IgM-positive, compared with 18 (41.9%) in 43 co-infected samples (p = 0.012) (Table 4 ).
Fig. 2.
Immunoblot assay for detection of antibodies via VP2 and four fragments of VP2. (a) VP2-expressing recombinant baculovirus-infected cells and purified VP2 fragments were subjected to SDS-PAGE and Coomassie blue staining. (b) HBoV proteins were analyzed by Western blot using an IgM-positive patient serum (dilution: 1:50). (c) HBoV proteins were analyzed by Western blot using an IgG-positive patient serum (dilution 1:100). M: Marker (kDa).
Table 4.
Antibody responses in sera (via Western blot) compared with viral loads in HBoV-positive samples.
| Viral load/Ab response | Total samples | IgM no. (%) | IgG no. (%) | Antibodies no. (%) |
|---|---|---|---|---|
| Viremia | 21 | 15 (71.4%) | 11 (52.4%) | 19 (90.5%) |
| With high viral load (>104 copies/ml) | 22 | 16 (72.7%) | 13 (59.1%) | 19 (86.4%) |
| Single HBoV infection | 36 | 26 (72.2%) | 19 (52.8%) | 33 (91.7%) |
| Multiple viral infections | 43 | 19 (44.2%) | 21 (48.8%) | 30 (69.8%) |
| Total HBoV-positive | 79 | 44 (55.7%) | 40 (50.6%) | 63 (79.7%) |
3.5. Analysis of anti-HBoV IgG antibodies by ELISA using VP2-derived P3 fragment
The sera of patients were tested in ELISA using a purified P3 antigenic fragment derived from the VP2 protein, and SARS-HCoV (HR2 spike protein fragment) as a negative control. At a serum dilution of 1:100, the mean OD450 value for negative controls using the SARS-HCoV spike HR2 fragment was 0.165. The assay was further confirmed by Western blots and the ELISA values were considered either negative (OD450 < 0.33 corresponding to twice the mean value of the negative control), equivocal (OD450 = 0.33–0.42), or positive (OD450 > 0.42).
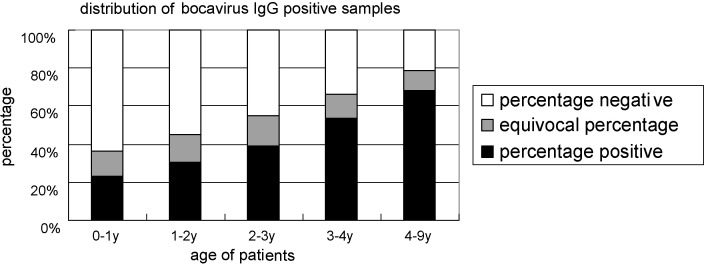
In all, 713 serum samples were available to test HBoV-specific IgG antibodies. 314 samples (44.0%) were seropositive, and 95 (13.3%) were equivocal. The number of IgG-positive individuals increased with their age, with a statistical significance for age distribution (p < 0.0001) (Fig. 3 ). Fewer children younger than 2 years old had anti-HBoV IgG (41.2%) than those older than 4 years old (67.93%) (p < 0.0001). In addition, 17 out of 29 sera showing an OD450 value above 0.85 were from children older than 4 years.
Fig. 3.
Anti-HBoV IgG-positive individuals categorized according to age group.
3.6. Persistent shedding of HBoV
Twelve HBoV-positive patients were called back around 2 weeks after the first sampling. The mean interval of specimen collection was 16 days (days 8–31). None of these children showed any symptoms of ARI, and obviously recovered from their previous infection. However, HBoV could still be detected in nose and throat swabs, serum or urine (Table 5 ) from 10 children with a viral load ranging from less than 102 to 104 copies/ml. In all four specimens, the virus loads were lower or similar in the second samples compared to the first samples, and two samples were found to be free of HBoV. No urine samples were collected during the first sampling, and three urine specimens were positive for HBoV in return-visit samples. In one case, virus genes were still detected in nose/throat swabs as well as in urine collected 31 days apart. Seven serum samples showed a seroconversion for IgG and three for IgM. All but two samples contained IgM in return samples. An anti-HBoV IgG antibody response was detected in all but two patients showing low virus titer during the second visit. Co-infecting viruses found in four children at the first visit were absent at the second visit but HRV or ADV were newly detected in three children showing low levels of persistent HBoV (Table 5).
Table 5.
Persistent HBoV shedding in patients, with the interval between two visits.
| Patient (n = 12) | Age*/gender** | HBoV genome (copies/ml of specimens) |
Antibody |
Co-infection | ||||
|---|---|---|---|---|---|---|---|---|
| Days interval | Nose swab† | Throat swab† | Serum | Urine | IgG | IgM | Virus | |
| R3071126003 | 17 m/F | 3.2 × 105 | 3.2 × 106 | 3.0 × 104 | NA | − | +++ | |
| 14 days apart | 0 | 1.2 × 102 | 3.3 × 102 | 2.3 × 103 | + | ++ | ||
| R3071203001 | 12 m/M | 2.3 × 104 | 4.6 × 104 | 9.6 × 104 | NA | + | ++ | |
| 14 days apart | 5.0 × 103 | 9.0 × 102 | 1.4 × 103 | 0 | +++ | ++ | HRV | |
| R3080218002 | 10 m/M | 4.4 × 102 | 2.7 × 102 | 0 | NA | − | + | |
| 14 days apart | 0 | 0 | 1.7 × 102 | 0 | + | + | ||
| R3080218005 | 5 y/M | 6.1 × 102 | 2.9 × 102 | 0 | NA | + | ++ | IAV |
| 15 days apart | 0 | 2.1 × 102 | 5.3 × 102 | 0 | + | + | AdV | |
| R3080303002 | 13 m/M | 0 | 2.4 × 102 | 0 | NA | − | − | HMPV |
| 8 days apart | 0 | 1.2 × 102 | 0 | 5.3 × 102 | − | + | ||
| R3080303004 | 5 y/M | 2.2 × 102 | 2.9 × 103 | 0 | NA | − | ++ | |
| 20 days apart | 4.9 × 102 | 4.4 × 102 | 0 | 0 | − | + | ||
| R3080320004 | 5 y/M | 0 | 4.6 × 102 | 7.9 × 102 | NA | − | − | |
| 31 days apart | 1.5 × 103 | 2.4 × 102 | 0 | 2.0 × 103 | + | + | HRV | |
| R3080508002 | 7 m/M | 1.6 × 102 | 3.4 × 102 | 0 | NA | + | + | HRV |
| 26 days apart | 1.4 × 102 | 4.7 × 102 | 0 | 0 | + | − | ||
| R3080508005 | 9 m/M | 2.9 × 102 | 1.9 × 102 | 0 | NA | − | − | PIV4/HRV |
| 26 days apart | 0 | 0 | 0 | 0 | ++ | − | ||
| R3080522002 | 5 y/M | 1.0 × 102 | 7.5 × 102 | 0 | NA | − | + | |
| 12 days apart | 6.0 × 102 | 0 | 0 | 0 | + | ++ | ||
| R3080811001 | 11 m/M | 2.5 × 105 | 2.4 × 105 | 5.6 × 104 | NA | − | + | |
| 20 days apart | 0 | 0 | 0 | 0 | + | ++ | ||
| R3080825005 | 2 y/M | 5.0 × 106 | 4.5 × 105 | 7.5 × 104 | NA | − | + | |
| 17 days apart | 5.4 × 102 | 4.6 × 102 | 1.2 × 103 | 0 | + | + | ||
NA: Not available.
m, Month; y, year.
F, Female; M, male.
HBoV load was normalized by GAPDH.
4. Discussion
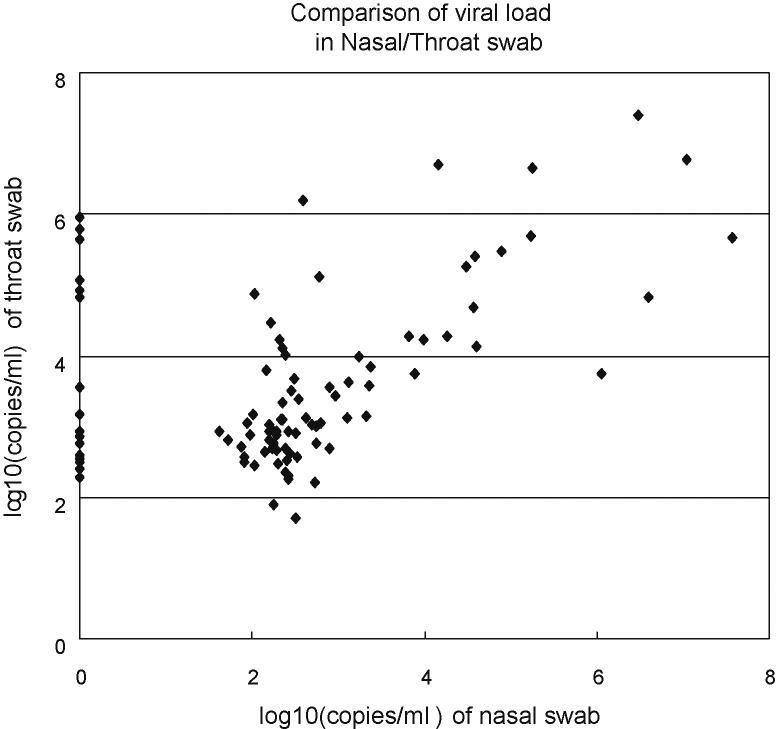
In this study, the prevalence rate of HBoV-positive non-hospitalized infants with ARI was as high as 11.8%, and infants less than 2-year-old were more frequently positive than older ones. This is similar to that reported by other studies,14, 15, 16 All the HBoV strains clustered in the ST2 genotype, as previously identified in mainland China and Hong Kong in children with LRTI.28, 29 The limited number of specimens studied, however, does not preclude the possible presence of the ST1 genotype of HBoV. Interestingly, higher HBoV positivity and load were observed in throat swabs (n = 96) than in nasal swabs (n = 78), suggesting an infection preferentially of the lower respiratory tract, as indicated by previous studies on bronchoalveolar lavage (Fig. 4 ).11, 30, 31 In addition, HBoV was detected in sera of infected children, as previously observed with other parvoviruses.1 Consistent with other reports, samples with high copies of HBoV detected in nasal and throat swabs were more frequently positive in sera, compared with those with low copies,5, 6, 20, 24 suggesting that viremia often occurs during acute infection.
Fig. 4.
The comparison of viral load in nasal and throat swabs. The points crossing the y axis indicate that nasal swabs were HBoV-negative. HBoV load was normalized by GAPDH.
A high multi-infection rate was also detected in HBoV-infected patients (51.0%), with RSV and HRV having the highest prevalence. The high ratio of these two co-infecting viruses may reflect their prevalence in children visiting the Pediatrics Department of Shanghai Nanxiang Hospital with ARI (Wang and Deubel, unpublished data). In addition, the percentage of HBoV-positive patients with co-infecting viruses was 47.6%, suggesting a correlation between the presence of HBoV and that of other respiratory viruses (p < 0.0001). Patients with lower copies of HBoV were more frequently associated with co-infection, as previously reported.32, 33 Conversely, data published in a recent paper25 reported a higher co-infection rate in patients with high HBoV load. It was probably due to a different pool of samples. Notably, all the six triple infections and one quadruple infection appeared in patients with low HBoV load in swabs (data not shown). HBoV infection may be maintained to low copies in some individuals by an as yet unresolved interfering process triggered by the presence of the co-infecting agent. Whether HBoV exacerbates other viral infections or appears in children infected with other viruses is difficult to address and would require further studies.
Previous studies reported that the majority of epitopes recognized by antibodies in HBoV-infected patients were located in the VP2 protein.20 In this study, we divided VP2 into 4 fragments (P1–P4) and tested their antigenic reactivity by Western blot. Sixteen of 20 sera contained IgG/IgM antibodies that reacted with VP2 and its P3 fragment, and four did not show reactivity with VP2 and any of the four fragments. Strong immunoreactivity with IgG and IgM was observed for fragment P3, consistent with results with the whole VP2 protein. However, preparation of large amounts of purified P3 antigen in E. coli is easier and cheaper than that of VP2, which requires a more tedious preparation in the baculovirus system. Therefore, the purified P3 fragment of VP2 represents an alternative antigen for IgG detection in ELISA to test for acute or recent infection. It remains to be determined, however, whether this polypeptide is still recognized by long lasting antibodies to prevent false negative result for past HBoV infection.20, 24, 33 The number of IgG-positive cases increased with the age of the children, suggesting that infection of HBoV likely occurred during childhood. The data validate other studies that showed a high number of IgG-positive serum samples in adults.23, 24 Since some previous studies reported the absence or only few cases of anti-HBoV IgM in virus-negative samples from children, and rare positive results in healthy adults,24, 25 only sera of patients confirmed with HBoV infection were tested by Western blot for IgM. Anti-HBoV IgM antibodies were likely detected more often in patients viremic for HBoV or with high copies of HBoV in nasal and throat swabs, as previously observed.20, 24, 25 In 11 patients with viremia, high copy number in swabs, and infected only by HBoV, 10 (90.9%) were IgM-positive, which is consistent with previous studies.20, 25 Patients infected singly with HBoV had comparatively more anti-HBoV IgM and IgG than multi-infection ones (p = 0.0119) (Table 4). In contrast, in 57 patients with low viral load in swab samples, the amount of IgG or IgM remained at a low level (data not shown).
Persistent HBoV shedding was reported in a few patients with symptoms lasting for several weeks,9, 25 and in immunocompromised children.4, 27, 34, 35 In this study, 12 patients were called for a second visit within an interval of 8–31 days (Table 5). No fever or symptoms of the respiratory tract were noted in these patients. Low copies of HBoV were still found in nasal and throat swabs, serum, and urine, indicating that infection of HBoV may last for a long period of time at a low level, and in the presence of anti-HBoV antibodies (Table 5). This situation was also observed in parvovirus B19 infection with systemic and long-term persistent infection, high IgG reactivity and activated CD8+ T cell responses.36 All ten patients with persistent PCR positivity of HBoV were found to be free of respiratory disease, but three were diagnosed with a secondary infection with ADV or HRV, suggesting that the persistence of HBoV was of less clinical significance, but may have played a role in promoting co-infection. Altogether, our study suggests that acute infection of HBoV causes systemic infection, induces immune responses, and may play a crucial role in respiratory disease of children, and is often associated with co-infection. Further studies are needed to understand the mechanism of the interactions between the host immune response, HBoV infection and prolonged virus shedding.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We thank Dr Jianming Qiu, University of Missouri-Columbia, USA, for sharing the plasmid pskHBoV and bacmid pFastBac1 containing HBoV genes, and Dr Yongjin Wang, East China University, Shanghai, China, for providing the plasmid pet30a-SARS spike containing the SARS-HCoV heptapeptide HR2 gene. This work was supported by the Li Ka-Shing Foundation (RESPARI project), the French Agency for Development (SISEA project), Chinese Academy of Sciences visiting professorship for senior international scientists, and the National Science and Technology Major Project 2009ZX10004-105 (the establishment of pathogen and immuno-response detection platforms for respiratory and central nervous system viral infections). WW is a recipient of a fellowship from the AREVA Foundation.
References
- 1.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander T. Human bocavirus. J Clin Virol. 2008;41:29–33. doi: 10.1016/j.jcv.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Lindner J., Karalar L., Schimanski S., Pfister H., Struff W., Modrow S. Clinical and epidemiological aspects of human bocavirus infection. J Clin Virol. 2008;43:391–395. doi: 10.1016/j.jcv.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tozer S.J., Lambert S.B., Whiley D.M., Bialasiewicz S., Lyon M.J., Nissen M.D. Detection of human bocavirus in respiratory, fecal, and blood samples by real-time PCR. J Med Virol. 2009;81:488–493. doi: 10.1002/jmv.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry A.M., Lu X., Chittaganpitch M., Peret T., Fischer J., Dowell S.F. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis. 2007;195:1038–1045. doi: 10.1086/512163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allander T., Jartti T., Gupta S., Niesters H.G., Lehtinen P., Osterback R. Human bocavirus and acute wheezing in children. Clin Infect Dis. 2007;44:904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neske F., Blessing K., Tollmann F., Schubert J., Rethwilm A., Kreth H.W. Real-time PCR for diagnosis of human bocavirus infections and phylogenetic analysis. J Clin Microbiol. 2007;45:2116–2122. doi: 10.1128/JCM.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicente D., Cilla G., Montes M., Pérez-Yarza E.G., Pérez-Trallero E. Human bocavirus, a respiratory and enteric virus. Emerg Infect Dis. 2007;13:636–637. doi: 10.3201/eid1304.061501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozo F., García-García M.L., Calvo C., Cuesta I., Pérez-Breña P., Casas I. High incidence of human bocavirus infection in children in Spain. J Clin Virol. 2007;40:224–228. doi: 10.1016/j.jcv.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kesebir D., Vazquez M., Weibel C., Shapiro E.D., Ferguson D., Landry M.L. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis. 2006;194:1276–1282. doi: 10.1086/508213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maggi F., Andreoli E., Pifferi M., Meschi S., Rocchi J., Bendinelli M. Human bocavirus in Italian patients with respiratory diseases. J Clin Virol. 2007;38:321–325. doi: 10.1016/j.jcv.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 12.García-García M.L., Calvo C., Pozo F., Pérez-Breña P., Quevedo S., Bracamonte T. Human bocavirus detection in nasopharyngeal aspirates of children without clinical symptoms of respiratory infection. Pediatr Infect Dis J. 2008;27:358–360. doi: 10.1097/INF.0b013e3181626d2a. [DOI] [PubMed] [Google Scholar]
- 13.Sloots T.P., McErlean P., Speicher D.J., Arden K.E., Nissen M.D., Mackay I.M. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X., Endo R., Ishiguro N., Ebihara T., Ishiko H., Ariga T. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006;44:1132–1134. doi: 10.1128/JCM.44.3.1132-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naghipour M., Cuevas L.E., Bakhshinejad T., Dove W., Hart C.A. Human bocavirus in Iranian children with acute respiratory infections. J Med Virol. 2007;79(5):539–543. doi: 10.1002/jmv.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning A., Russell V., Eastick K., Leadbetter G.H., Hallam N., Templeton K. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J Infect Dis. 2006;194:1283–1290. doi: 10.1086/508219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi E.H., Lee H.J., Kim S.J., Eun B.W., Kim N.H., Lee J.A. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000–2005. Clin Infect Dis. 2006;43:585–592. doi: 10.1086/506350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan B.H., Lim E.A., Seah S.G., Loo L.H., Tee N.W., Lin R.T. The incidence of human bocavirus infection among children admitted to hospital in Singapore. J Med Virol. 2009;81:82–89. doi: 10.1002/jmv.21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endo R., Ishiguro N., Kikuta H., Teramoto S., Shirkoohi R., Ma X. Seroepidemiology of human bocavirus in Hokkaido prefecture, Japan. J Clin Microbiol. 2007;45:3218–3223. doi: 10.1128/JCM.02140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kantola K., Hedman L., Allander T., Jartti T., Lehtinen P., Ruuskanen O. Serodiagnosis of human bocavirus infection. Clin Infect Dis. 2008;46:540–546. doi: 10.1086/526532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin F., Guan W., Cheng F., Yang N., Pintel D., Qiu J. ELISAs using human bocavirus VP2 virus-like particles for detection of antibodies against HBoV. J Virol Methods. 2008;149:110–117. doi: 10.1016/j.jviromet.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahn J.S., Kesebir D., Cotmore S.F., D’Abramo A., Jr., Cosby C., Weibel C. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. J Infect Dis. 2008;198:41–50. doi: 10.1086/588674. [DOI] [PubMed] [Google Scholar]
- 23.Indner J., Zehentmeier S., Franssila R., Barabas S., Schroeder J., Deml L. CD4+ T helper cell responses against human bocavirus viral protein 2 viruslike particles in healthy adults. J Infect Dis. 2008;198:1677–1684. doi: 10.1086/592985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindner J., Karalar L., Zehentmeier S., Plentz A., Pfister H., Struff W. Humoral immune response against human bocavirus VP2 virus-like particles. Viral Immunol. 2008;21:443–449. doi: 10.1089/vim.2008.0045. [DOI] [PubMed] [Google Scholar]
- 25.Soderlund-Venermo M., Lahtinen A., Jatti T., Hedman L., Kemppainen K., Lehtnen P. Clinical assessment and improved diagnosis of bocavirus-induced wheezing in children, Finland. Emerg Infect Dis. 2009;15:1423–1429. doi: 10.3201/eid1509.090204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X., Chittaganpitch M., Olsen S.J., Mackay I.M., Sloots T.P., Fry A.M. Real-time PCR assays for detection of bocavirus in human specimens. J Clin Microbiol. 2006;44:3231–3235. doi: 10.1128/JCM.00889-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dina J., Vabret A., Gouarin S., Petitjean J., Lecoq J., Brouard J. Detection of human bocavirus in hospitalised children. J Paediatr Child Health. 2009;45:149–153. doi: 10.1111/j.1440-1754.2008.01442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau S.K., Yip C.C., Que T.L., Lee R.A., Au-Yeung R.K., Zhou B. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J Infect Dis. 2007;196:986–993. doi: 10.1086/521310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J.M., Li D.D., Xu Z.Q., Cheng W.X., Zhang Q., Li H.Y. Human bocavirus infection in children hospitalized with acute gastroenteritis in China. J Clin Virol. 2008;42:280–285. doi: 10.1016/j.jcv.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 30.Garbino J., Soccal P.M., Aubert J.D., Rochat T., Meylan P., Thomas Y. Respiratory viruses in bronchoalveolar lavage: a hospital-based cohort study in adults. Thorax. 2009;64:399–404. doi: 10.1136/thx.2008.105155. [DOI] [PubMed] [Google Scholar]
- 31.Kleines M., Scheithauer S., Rackowitz A., Ritter K., Häusler M. High prevalence of human bocavirus detected in young children with severe acute lower respiratory tract disease by use of a standard PCR protocol and a novel real-time PCR protocol. J Clin Microbiol. 2007;45:1032–1034. doi: 10.1128/JCM.01884-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacques J., Moret H., Renois F., Lévêque N., Motte J., Andréoletti L. Human Bocavirus quantitative DNA detection in French children hospitalized for acute bronchiolitis. J Clin Virol. 2008;43:142–147. doi: 10.1016/j.jcv.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christensen A., Nordbø S.A., Krokstad S., Rognlien A.G., Døllner H. Human bocavirus commonly involved in multiple viral airway infections. J Clin Virol. 2008;41:34–37. doi: 10.1016/j.jcv.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corcoran A., Doyle S. Advances in the biology, diagnosis and host-pathogen interactions of parvovirus B19. J Med Microbiol. 2004;53:459–475. doi: 10.1099/jmm.0.05485-0. [DOI] [PubMed] [Google Scholar]
- 35.Koskenvuo M., Möttönen M., Waris M., Allander T., Salmi T.T., Ruuskanen O. Human bocavirus in children with acute lymphoblastic leukemia. Eur J Pediatr. 2008;167:1011–1015. doi: 10.1007/s00431-007-0631-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isa A., Kasprowicz V., Norbeck O., Loughry A., Jeffery K., Broliden K. Prolonged activation of virus-specific CD8+ T cells after acute B19 infection. PLoS Med. 2005;2:e343. doi: 10.1371/journal.pmed.0020343. [DOI] [PMC free article] [PubMed] [Google Scholar]