Highlights
-
•
Viral nucleic acids (NAs) are targeted by cellular proteins with diverse functions.
-
•
NA sensing proteins are forming a three-layered defence system.
-
•
NA localisation and modifications synergistically activate defence systems.
Abstract
Viruses are the most abundant pathogens on earth. A fine-tuned framework of intervening pathways is in place in mammalian cells to orchestrate the cellular defence against these pathogens. Key for this system is sensor proteins that recognise specific features associated with nucleic acids of incoming viruses. Here we review the current knowledge on cytoplasmic sensors for viral nucleic acids. These sensors induce expression of cytokines, affect cellular functions required for virus replication and directly target viral nucleic acids through degradation or sequestration. Their ability to respond to a given nucleic acid is based on both the differential specificity of the individual proteins and the downstream signalling or adaptor proteins. The cooperation of these multiple proteins and pathways plays a key role in inducing successful immunity against virus infections.
Current Opinion in Virology 2015, 11:31–37
This review comes from a themed issue on Viral pathogenesis
Edited by Luca G Guidotti and Matteo Iannacone
For a complete overview see the Issue and the Editorial
Available online 7th February 2015
http://dx.doi.org/10.1016/j.coviro.2015.01.012
1879-6257/© 2015 Elsevier Ltd. All rights reserved.
General properties of virus sensors
Almost all cells express germ-line encoded sensors with the ability to recognise virus infections and to initiate defence systems necessary to limit virus spread and pathogenicity. In technical terms, a sensor is ‘a device that detects events or changes in quantities and provides a corresponding output without affecting the original trigger’. Sensors follow certain rules that include selective sensitivity to a specific measured property and insensitivity to other properties likely to be encountered. In analogy to technical terms, virus sensors convert a signal (virus infection) to an output that instructs the cell to take further actions. The magnitude of its activation is characterised by properties related to the exact nature and the quantity of the trigger. The targets of these sensors can be incoming virus particles [1], particular viral proteins [2] as well as general integrity of the cell [3]. However, the yet best understood sensors involved in antiviral defence are activated by viral nucleic acids [4]. Endosomal Toll-like receptors sample the extracellular milieu or cytoplasmic contents that are delivered into endosomes through autophagy. In this review we concentrate on intracellular nucleic acid sensors and effector proteins that evolved to mediate specialised tasks including, firstly, expression of cytokines such as type I interferons (IFN-α/β); secondly, modulation of cellular machineries required for virus replication and thirdly, direct inhibition of virus growth (Figure 1 ). Induction of cytokines utilises at least two distinct pathways either involving the adaptor proteins mitochondrial antiviral-signalling protein (MAVS) or stimulator of interferon genes (STING). Activation of either pathway regulates transcription of cytokines, which are key signals to shape adaptive immunity to induce an intracellular ‘antiviral state’ characterised by expression of antiviral defence proteins. Some of the latter proteins are activated by viral nucleic acids and in turn re-wire cellular machineries to limit virus spread. Other proteins directly bind viral nucleic acid and impair functionality through steric hindrance or degradation.
Figure 1.
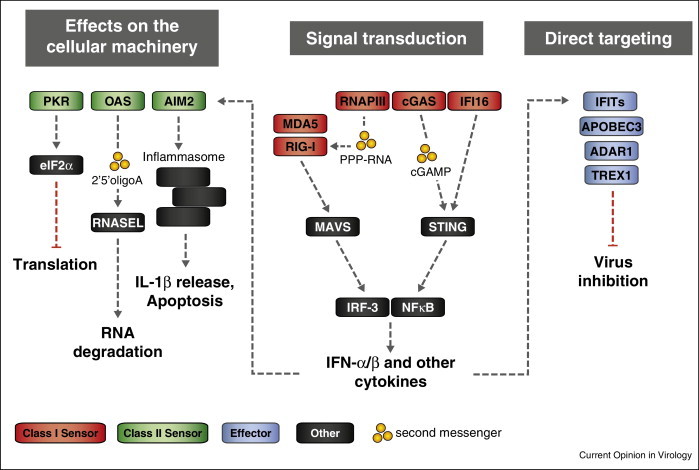
Viral nucleic acid sensor and effector proteins and their primary antiviral properties. Engagement of a particular set of nucleic acid sensors (Class I sensors, red) results in signal transduction events, leading to expression of the type I interferons IFN-α/β and other cytokines. These in turn upregulate additional sensors (Class II sensors, green) with the ability to modulate the cellular machinery. In addition cytokines induce expression of effector proteins (blue) directly targeting viral nucleic acids. Transmission of signals in some pathways occurs through second messengers (yellow). Class II sensors include PKR, OAS and AIM2. PKR phosphorylates translation initiation factor eIF2α and consequently inhibits translation. Activated OAS synthesises the second messenger 2′5′ oligoA, which then binds to and activates the latent endoribonuclease RNASEL. Activation of the inflammasome, a large multimeric complex including pro-caspase-1, is mediated by the DNA sensor AIM2. Caspase-1 cleaves its substrates pro-IL-1b and IL-18 for extracellular release. For signal transduction, MDA5 and RIG-I (either activated directly or through binding of RNAPIII-synthesised PPP-RNA) engage the adaptor protein MAVS. cGAS and IFI16 transmit their signal to the adaptor STING. Both pathways culminate in phosphorylation and dimerization of IRF-3 as well as release of active NFκB into the nucleus, where they cooperate to form an enhanceosome to turn on transcription of cytokine genes.
Abbreviations: OAS, 2′5′ oligoadenylate synthetase; PKR, dsRNA-dependent protein kinase R; AIM2, absent in melanoma 2; eIF2α, eukaryotic initiation factor 2 alpha subunit; RNASEL, 2-5A-dependent ribonuclease L; MDA5, melanoma differentiation-associated protein 5; RIG-I, retinoic acid inducible gene I; RNAPIII, RNA polymerase III; cGAS, cyclic GMP–AMP synthase; IFI16, interferon gamma-inducible protein 16; MAVS, mitochondrial antiviral-signalling protein; STING, stimulator of interferon genes; IRF-3, interferon regulatory factor 3; NFκB, nuclear factor κ-light-chain enhancer of activated B cells; IFIT, interferon-induced protein with tetratricopeptide repeats; APOBEC3, apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3; ADAR1, RNA-specific adenosine deaminase 1; TREX1, three prime repair exonuclease 1; PPP-RNA, 5′ triphosphorylated RNA.
Differences between cellular and viral nucleic acids
To understand how viral nucleic acids are sensed by the innate immune system it is important to consider the different types of nucleic acids generated after virus infection. Viruses are intracellular pathogens that require cellular translation and host metabolism, but provide their own replication machinery. Independence of the host for multiplication of viral genomes allows high replication rates, which is often associated with pathogenicity [5]. 24–48 hours after infection approximately 25% of RNA can be of viral origin (P. Hubel and A. Meiler, unpublished). Viral nucleic acids accumulate in compartments typically devoid of cellular nucleic acids and often possess or lack modifications or physical properties that are not normally associated with cellular RNA or DNA (Figure 2 ). RNA polymerases commonly generate RNA with a 5′ triphosphate group (PPP-RNA). Cellular RNA polymerases co-transcriptionally modify newly synthesised RNA at the 5′ terminus. In case of mRNA an inverted guanine nucleotide cap is added and methylated at the N7-position as well as the 2′O position of the first ribose of the RNA strand (Cap1 mRNA) (Figure 2) [6]. These modifications are necessary to mark mRNA for further processing and export into the cytoplasm, where translation takes place. Other cellular RNAs are cleaved and have a 5′ monophosphate in case of transfer (t)RNA, most ribosomal (r)RNAs and small nucleolar (sno) RNAs [7]. Some small RNAs bear a terminally methylated 5′ triphosphate (U6 snRNA, 7SK RNA) or are further processed to a hypermethylated 2,2,7-trimethylguanosine cap (TMG) cap (snRNAs). In addition, more than 100 modifications on internal nucleotides of cellular RNAs have been described, some of which are critical to tame activation of the innate immune system. Total cellular RNA isolated from cells and transfected into indicator cells does not activate the innate immune system, whereas the products of most viral RNA polymerases are strong stimuli of antiviral responses [8]. Negative strand RNA viruses such as orthomyxo-viruses, paramyxo-viruses and bunyaviruses commonly generate full-length genomic PPP-RNA and short 5′ PPP subgenomic RNA, which have strong immunostimulatory potential [4]. To avoid the cellular defence system many viruses mimic cellular mRNA-like cap structures by encoding capping enzymes (e.g. Flaviviruses, Coronaviruses, Poxviruses, and Reoviruses), ‘steal’ cap structures from cellular mRNAs for their transcripts (e.g. Orthomyxoviruses, Bunayviruses) or trim their genomic RNA to display only monophosphorylated termini (Bunyaviruses, Bornaviruses) [9, 10]. Picornaviruses and Caliciviruses mask their RNA with a covalently 5′ genome-linked viral protein (VpG). In addition to the cap itself, 2′O methylation of the first ribose of mRNAs is an additional modification that is highly conserved between viruses and their hosts, evidenced by the presence of dedicated viral proteins that catalyse this reaction [10]. Lack of 2′O methylation renders viruses highly vulnerable to the antiviral activity of the interferon system [11, 12].
Figure 2.
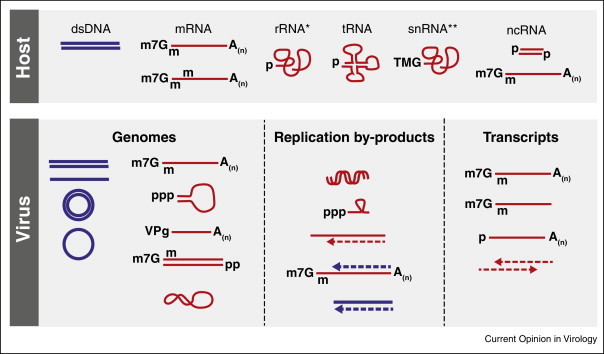
Differences between cellular and viral nucleic acids. Synthesis of host RNA (red) from nuclear dsDNA (blue) is achieved by three cellular RNA polymerases. RNA polymerase II synthesises mRNA, ncRNA and some snRNAs, whereas RNA polymerase III generates tRNA and 5S rRNA. rRNAs are produced by RNA polymerase I. Virus-derived DNA and RNA are present in the cell either as genomes, transcripts or replication by-products. Indicated are particular differences at the RNA 5′ and 3′ end, such as cap structures and methylations (e.g. cellular mRNA harbouring an N7-methylated guanine cap structure and 2′O-methylation at the first and/or second ribose). *5S rRNA harbours a 5′ triphosphate group; **U6 and 7SK RNA both have a 5′ gamma-monomethyl phosphate, and SRP RNA has a 5′ triphosphate. Abbreviations: ds, double-stranded; mRNA, messenger RNA; rRNA, ribosomal RNA; tRNA, transfer RNA; snRNA, small nuclear RNA; ncRNA, non-coding RNA; m7G, N7-methylated guanine cap; m, 2′O-methylation; p, phosphate group; TMG, hypermethylated 2,2,7-trimethylguanosine cap; VPg, viral protein genome-linked; A(n), poly(A) tail.
A type of RNA often associated with viral replication is double-stranded RNA (dsRNA). dsRNA could be either the result of replication intermediates (for RNA viruses), generation of genomic RNA (for dsRNA viruses), convergent transcription (for DNA viruses), or of the presence of secondary structures found in viral RNAs (e.g. the IRES structure of ssRNA viruses) [5]. However, the definition and exact nature of dsRNA still remains enigmatic. Using an antibody raised against dsRNA, it was found that such RNA is produced in cells infected with DNA viruses as well as some RNA viruses, such as Flavi and Picornaviruses [8]. However, although double-strandedness is an important feature recognised by virus sensors, it does not seem to be the only important determinant to stimulate fulminant antiviral responses since different double-stranded homopolymers vary considerably in their ability to induce IFN-α/β. Furthermore, dsRNA is commonly generated by convergent transcription of cellular RNA polymerases and is involved in transcriptional and post-transcriptional gene silencing [13•, 14]. Despite the presence of cell-generated dsRNA no spontaneous synthesis of IFN-α/β is apparent nor is transfection of total cellular RNA containing detectable dsRNA molecules or plasmid-based convergent transcription capable to induce significant levels of IFN-α/β [13•]. A possible explanation for the lack of stimulatory activity of cellular RNA may be insufficient concentration of dsRNA as proposed by a recent study showing that nuclear dsRNA is digested by the endonuclease Dicer [15]. It may be that the latter function is used by orthomyxoviruses that replicate in the nucleus to reduce the abundance of viral dsRNA in the cytoplasm.
Since most virus sensors and signalling molecules are localised in the cytoplasm, the cellular nucleus is considered not to promote sensing and signalling of virus infection. Indeed, cellular DNA, present in the nucleus does not elicit IFN-α/β whereas double-stranded DNA (dsDNA) introduced into the cytoplasm through transfection or virus infection induces an innate immune response [4]. However, simple compartmentalisation is insufficient to explain the ability of the innate immune system to recognise DNA viruses that replicate the nucleus [16•]. It is therefore likely that additional yet unknown features of viral DNA can be sensed by the innate immune system.
Sensors that drive expression of cytokines
Among the best characterised cytoplasmic proteins involved in virus sensing are RIG-I-like receptors (RLRs), a family of DExD/H-box helicases which specifically identify viral RNAs and have the ability to stimulate expression of IFN-α/β and other cytokines (Figure 3 ) [4, 17]. The founding member of this family, Retinoic acid inducible gene-I (RIG-I) bears two N-terminal Caspase activation and recruitment domains (CARDs) required for signalling, a central helicase domain that mediates binding to dsRNA and a C-terminal repressor domain, which binds 5′ tri-phosphorylated, di-phosphorylated or dephosphorylated RNA ends [18, 19, 20]. RIG-I forms oligomers along the bound RNA in an ATP-dependent manner, the CARDs oligomerize and allow signalling through CARD–CARD interactions with MAVS [21, 22]. Activation in addition requires dephosphorylation and ubiquitination of RIG-I [23]. Optimal RIG-I ligands are consisting of blunt dsRNA formed by two complementary RNAs (e.g. Reovirus) or generated by intramolecular base pairing as is proposed for the sensing of influenza A virus ribonucleoprotein complexes [20].
Figure 3.
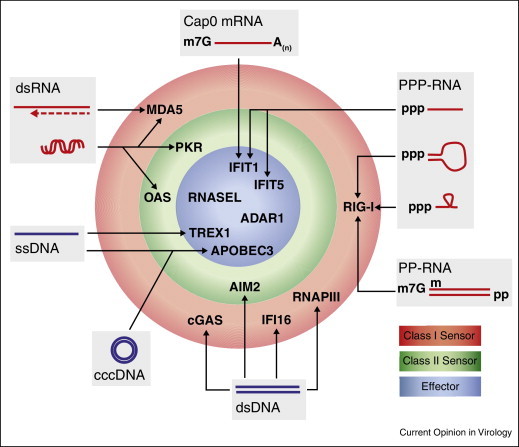
Cellular proteins involved in sensing and engagement of viral nucleic acids. Antiviral nucleic acid sensors operate in three layers, including viral sensing leading to signal transduction (red), modulation of the cellular machinery (green), and direct targeting of viral nucleic acids (blue). The exact mechanisms of particular cellular proteins engaging specific viral RNA and DNA structures are described in detail in the text. Abbreviations: ss, single-stranded; ds, double-stranded; Cap0, m7G cap structure lacking 2′O-methylation at the first and/or second ribose; PPP-RNA, 5′ triphosphorylated RNA; PP-RNA, 5′ dephosphorylated RNA; cccDNA, covalently closed circular DNA.
Other proteins belonging to RLR helicases are MDA5 and LGP2. LGP2 lacks functional CARD domains and therefore cannot induce signalling. However, LGP2 appears to be an important co-factor to facilitate sensing of some viruses [24]. Recently, L-antisense RNA expressed by encephalomyocarditis virus (EMCV) has been identified to associate with LGP2 [25]. L-antisense RNA activates MDA5, raising the possibility that LGP2 prepares ligands for sensing through other RLRs. MDA5 appears to be activated by structural properties of viral RNAs, but there is no unifying feature known that could generally explain MDA5 activation. Instead, a number of different RNAs are proposed to activate MDA5. Firstly, long synthetic dephosphorylated dsRNA stimulates MDA5 whereas shorter dsRNA loses this ability [26, 27]. Secondly, replication intermediates consisting of dsRNA and generated by picornaviruses [28, 29]. Thirdly, high molecular weight RNA generated during replication and likely bearing branched RNA molecules activates MDA5 [8]. The most commonly used synthetic MDA5 stimulus, poly-I:C would most likely form such structures. Fourthly, a specific sequence in the L-region present on the antisense single-stranded genomic RNA of EMCV appears to stimulate MDA5 [25]. Fifthly, for measles virus a sequence bias towards AU-rich regions was proposed to be associated with MDA5 activating activity [30]. Sixthly, mutant coronaviruses that generate RNA lacking 2′O methylation on the first ribose are a stronger MDA5 agonists than corresponding wild-type viruses highlighting the possibility that MDA5 may sense a chemical modification on the RNA 5′ end [31]. Similarly to RIG-I, N-terminal CARDs of MDA5 are required for downstream signalling but unlike RIG-I, MDA5 oligomerizes along dsRNA in a head to tail manner and positions the CARDs in an elongated structure that activates signalling through MAVS [32].
In addition to cytoplasmic RNA sensors, cells are equipped with sensors of cytoplasmic DNA. DNA sensing shows considerable cell-type specificity but in general two different concepts of DNA sensing seem to emerge: firstly, direct activation of IFN-α/β and secondly, generation of second messengers that are activating other proteins to induce IFN-α/β. Proteins directly activating IFN-α/β via the STING pathway are DNA-dependent activator of IRFs (DAI) [33•] and the more recently identified interferon-inducible protein 16 (IFI16) [34]. IFI16 belongs to the family of PYHIN proteins and contains a pyrin domain and two DNA-binding HIN domains. IFI16 is able to induce IFN-α/β after infection with Herpes simplex virus 1 (HSV-1) and Human immunodeficiency virus 1 (HIV-1) as well as transfected DNA [34, 35]. Proteins generating a second messenger include RNA polymerase-III (RNAPIII) and cyclic GMP–AMP synthase (cGAS). RNAPIII binds AT-rich regions in viral DNA genomes to produce PPP-RNA, serving as ligand for RIG-I [36, 37]. cGAS belongs to the nucleotidyltransferase family and upon dsDNA-binding generates cyclic 2′–5′ GMP–AMP (cGAMP) from ATP and GTP [38, 39••, 40, 41, 42]. cGAMP binds and directly activates STING and can also cross cell barriers to activate innate immune responses in adjacent cells [43]. Although the exact viral ligand has not yet been defined, lack of cGAS in human or mouse cells impairs interferon responses to DNA viruses and transfected DNA [44••].
Nucleic acid sensors with direct effects on the cellular machinery
Another set of nucleic acid sensors that activate transcription another subset of sensors directly affects cellular machineries to impair virus growth. These proteins include 2′5′ oligoadenylate synthetase (OAS), dsRNA-dependent protein kinase R (PKR) and absent in melanoma 2 (AIM2). DsRNA binding to OAS catalyses the conversion of ATP to 2′5′-linked oligoadenylates, which activate the latent ribonuclease RNASEL to degrade cellular and viral RNAs [42]. RNASEL cleavage products have been demonstrated to stimulate the MAVS pathway but the exact mechanism is not known. PKR is a serine/threonine kinase that is activated either by dsRNA of at least 30 bp in length or by PPP-RNA and suppresses general translation by phosphorylating eukaryotic initiation factor 2 alpha (eIF2-α) [45, 46]. In addition PKR induces apoptosis and regulates cytokine expression, most likely by modulating mRNA stability. Some nucleic acid binding proteins, such as the PYHIN family member AIM2, regulate post-translational processing and cell death [47]. AIM2 binds DNA and triggers the activation of the inflammasome, a molecular platform responsible for the maturation of interleukin 1β (IL-1β) and IL18 as well as triggering cell death. The RNA-binding helicases RIG-I [48] and DHX33 [49] have also been implicated in inflammasome activation, but the precise molecular details remain to be determined.
Cellular effector proteins directly targeting viral nucleic acids
Innate sensing leads to expression of effector proteins with the ability to sequester, modify or degrade viral nucleic acid (Figure 1). Sequestration of viral RNAs can be achieved by interferon-induced proteins with tetratricopeptide repeats (IFITs). Although combinations of IFITs are expressed in a species-specific manner most IFITs are highly induced in expression after virus infection. IFITs bind viral RNA through a deep binding cleft formed by a complex arrangement of tetratricopeptide repeats [50, 51]. IFIT1 preferentially binds single-stranded capped non-2′O-methylated (Cap0) or 5′ triphosphorylated (PPP) RNA, IFIT5 exclusively binds PPP-RNA [12, 52•]. IFITs compete with the function of other RNA-binding proteins, such as cellular translation initiation factors and/or viral proteins. Since RNA-binding by IFITs is highly specific, translation or localisation of cellular mRNAs is not affected by IFIT proteins [12].
An alternative strategy to directly target viral nucleic acids is to modify or degrade them. RNA-specific adenosine deaminase 1 (ADAR1) or members of the DNA-specific apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like (APOBEC) family deaminate nucleotides to introduce mutations, which potentially impacts RNA secondary structure, stability and protein-coding capacity [45, 53]. APOBEC3A and 3B recognise the Hepatitis B virus core protein and target core-associated DNA to impair virus growth [54]. Viral nucleic acids are directly targeted for degradation by 2′5′oligoadenylate-activated ribonuclease RNASEL [45] and by Zinc-finger antiviral protein (ZAP), which specifically targets viral mRNAs for degradation through recruitment of the cellular exosome machinery [55•]. DNA degradation through Three prime repair exonuclease 1 (TREX1) is required to restrict endogenous retroviruses [56].
Mutations in TREX1 have been linked to autoimmune diseases, clearly highlighting the importance of nucleic acid metabolising enzymes to reduce the abundance of stimulatory nucleic acids. More recently it has also been shown that the SKIVL2 exosome is important to reduce stimulatory RNA [57•].
Concluding remarks
Virus infection activates a restricted set of sensor and effector proteins that modulate cellular pathways and directly target viral nucleic acid, thereby shaping the innate immune response. Despite remarkable progress in the last few years to uncover modifications that are sensed by the innate immune system, many questions still remain to be answered. The natural ligand of cytoplasmic sensors, for instance, is often not well understood, nor do we know the exact localisation of virus sensing in the cytoplasm. Furthermore, numerous cellular pathways and second messengers contribute to innate immunity to viral pathogens and cell biological processes are similarly prominent in contributing to virus defence. We thus anticipate that even more entangled relationships between viruses and hosts are likely to be uncovered in the future.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The authors want to thank Friedemann Weber, Gareth Brady and the members of the Innate Immunity Laboratory for critical input and suggestions. MH is funded by an Alexander von Humboldt fellowship. AP is funded by the Max-Planck Free Floater programme, an ERC starting grant (iVIP), Infect-ERA (ERASE) and the German Research Foundation (PI 1084/2-1).
References
- 1.Hare D., Mossman K.L. Novel paradigms of innate immune sensing of viral infections. Cytokine. 2013;63:219–224. doi: 10.1016/j.cyto.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Rassa J.C., Ross S.R. Viruses and Toll-like receptors. Microbes Infect. 2003;5:961–968. doi: 10.1016/s1286-4579(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 3.Sancho D., Reis e Sousa C. Sensing of cell death by myeloid C-type lectin receptors. Curr Opin Immunol. 2013;25:46–52. doi: 10.1016/j.coi.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goubau D., Deddouche S., Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Resa-Infante P., Jorba N., Coloma R., Ortin J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 2011;8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topisirovic I., Svitkin Y.V., Sonenberg N., Shatkin A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 7.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. fourth edition. Garland Publishing; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- 8.Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habjan M., Andersson I., Klingstrom J., Schumann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Muhlberger E. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habjan M., Hubel P., Lacerda L., Benda C., Holze C., Eberl C.H., Mann A., Kindler E., Gil-Cruz C., Ziebuhr J. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog. 2013;9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Gullerova M., Proudfoot N.J. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat Struct Mol Biol. 2012;19:1193–1201. doi: 10.1038/nsmb.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]; IFIT1 specifically binds viral RNA and thereby provides a mechanism allowing very specific targeting of viral nucleic acids without harming cellular functions.
- 14.Capshew C.R., Dusenbury K.L., Hundley H.A. Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing. Nucleic Acids Res. 2012;40:8637–8645. doi: 10.1093/nar/gks590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White E., Schlackow M., Kamieniarz-Gdula K., Proudfoot N.J., Gullerova M. Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat Struct Mol Biol. 2014;21:552–559. doi: 10.1038/nsmb.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Orzalli M.H., DeLuca N.A., Knipe D.M. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dicer degrades dsRNA in the nucleus to avoid activation of cytoplasmic dsRNA receptors that activate the innate immune system.
- 17.Kato H., Takahasi K., Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 18.Cui S., Eisenacher K., Kirchhofer A., Brzozka K., Lammens A., Lammens K., Fujita T., Conzelmann K.K., Krug A., Hopfner K.P. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peisley A., Wu B., Yao H., Walz T., Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Wu B., Peisley A., Tetrault D., Li Z., Egelman E.H., Magor K.E., Walz T., Penczek P.A., Hur S. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wies E., Wang M.K., Maharaj N.P., Chen K., Zhou S., Finberg R.W., Gack M.U. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh T., Kato H., Kumagai Y., Yoneyama M., Sato S., Matsushita K., Tsujimura T., Fujita T., Akira S., Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deddouche S., Goubau D., Rehwinkel J., Chakravarty P., Begum S., Maillard P.V., Borg A., Matthews N., Feng Q., van Kuppeveld F.J. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binder M., Eberle F., Seitz S., Mucke N., Huber C.M., Kiani N., Kaderali L., Lohmann V., Dalpke A., Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triantafilou K., Vakakis E., Kar S., Richer E., Evans G.L., Triantafilou M. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 29.Feng Q., Hato S.V., Langereis M.A., Zoll J., Virgen-Slane R., Peisley A., Hur S., Semler B.L., van Rij R.P., van Kuppeveld F.J. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Runge S., Sparrer K.M., Lassig C., Hembach K., Baum A., Garcia-Sastre A., Soding J., Conzelmann K.K., Hopfner K.P. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 33•.Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]; Multiple MDA5 molecules assembles along dsRNA to arrange CARD domains for downstream signalling.
- 34.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsen M.R., Bak R.O., Andersen A., Berg R.K., Jensen S.B., Tengchuan J., Laustsen A., Hansen K., Ostergaard L., Fitzgerald K.A. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci USA. 2013;110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu Y.H., Macmillan J.B., Chen Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K.A., Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Witte G., Hornung V., Hopfner K.P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this landmark paper the authors discovered cGAS as DNA sensor that generates the cyclic dinucleotide cGAMP as signalling molecule.
- 40.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Rohl I., Hopfner K.P., Ludwig J., Hornung V. cGAS produces a 2′–5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao P., Ascano M., Wu Y., Barchet W., Gaffney B.L., Zillinger T., Serganov A.A., Liu Y., Jones R.A., Hartmann G. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP–AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hornung V., Hartmann R., Ablasser A., Hopfner K.P. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E., Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Li X.D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS–cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes a novel type of signal distribution utilising cGAMP, a small dinucleotide, which can cross cell barriers through gap junctions. The use of cGAMP as signalling molecule allows immediate response to virus infections in bystander cells, bypassing type I interferon signalling.
- 45.Pfaller C.K., Li Z., George C.X., Samuel C.E. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol. 2011;23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nallagatla S.R., Hwang J., Toroney R., Zheng X., Cameron C.E., Bevilacqua P.C. 5’-Triphosphate-dependent activation of PKR by RNAs with short stem–loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 47.Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., Alnemri E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poeck H., Bscheider M., Gross O., Finger K., Roth S., Rebsamen M., Hannesschlager N., Schlee M., Rothenfusser S., Barchet W. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 49.Mitoma H., Hanabuchi S., Kim T., Bao M., Zhang Z., Sugimoto N., Liu Y.J. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pichlmair A., Lassnig C., Eberle C.A., Gorna M.W., Baumann C.L., Burkard T.R., Burckstummer T., Stefanovic A., Krieger S., Bennett K.L. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 51.Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp-mRNAs. Nucleic Acids Res. 2014;42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate that viral RNA is bound in a groove formed by structural elements of IFIT proteins. This binding mechanism is potentially flexible to accommodate additional ligands with high specificity in a similar manner.
- 53.Malim M.H. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucifora J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X., Sprinzl M.F., Koppensteiner H., Makowska Z., Volz T. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Guo X., Ma J., Sun J., Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]; A long-sought issue on APOBEC-mediated virus targeting was how these proteins detect viral nucleic acid. It appears that APOBEC3A and APOBEC3B sense the HBV core protein associated to viral DNA and this is required for specific targeting.
- 56.Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Eckard S.C., Rice G.I., Fabre A., Badens C., Gray E.E., Hartley J.L., Crow Y.J., Stetson D.B. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol. 2014;15:839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]; Several mechanisms are in place to reduce endogenous RNA that may trigger autoinflammation or autoimmunity. The authors show that the SKIV2L exosome is contributing to degradation of endogenous ligands that would otherwise activate the RLR pathway.


