Abstract
Semliki Forest virus (SFV, genus Alphavirus) has a broad host range, high efficiency of viral protein expression, and the ability to stimulate an immune response. These properties have made SFV an attractive tool for development of expression vectors, and plasmid clones containing cDNA of the SFV genome often are used. However, instability of these plasmids resulting from cryptic expression of SFV envelope proteins in Escherichia coli represents a problem both for the development of SFV-based vectors and for SFV research. In this study, an infectious plasmid of SFV, pCMV-SFV4, was constructed; its toxic effect was eliminated by intron insertion in the capsid protein encoding region. When transfected into mammalian cells, the plasmid clone was highly infectious and produced virus with properties identical to those of wild-type SFV. The inserted intron was efficiently and properly removed from the RNA genome of SFV. Therefore, this novel and stabilized infectious SFV plasmid represents a superior tool for basic studies of SFV as well as for biotechnological applications.
Keywords: Alphavirus, Intron, Ribozyme, Infectious cDNA, CMV promoter
Semliki Forest virus (SFV) belongs to the genus Alphavirus (family Togaviridae). It infects both vertebrate hosts and insect vectors and replicates in the cytoplasm of infected cells. The virus-specific part of the replicase complex consists of the non-structural (ns) proteins (nsP) 1–4, which are expressed directly from the 5′ two-thirds of the 11.5 kb RNA genome. The structural proteins are encoded in the 3′ one-third of the genome and are expressed by subgenomic RNA, which is synthesized in infected cells. Both the nsPs and the structural proteins are expressed in the form of polyprotein precursors, which are subsequently processed by viral and host cell proteases. Five structural proteins of SFV – capsid protein C; envelope proteins E3, E2, and E1; and 6K peptide – are not essential for replication (Strauss and Strauss, 1994). Its broad host range, ability to express its own as well as foreign genes at high levels, and capacity to activate an immune response make SFV attractive for the development of expression vectors and for gene therapy (recently reviewed by Lunstrom, 2005, Liniger et al., 2007).
Basic research about SFV and the development of SFV-based vectors both utilize the cDNA of the SFV genome and/or its fragments, cloned into bacterial plasmids. The full-length infectious cDNA (icDNA) clone of SFV, pSFV4 (Liljestrom et al., 1991), has been the main tool used to construct recombinant SFV genomes and genome-based expression vectors. However, pSFV4 is rather unstable in Escherichia coli cells: it is tolerated only by a few strains and can be propagated only under certain conditions, which makes such work both inconvenient and costly. In addition, obtaining high quality plasmid preparations is very difficult. Thus, the use of pSFV4-based constructs as vaccines or gene therapy vectors is problematic.
Furthermore, the instability of pSFV4 and its derivatives does not allow its usage for several approaches. For example, the construction of libraries generated by random mutagenesis, which have been successfully used for studies of the related Sindbis virus genome (Frolova et al., 2006, Atasheva et al., 2007), is virtually impossible for pSFV4 because the recombinant libraries are rapidly overgrown by clones containing plasmids with deletions in the SFV sequence. Similarly, often no recombinant clones can be obtained when internal ribosome entry site (IRES) elements are inserted into pSFV4-based vectors (our unpublished results). In contrast, the plasmids pSFV1 and pHelper1 (Liljestrom and Garoff, 1991), which contain complete cDNAs of ns- and structural regions of SFV4, respectively, were found to be stable in all tested E. coli strains. Thus, the instability only appears when ns- and structural parts are joined together, suggesting that the instability may result from the toxic effects of virus-encoded membrane proteins that are cryptically expressed in bacterial cells. However, no bacterial promoter that might be responsible for synthesis of the SFV RNA in bacterial cells occurs in the non-viral part of the pSFV4. Based on these data, it was hypothesized that the existence of a cryptic promoter inside the ns-protein encoding sequences might be responsible for expression of the mRNA used for translation of SFV membrane proteins in bacterial cells.
To test this hypothesis, the sequence of pSFV4 was analyzed for the presence of E. coli promoter-like sequences using the SoftBerry server (http://softberry.com). Several putative promoter sequences were found, with a promoter-like region located inside the nsP2 encoding region (–35 box: residues 3176-3181 TTGACA of SFV4; –10 box: residues 3199-3204 ATAAT of SFV4) showing the highest probability. However, all attempts to detect the expression of SFV-specific proteins located downstream of this promoter (nsP3, nsP4, or capsid protein) in bacterial cells using Western blotting failed (data not shown). Because this negative result may reflect the high toxicity of SFV proteins, the scanning of pSFV4 by introduction of nonsense codons into the reading frame of structural proteins was conducted. Stop codons were introduced using PCR mutagenesis immediately downstream of the initiation codon (position 7423 in the pSFV4 sequence), 99 bp downstream (position 7522 in the pSFV4 sequence), at the end of the capsid protein coding sequence (position 8221 in the pSFV4 sequence), and at the end of the coding sequence of the E2 protein (position 9685 in the pSFV4 sequence) (Fig. 1 ). These plasmids, together with non-mutated pSFV4, were transformed into the DH5α strain of E. coli. Very small colonies appeared in bacteria transformed with pSFV4, whereas transformants harboring the mutant plasmids formed significantly larger colonies (data not shown). The differences in growth properties were even more drastic in samples grown on liquid SOY-broth (Difco) containing 100 μg/ml ampicillin. While the bacteria transformed with pSFV4 failed to grow in liquid selective media, all bacteria transformed with mutated plasmids grew to high densities. Bacteria harboring plasmids with nonsense mutations immediately downstream of the initiation codon of structural proteins grew slightly slower than did the other three transformants (Fig. 1). Restriction analysis of the plasmid DNAs purified from these overnight cultures (four independent colonies per construct were analyzed) did not reveal any differences from the predicted restriction map (data not shown), which indicates that all mutated plasmids were stable in the DH5α strain of E. coli. Based on these data, it can be concluded that the toxic effect of pSFV4 on the bacteria is mediated by expression of the 6K-E1 region. This conclusion agrees with the finding that when the E1 protein is expressed alone in E. coli strain BL21(DE3)pLysS, it has toxic effects, binds to the bacterial membranes, and permeabilizes the cells (Nieva et al., 2004). However, the toxic effect of the E1 envelope protein observed by Nieva et al. was moderate and the bacteria tolerated significant levels of E1 expression. Attempts to propagate pSFV4 in E. coli strain BL21(DE3)pLysS were not successful, which suggests that for pSFV4, the toxic effect of E1 on bacterial cells is significantly enhanced by co-expression with the 6K peptide and possibly also with E3 and E2 envelope proteins.
Fig. 1.
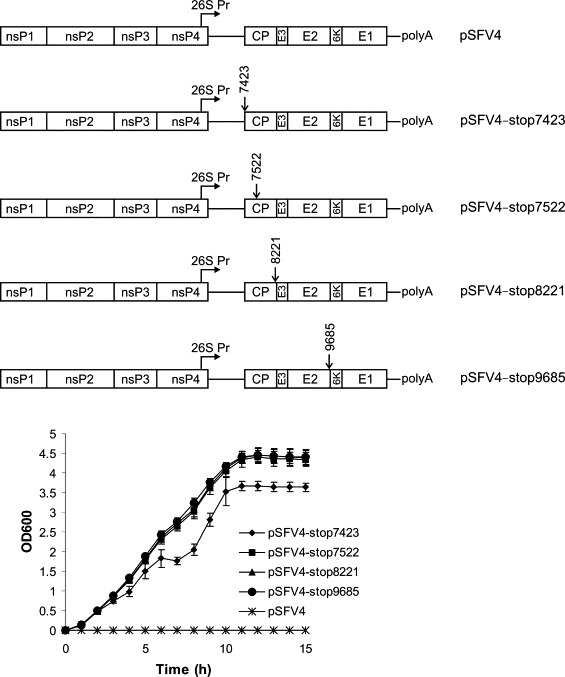
Positions of nonsense mutations introduced into the structural region SFV4 and their effect on the growth of E. coli DH5α cultures transformed using mutant versions of pSFV4. The growth of E. coli harbouring pSFV4 or its derivatives was measured in SOY medium supplemented with ampicillin (50 μg/ml). Optical density of the cultures was determined at 600 nm. The average of four parallel measurements are shown; error bars represent the standard deviation.
The instability of the plasmids containing cDNAs of RNA viruses can be reduced by preventing expression of the toxic protein either through elimination of the cryptic promoter activity or by disrupting the open reading frame of the toxic protein(s). The latter is more commonly used because the elimination of promoter activity requires extensive silent mutagenesis and the possible side effects of this treatment cannot be predicted. The disruption of viral sequences by intron insertion has been used successfully in several plant- and animal-infecting viruses, including potyviruses (Johansen, 1996, Yang et al., 1998, Lopez-Moya and Garcia, 2000), tobamoviruses (Marillonnet et al., 2005), and coronaviruses (Gonzalez et al., 2002). In addition to stabilizing the plasmids in E. coli, this approach also has potential to increase the infectivity of cloned sequences and to enhance virus-mediated gene expression (Marillonnet et al., 2005).
Insertion of a functional intron requires the use of expression plasmids that contain elements necessary for eukaryotic transcription. The plasmids encompassing full-length icDNA have been described for Sindbis virus (Dubensky et al., 1996) but not for SFV. However, numerous clones containing either the SFV non-structural portion or the structural region that are under the control of eukaryotic transcription elements have been reported (Berglund et al., 1998, DiCommo and Bremner, 1998, Kohno et al., 1998, Nordström et al., 2005).
In this work, the sequence of the full-length icDNA of SFV under the control of a CMV promoter (pCMV-SFV4) was constructed by combining restriction fragments from pBK-SFV-E (Berglund et al., 1998), pSFV1, and pHelper1 (Liljestrom and Garoff, 1991). The hepatitis delta virus antisense ribozyme sequence, synthesized by Geneart AG (Germany), was placed immediately downstream of the polyA sequence of SFV, essentially as described by Nordström et al. (2005). A large (573 bp long) intron from the rabbit beta-globin gene was PCR amplified and inserted between nucleotides 7864 and 7865 of the SFV4 genome. Sequences of all primers used are available from the authors upon request. The selection of the intron-insertion region in the middle of the sequence that encodes the SFV capsid protein was based on the results of a nonsense codon insertion experiment (Fig. 1) and on the assumption that possible expression of any SFV envelope protein, not just the 6K-E1 region, can be toxic to bacteria under some conditions. The precise point of insertion was selected using a prediction made by the NetGene2 server (http://www.cbs.dtu.dk/services/NetGene2), which indicated a very high probability of the splicing of the intron inserted in that position.
An infectious center assay in BHK-21 cells was conducted as described by Gorchakov et al. (2004). The infectivity of the pCMV-SFV4 plasmid was approximately 1 × 105 plaques/μg of DNA (electroporation with Bio-Rad Gene Pulser II; 200 V, 975 μF, one pulse in cuvette with 0.4 cm electrode cap, 50 μg herring sperm DNA added to sample), and the infectivity of transcripts from pSFV4 was 5 × 105 plaques/μg of RNA (electroporation with Bio-Rad Gene Pulser II; 850 V, 25 μF, two pulses in cuvette with 0.4 cm electrode cap). The infectivity of pCMV-SFV4 was nearly 10-fold higher than that previously reported for the Sindbis virus (Dubensky et al., 1996). The higher infectivity observed in this study might be due to the intron insertion in pCMV-SFV4, although it also could be the result of the different protocols used in the two studies.
The growth curves in BHK-21 cells were constructed and compared. 5 × 106 BHK-21 cells were transfected with 1 × 106 infectious units of RNA transcripts or pCMV-SFV4, and the titers of collected viruses were determined by plaque titration. The release of infectious virus from pCMV-SFV4 infected cells was slightly delayed and the virus titer was 5- to 10-fold lower than when the cells were transfected with RNA transcripts (Fig. 2 ). The initial delay (less than 1 h) in virus production may be explained as follows: unlike infectious RNA, which can start the infection process immediately after entering the cell cytoplasm, pCMV-SFV4 must be transported first into the nucleus where its transcription with cellular RNA polymerase occurs. Differences in the final titers of collected stocks might have resulted from the different electroporation conditions. Alternatively, the overall particle production could be similar in both cases, but the infectious unit/particle ratio could be reduced in the case of transfection with pCMV-SFV4.
Fig. 2.
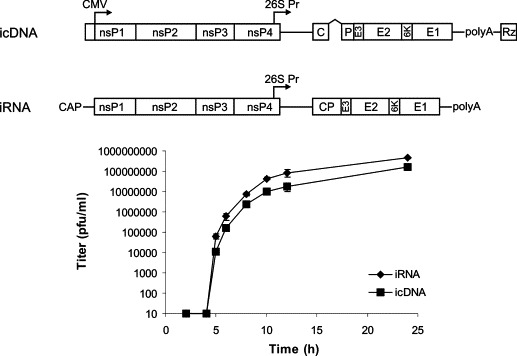
Comparison of the virus production in BHK-21 cultures transfected with infectious RNA transcripts from pSFV4 (iRNA) or pCMV-SFV4 plasmids (icDNA). 5 × 106 BHK-21 cells were electroporated with 1 × 106 infectious units of iRNA (2 μg; 850 V, 25 μF, two pulses in cuvette with 0.4 cm electrode cap) or icDNA (10 μg; 200 V, 975 μF, one pulse in cuvette with 0.4 cm electrode cap, 50 μg herring sperm DNA added to sample) using Gene Pulser II equipment (Bio-Rad). Transfected cells were incubated at 37 °C, aliquots of growth media were collected at selected time points, and the amount of released virions was detected by plaque titration. Error bars represent the standard deviation; results of one from two reproducible experiments are shown.
A common reason for the reduced infectious unit/particle ratio is the formation and packaging of defective interfering (DI) genomes. In cells transfected with pCMV-SFV4, two putative specific mechanisms for generation of DI-genomes can be proposed. First, the intron inserted into the CP gene may be inefficiently spliced, which would allow the resulting viral genomes to replicate but not to express envelope proteins or full-length capsid protein. The latter would be composed of 153 aa residues instead of 267 aa residues. When co-replicating with correctly spliced genomes, the non spliced RNAs can be packed into virions. Second, the genomic sequence of SFV contains a number of cryptic splicing sites. The NetGene2 server analysis identified 35 possible donor splice sites and 129 possible acceptor splice sites in the positive strand of SFV4 RNA. Furthermore, seven possible donor splice sites were predicted as highly confident splice sites. Thus, splicing between them or between native and introduced splicing sites could occur and, as a result, different DI-RNAs could be generated. Because these processes may seriously affect the use of pCMV-SFV4 for different applications, the formation of putative splicing products was studied.
First, the genomic RNA of SFV was purified from the collected virions and subjected to RT–PCR with primers that flanked the inserted intron. This analysis, as well as RT–PCR with intron-specific primers, failed to reveal any packed genomes with unspliced introns (data not shown). Sequencing of the obtained RT–PCR products revealed that the intron always was removed correctly. Second, RT–PCR analysis was performed using pairs of primers, of which one was located close to the inserted intron and the other close to a predicted cryptic splicing site. Seven different pairs of primers failed to detect any incorrect splicing products (data not shown). Taken together, these data indicate that the inserted intron was removed efficiently and correctly from the RNA genome of SFV.
Generally, the DI-genomes of viruses have a growth advantage over full-length genomes and, as a result, their numbers rapidly increase in the population when the virus multiplies at a high multiplicity of infection (m.o.i.). Therefore, the expression of the nsPs and capsid protein of SFV in cells transfected with pCMV-SFV4 or infected with collected virus stock at different m.o.i.s was analyzed. Under all tested conditions, the correct sized proteins were the major products (Fig. 3 ). Proteins of smaller sizes were detected using antibodies against nsP2 and nsP4; they most likely were degradation products of these proteins, as they also were detected in cells infected with control stock of SFV4. Some smaller proteins also were detected with antibodies against capsid protein (Fig. 3). These proteins were not found in the control sample; at the same time, the molar ratio of truncated products and full-size products in cells infected with collected virus stock did not depend on the amount of plasmid-derived virus used for infection (Fig. 3). Thus, the origin of the shorter proteins recognized by antibody against capsid protein remains unclear, but it cannot be excluded that they were expressed from minor non-spliced and/or incorrectly spliced RNAs. Therefore, standard precautions, such as propagation of SFV stocks under low m.o.i. conditions, should be used for viruses generated using pCMV-SFV4 or its derivatives. This may be the only possible option in the case of propagation of large SFV4-based libraries; in the case of propagation of individual mutant genomes, plaque purification also can be used.
Fig. 3.
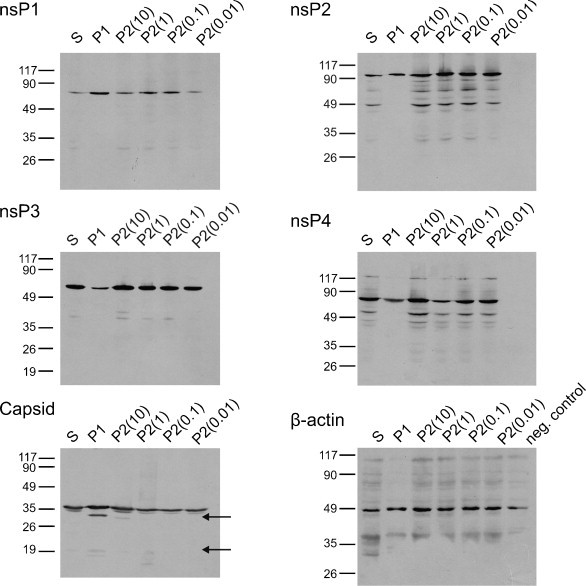
Western blot analysis of the nsP1–4 and capsid protein expressed in transfected (P1) or infected (P2) BHK-21 cells. The BHK-21 cells transfected with 1 × 106 infectious units of pCMV-SFV4 or infected with collected virus stocks at the indicated m.o.i. were lysed in Laemmli's buffer at 16 h post-infection and analyzed by SDS–PAGE in 10% (nsPs) or 15% (capsid protein) gels. The amount of lysate corresponding to 50,000 cells was loaded on each line; the lysate from standard SFV4 stock-infected cells (S, m.o.i. 10) was used as the control. Proteins were transferred to nitrocellulose membrane and probed with corresponding mono-specific antibodies and HRP-conjugated secondary antibody. The blots were developed using the ECL procedure (Amersham). The Western blot of the same extracts made using antibody against beta-actin is presented as the control. Positions of the molecular mass standards are indicated; the arrows indicate the truncated forms of the capsid protein.
Taken together, these data demonstrate that interruption of the reading frame of structural proteins by intron insertion drastically increases the stability of plasmids containing full-length icDNA of SFV4. The plasmid pCMV-SFV4, which encompasses the SFV4 sequence flanked by the CMV promoter, the hepatitis delta virus ribozyme, and the eukaryotic transcription terminator sequence, was highly infectious. The virus stock collected from transfected cells did not contain detectable amounts of non-spliced or incorrectly spliced RNAs, and BHK-21 cells infected with that stock expressed nsPs and capsid proteins of correct sizes. Therefore, the new clone represents a useful instrument that can be used for basic studies of SFV as well as for biotechnological applications. As an example of the latest developments, it already has been demonstrated that pCMV-SFV4 (in contrast to pSFV4) tolerates insertion of different IRES sequences between ns- and structural regions (our unpublished results).
Acknowledgements
The authors are grateful to Margus Varjak for his help. This research was supported by Enterprise Estonia project “Ekspress”.
Contributor Information
Liane Ülper, Email: liane.ylper@mail.ee.
Inga Sarand, Email: isarand@ebc.ee.
Kai Rausalu, Email: kaira@ebc.ee.
Andres Merits, Email: andres.merits@ut.ee.
References
- Atasheva S., Gorchakov R., English R., Frolov I., Frolova E. Development of Sindbis viruses encoding nsP2/GFP chimeric protein and their application for studying nsP2 functioning. J. Virol. 2007;81:5046–5057. doi: 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund P., Smerdou C., Fleeton M.N., Tubulekas I., Liljestöm P. Enhancing immune response using suicidal DNA vaccines. Nat. Biotechnol. 1998;16:562–565. doi: 10.1038/nbt0698-562. [DOI] [PubMed] [Google Scholar]
- DiCommo D.P., Bremner R. Rapid, high level protein production using DNA-based Semliki Forest virus vectors. J. Biol. Chem. 1998;273:18080–18086. doi: 10.1074/jbc.273.29.18060. [DOI] [PubMed] [Google Scholar]
- Dubensky T.W., Jr., Driver D.A., Polo J.M., Belli B.A., Latham E.M., Ibanez C.E., Chada S., Brumm D., Banks T.A., Mento S.J., Jolly D.J., Chang S.M.W. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J. Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova E., Gorchakov R., Garmashova N., Atasheva S., Vergara L.A., Frolov I. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 2006;80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.M., Penzes Z., Almazan F., Calvo E., Enjuanes L. Stabilization of a full-length infectious cDNA clone of transmissible gastroenterits coronavirus by insertion an intron. J. Virol. 2002;76:4644–4661. doi: 10.1128/JVI.76.9.4655-4661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R., Frolova E., Williams B.R., Rice C.M., Frolov I. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 2004;78:8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen I.E. Intron insertion facilitates amplification of clones virus cDNA in Escherichia coli while biological activity is re-established after transcription in vivo. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12400–12405. doi: 10.1073/pnas.93.22.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno A., Emi N., Kasai M., Tanimoto M., Saito H. Semliki Forest virus-based expression vector: transient protein production followed by cell death. Gene Ther. 1998;5:415–418. doi: 10.1038/sj.gt.3300589. [DOI] [PubMed] [Google Scholar]
- Liljestrom P., Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (NY) 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- Liljestrom P., Lusa S., Huylebroeck D., Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liniger M., Zuniga A., Naim H.Y. Use of viral vectors for the development of vaccines. Expert Rev. Vac. 2007;6:255–266. doi: 10.1586/14760584.6.2.255. [DOI] [PubMed] [Google Scholar]
- Lopez-Moya J.J., Garcia J.A. Construction of a stable and highly infectious intron-containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 2000;68:99–107. doi: 10.1016/s0168-1702(00)00161-1. [DOI] [PubMed] [Google Scholar]
- Lunstrom K. Biology and application of alphavirus in gene therapy. Gene Ther. 2005;12(Suppl 1):S92–S97. doi: 10.1038/sj.gt.3302620. [DOI] [PubMed] [Google Scholar]
- Marillonnet S., Thoeringer C., Kandzia R., Klimyuk V., Gleba Y. Systemic Agrobacterium tumifaciens-mediated trnsfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 2005;23:718–723. doi: 10.1038/nbt1094. [DOI] [PubMed] [Google Scholar]
- Nieva J.L., Sanz M.A., Carrasco L. Membrane permeabilizing motif in Semliki Forest virus E1 glycoprotein. FEBS Lett. 2004;576:417–422. doi: 10.1016/j.febslet.2004.09.061. [DOI] [PubMed] [Google Scholar]
- Nordström E.K.L., Forsell M.N.E., Barnfield C., Bonin E., Hanke T., Sundström M., Karlsson G.B., Liljeström P. Enhanced immunogenicity using an alphavirus replicon DNA vaccine against human immunodeficiency virus type 1. J. Gen. Virol. 2005;86:349–354. doi: 10.1099/vir.0.80481-0. [DOI] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.J., Revers F., Souche S., Lot H., Le Gall O., Candresse T., Dunez J. Construction of full-length cDNA clones of lettuce mosaic virus (LMV) and the effects of intron-insertions on their viability in Escherichia coli and their infectivity to plants. Arch. Virol. 1998;143:2443–2451. doi: 10.1007/s007050050474. [DOI] [PubMed] [Google Scholar]


