Abstract
Yellow head virus (YHV) is an invertebrate nidovirus that is highly pathogenic for marine shrimp. Nucleotide sequence analysis indicated that the YHV ORF2 gene encodes a basic protein (pI = 9.9) of 146 amino acids with a predicted molecular weight of 16,325.5 Da. The deduced amino acid sequence indicated a predominance of basic (15.1%), acidic (9.6%) and hydrophilic polar (34.3%) residues and a high proportion proline and glycine residues (16.4%). The ORF2 gene was cloned and expressed in Escherichia coli as a Mr = 21 kDa His6-protein that reacted with YHV nucleoprotein (p20) monoclonal antibody. Segments representing the four linear quadrants of the nucleoprotein were also expressed in E. coli as GST-fusion proteins. Immunoblot analysis using YHV polyclonal rabbit antiserum indicated the presence of linear epitopes in all except the V37–Q74 quadrant. Immunoblot analysis of the GST-fusion proteins and C-terminally truncated segments of the nucleoprotein allowed mapping of YHV monoclonal antibodies Y19, Y20 and YII4 to linear epitopes in the acidic domain between amino acids I116 and E137. The full-length nucleoprotein was expressed at high level in E. coli and was easily purified in quantity from the soluble cell fraction by Ni+-NTA affinity chromatography.
Keywords: Yellow head virus, Shrimp virus, Nucleoprotein, Bacterial expression
1. Introduction
Yellow head virus (YHV) is a virulent pathogen of the black tiger shrimp (Penaeus monodon). The disease was first reported to occur in 1990 in shrimp farms in central Thailand (Limsuwan, 1991). Typical signs of yellow head disease (YHD) include cessation of feeding and erratic swimming, and moribund shrimp often display a pale yellowish hue of the cephalothorax due to discoloration of the underlying hepatopancreas (Chantanachookin et al., 1993). Although the impact of YHD in Asia was quickly overshadowed by the subsequent emergence and explosive spread on white spot disease, yellow head outbreaks remain a serious concern for farmers, commonly resulting in total crop loss within 3 days of the first appearance of dead or moribund shrimp at the pond edge (Flegel et al., 1997, Walker et al., 2001). However, this important pathogen is poorly understood with relatively little information yet available on either the epidemiology of YHD or the molecular biology of YHV infection.
YHV is (+) ssRNA virus that, together with closely related Gill-associated virus (GAV) from Australia, has recently been classified in the family Roniviridae, genus Okavirus within the order Nidovirales (Walker et al., 2004). YHV virions are rod-shaped, enveloped particles (approximately 70 nm × 180 nm) with prominent surface spikes and an internal helical nucleocapsid (Wongteerasupaya et al., 1995, Nadala et al., 1997). Three YHV structural proteins have been described, transmembrane glycoproteins gp116 and gp64, which are located in the viral envelope, and the nucleoprotein p20 which is the only protein known to be present in nucleocapsids (Jitrapakdee et al., 2003, Soowannayan et al., 2003). For GAV, the 26,235 nt genome has been sequenced and reported to contain at least four long open reading frames (ORFs). ORF1a and ORF1b are in overlapping frames and encode non-structural proteins, including a 3C-like proteinase and enzymes of the replication complex (Cowley et al., 2000, Ziebuhr et al., 2003). ORF1b is expressed from a genome-length polyadenylated mRNA only following a −1 ribosomal frame-shift at a putative pseudoknot structure in the ORF1a/1b overlap region (Cowley et al., 2000). ORF3 has been shown to encode a long polyprotein with multiple N-linked glycosylation sites and membrane-spanning domains (Cowley and Walker, 2002). GAV ORF2 has recently been shown to encode the GAV nucleoprotein—a remarkable observation as, in all other known nidoviruses (including coronaviruses, toroviruses and arteriviruses), the nucleoprotein gene is located downstream (5′-proximate) rather than upstream (3′-proximate) of the glycoprotein genes (Cowley et al., 2004).
The YHV genome is less well characterized. ORF1b has been shown to share 80.5% overall nucleotide sequence identity with the cognate gene of GAV and encodes a polypeptide containing all recognized functional domains including polymerase, helicase and metal ion-binding sequence motifs (Sittidilokratna et al., 2002). YHV structural glycoproteins gp116 and gp64 have been shown to be encoded in ORF3 and generated by post-translational proteolytic cleavage of a long polyprotein (Jitrapakdee et al., 2003). In this paper, we describe the nucleotide sequence and deduced amino acid sequence of YHV ORF2 and show that, like the cognate gene in GAV, it encodes the nucleocapsid protein p20. We also show that recombinant YHV nucleoprotein is expressed at high level E. coli and contains linear epitopes in the C-terminal domain that react with available nucleoprotein monoclonal antibodies.
2. Materials and methods
2.1. Virus purification, RT-PCR amplification and sequencing
The YHV isolate used in this work was obtained from moribund P. monodon showing signs of yellow head disease that were collected from a farm in Chachoengsao Province, Thailand, in July 1998 and was purified using the protocol described previously (Sittidilokratna et al., 2002). Genomic RNA was extracted from purified YHV using Trizol™ reagent (Invitrogen), dissolved in diethyl pyrocarbonate-treated, sterile water and stored at −80 °C.
The generation of a 6.7 kb RT-PCR product extending from within the ORF1b gene to the 3′-poly[A] tail was described previously (Jitrapakdee et al., 2003). The 6.7 kb product was purified using a QIAquick™ gel-affinity column (QIAGEN) and sequenced directly using the BIG Dye™ (ABI) dye-terminator system and the ABI Model 377 automated sequencer at the Bioservice Unit (BSU), National Science and Technology Development Agency (NSTDA), Bangkok, Thailand. Sequences downstream of the ORF1b gene were obtained by walking from the 5′-end of the 6.7 kb product using sequence-specific primers. Overlapping sequence contigs were complied using SeqEd 1.0.3 (ABI) software. The entire nucleotide sequence of the ORF2 region was analyzed using the MacVector 6.0.1 software. Potential phosphorylation sites in the deduced amino acid sequence were predicted using NetPhos 2.0 server of the Technical University of Denmark by imposing a threshold prediction score of 0.9.
2.2. Amplification and cloning of full-length YHV ORF2
ORF2 was generated by RT-PCR from YHV genomic RNA using forward primer NS2 and reverse primer NA2 (Table 1 ). The forward primer included a NheI restriction site immediately upstream of the ORF2 initiation codon. The reverse primer included a XhoI restriction site, a termination codon and a sequence encoding a His6 tag. The RT-PCR reaction was conducted using ∼20 pg of YHV RNA, a primer set containing 500 nM total primers and 8 U RNasin (Promega) in a total reaction volume of 25 μl. Amplification was conducted in a Perkin-Elmer 2400 thermal cycler using the following temperature cycles: 1 × 50 °C/30 min, 94 °C/2 min (for cDNA synthesis), 35 × 94 °C/15 s, 58 °C/15 s, 72 °C/30 s (for amplification) and 1 × 72 °C/10 min (for end-filling). The PCR products were fractionated in agarose gels, affinity purified (QIAGEN) and cloned into pGEM-T Easy vector (Promega). The nucleotide sequence of the inserts was confirmed. Plasmid pGEM-T-FL containing the amplified full-length ORF2 (M1–S146) and expression vector pET17b were digested with NheI and XhoI, ligated and transformed into E. coli DH5α for amplification.
Table 1.
Primers used for PCR amplification and cloning of segments of the YHV ORF2 gene
| Primer pair | Sequence | Amplified region | Recombinant plasmid |
|---|---|---|---|
| NS2 NA2 | 5′-ACTTCTGCTAGCATGAACCGTCGTACACGCACCGCA-3′a 5′-ATTTAGCTCGAGTTA(ATG)6TGATTGTGTTTCCATGGGTTC-3′ | M1–S146 | pET17b-FL |
| NS5 NA2 | 5′-ACTGGGATCCCCATGAACCGTCGTACACGCACCGCA-3′ | M1–E73 | pGEX-5X-1-N |
| NS4 NA4 | 5′-CCATTCATTGGGATCCCCCATATGCAATCCCTCCAAGTTGTCA-3′. 5′-CCATCCTCGAGCTA(ATG)6TTCTTGAAGTCCTTGAATGGATGG-3′ | Q74–S146 | pGEX-5X-1-C |
| NS5 NA6 | 5′-TATGCCTCGAGCTA(ATG)6TGCGAAGGATTGAGGAATCTCGAT-3′ | M1–A36 | pGEX-5X-1-NN |
| NS7 NA4 | 5′CCTCGGATCCCCATGGTCGAACGCGGTAATGGATGGACT-3′ | V37–E73 | pGEX-5X-1-CN |
| NS4 NA5 | 5′-GATCCTCGAGCTA(ATG)6CTTTGGCAGAATGGGATCGTTGGG-3′ | Q74–K109 | pGEX-5X-1-NC |
| NS6 NA2 | 5′-GATGGGATCCCCCATATGCGTTCAACACAGAAGTCAATCGTTCC-3′ | R110–S146 | pGEX-5X-1-CC |
| NS2 NA9 | 5′-CATGGGCTCGAGCTATTCAAGGCTCATTGCGTGGATACC-3′ | M1–E137 | pET17b-FL-Δ9 |
| NS2 NA8 | 5′-TGCGTGCTCGAGCTAAGCTAGGTTCTCTATGTCAAGGGA-3′ | M1–A128 | pET17b-FL-Δ18 |
| NS2 NA7 | 5′-TATGTCCTCGAGCTAATCGGGAACGATTGACTTCTGTGT-3′ | M1–D119 | pET17b-FL-Δ27 |
Restriction endonuclease sites are underlined and transcription initiation and termination codons are in bold.
2.3. Amplification and cloning of ORF2 segments
Segments encoding the N-terminal half (M1–E73) and C-terminal half (Q74–S146) of the ORF2 protein and peptides NN (M1–A36), CN (V37–E73), NC (Q74–K109) and CC (R110–S146) were generated by PCR using plasmid pGEM-T-FL as template and primer pairs NS5/NA4, NS4/NA2, NS5/NA6, NS7/NA4; NS4/NA5 and NS6/NA2 (Fig. 1 ). PCR was performed and the PCR were products gel purified and cloned into pGEM-T Easy vector (Promega) as described above. The selected clones were digested with BamHI and XhoI and the inserts gel purified and ligated into the multiple cloning site of pGEX-5X-1 (Amersham Pharmacia Biotech) to generate plasmids pGEX-5X-1-N, pGEX-5X-1-C, pGEX-5X-1-NN, pGEX-5X-1-CN, pGEX-5X-1-NC and pGEX-5X-1-CC (Table 1). Segments of the His6-ORF2 protein truncated at the C-terminus by 9, 18 and 27 amino acids were also generated by PCR using pGEM-T-FL as a template and primer pairs NS2/NA9, NS2/NA8 and NS2/NA7 (Fig. 1), and cloned into pGEM-T Easy vector as above. Clones encoding the required terminally truncated segments were digested with NheI and XhoI, gel purified and ligated into the multiple cloning site of expression vector pET17b to generate plasmids pET17b-FL-Δ9, pET17b-FL-Δ18 and pET17b-FL-Δ27 (Table 1). All recombinant plasmids were transformed into E. coli DH5α for amplification.
Fig. 1.

(a) Clustal W alignment of YHV and GAV ORF2 deduced amino acid sequences indicating predicted phosphorylation sites (underlined) and predicted epitopes for monoclonal antibodies Y19, Y20 and YII4 (boxed); (b) Schematic representation of YHV ORF2 gene segments expressed in E. coli and immunoblot reactions of the expressed segments with polyclonal rabbit antiserum and mouse monoclonal antibodies Y19, Y20 and YII4.
2.4. Expression in E. coli of recombinant ORF2 protein and partial ORF2 segments
Plasmids pET17b-FL, pGEX-5X-1-N, pGEX-5X-1-C, pGEX-5X-1-NN, pGEX-5X-1-CN, pGEX-5X-1-NC, pGEX-5X-1-CC, pET17b-FL-Δ9, pET17b-FL-Δ18 and pET17b-FL-Δ27 were purified and the in-frame orientation of inserts was confirmed by PCR. E. coli BL21cells were transformed with the plasmids selected clones were incubated overnight at 37 °C in LB medium containing 100 μg/ml ampicillin and 25 μg/ml chloramphenicol to an optical density of 0.6–0.8 A600nm. A 2 ml aliquot of each culture was induced with 1 mM isopropyl-β-d-thiogalactoside (IPTG) for 0, 1 and 4 h at 37 °C shaking incubator. Cells were collected by centrifugation, resuspended in 100 μl of 1X protein loading buffer (Laemmli, 1970) and 10 μl of each induction was used to analyze directly by SDS-PAGE using a 12.5% gel. Purified YHV was used as positive control and E. coli BL21 containing pET17b and pGEX-5X-1 with no inserts were used as negative controls. Gels were stained with Coomassie brilliant blue R250 and de-stained by standard procedures. Broad range protein molecular weight markers (2–212 kDa) were used to estimate molecular weights of expressed proteins (New England Biolabs).
2.5. Antibodies
Polyclonal YHV antibody was prepared in rabbits at the Institute of Medical and Veterinary Services, Adelaide, South Australia using YHV purified as described previously (Sittidilokratna et al., 2002). Rabbits were injected with 100 μg of purified virus in Freund's complete adjuvant followed at intervals of 10–14 days by three doses of 100 μg in Freund's incomplete adjuvant. Mouse monoclonal antibodies were kindly provided by Dr Paisarn Sithigorngul, Srinakarinwirot University, Bangkok, Thailand. Clone Y20 was specific for the YHV nucleocapsid protein p20; clones Y19 and YII4 were also specific for YHV p20 but cross-reacted with GAV. Clone Y3 (V3-2B) was specific for YHV structural glycoprotein gp116 (Sithigorngul et al., 2000, Sithigorngul et al., 2002, Soowannayan et al., 2003).
2.6. Immunoblot analysis
Proteins separated by SDS-PAGE were electroblotted to nitrocellulose membranes (Protran, Schleicher and Schuel) using a Semi Phor™ Semi-Dry Transfer Unit apparatus (Amersham Pharmacia Biotech). Blots were blocked with 5% skim milk powder in TBS-T buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.05% Tween-20) at room temperature overnight. Membranes were probed with mouse monoclonal antibody or rabbit polyclonal antibody (1:200) in 5% skim milk powder for 2 h and then washed three times with TBS-T. Membranes were then incubated with gentle agitation for 1 h at room temperature in alkaline phosphatase-conjugated goat anti-mouse IgG (Zymed) (1:3000) or goat anti-rabbit IgG (Sigma) (1:30,000) in TBS-T buffer. Membranes were washed three times with TBS-T buffer and developed in the dark in a solution containing 45 μl of 100 mg/ml NBT (p-nitro blue tetrazolium chloride) and 35 μl of 50 mg/ml BCIP (5-bromo-4-chloro-3-indolyl phosphate, p-toluidine salt) in 10 ml of TBS-T. The reaction was stopped by adding 0.5 M EDTA and immunoreactive bands were detected visually. PageRuler™ Prestained Protein Ladder (Fermentas Life Sciences) was used to confirm the size of reactive bands.
2.7. Purification of recombinant ORF2 protein
Following induction for 4 h with IPTG, E. coli BL21cells transformed with plasmid pET17b-FL and expressing full-length His6-ORF2 protein were suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM PMSF [phenyl-methyl-sulphonyl fluoride], 20 mM imidazole, pH 8.0) and 1 mg/ml lysozyme. The cells were lysed by three cycles of sonication and centrifuged at 10,000 × g for 30 min. The supernatant fraction was applied to a 0.5 ml Ni-NTA column equilibrated with lysis buffer, washed with two column volumes of lysis buffer containing 30 mM imidazole, and then with four column volumes of lysis buffer containing increasing concentrations up to 100 mM imidazole. The bound His6-ORF2 protein was then eluted by washing with lysis buffer containing 250 mM imidazole. Protein determinations were conducted using the Quick Start Bradford Protein Assay Kit (Bio-Rad Laboratories). The relative intensity of protein bands in Coomassie-stained gels was determined using an ImageScanner™ flatbed scanner (Pharmacia Amersham) and analysed using ImageMaster TotalLab software.
3. Results
3.1. Nucleotide and deduced amino acid sequences of YHV ORF2
The nucleotide sequence of the YHV ORF2 gene is deposited in GenBank under accession number DQ067891. Analysis of the sequence indicated that the ORF2 gene initiation codon is located 352 nucleotides downstream of the termination codon of ORF1b. The characteristics of this long intergenic region have been described previously (Sittidilokratna et al., 2002). The ORF2 gene comprises 441 nucleotides and encodes a polypeptide of 146 amino acids with predicted molecular weight of 16 325.5 Da. The ORF2 amino acid sequence indicates a highly hydrophilic, basic polypeptide (pI = 9.9) with a predominance of basic (15.1%), acidic (9.6%) and hydrophilic polar (34.3%) residues. The linear distribution of charged amino acids is highly polar. The extreme N-terminal domain of 27 amino acids contains 9 of the 22 basic residues and no acidic residues; the extreme C-terminal domain contains 9 of the 14 acidic residues and only one basic residue. The ORF2 amino acid sequence also features a high proportion P and G residues (16.4%), which are commonly associated with angular turns in the secondary structure, and 6 potential phosphorylation sites (T9, S19, Y86, T112, S115 and S135) (Fig. 1a).
Clustal W alignment of the YHV sequences with GAV ORF2 (Cowley et al., 2004) indicates 79% nucleotide and 84% amino acid identity. Variations in the deduced amino acid sequences occur primarily in the highly charged N-terminal and C-terminal domains. The central core of the polypeptide is highly conserved with only five conservative and one non-conservative substitutions in 110 amino acid residues, and four of six predicted phosphorylation sites are conserved (Fig. 1a). The overall level of amino acid sequence identity for the YHV and GAV ORF2 proteins is similar to that previously reported for the ORF1b polyprotein (88.9%) (Sittidilokratna et al., 2002).
3.2. Identification of ORF2 as the YHV nucleocapsid protein gene
E. coli BL21 cells transformed with recombinant plasmid pET17b-FL containing full-length YHV ORF2 gene were cultured to mid-log phase and induced with IPTG for 0, 1 and 4 h. Following induction, His6-ORF2 protein was expressed at high level, migrating in SDS-PAGE gels as a M r = 21 kDa protein (Fig. 2a). Immunoblot analysis (Fig. 2b) indicated that both the YHV virion p20 and the expressed recombinant ORF2 protein reacted with monoclonal antibody Y19 which is known to be specific for the YHV nucleoprotein (Sithigorngul et al., 2002, Soowannayan et al., 2003).
Fig. 2.
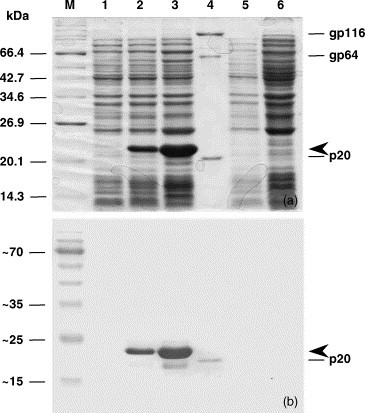
Full-length recombinant YHV His6-ORF2 protein expressed in E. coli and analysed by SDS-PAGE (panel a) and immunoblot using YHV monoclonal antibody Y19 (panel b). Molecular weight markers (lane M); pET17b-FL uninduced (lane 1); pET17b-FL after 1 h induction with 1 mM IPTG (lane 2), pET17b-FL after 4 h induction with 1 mM IPTG (lane 3), purified virus (lane 4), pET17b uninduced (lane 5), pET17b after 4 h induction with 1 mM IPTG (lane 6). Arrowheads indicate the expressed His6-tagged recombinant nucleoprotein.
3.3. Location of linear B cell epitopes
E. coli BL21 cells were transformed with recombinant plasmids pGEX-5X-N and pGEX-5X-C, induced with IPTG and analyzed by SDS-PAGE and immunoblotting. Following induction, GST-fusion proteins incorporating the N-terminal half (M1–E73) and C-terminal half (Q74–S146) of the YHV ORF2 nucleoprotein were expressed as 32 and 35 kDa proteins, respectively (Fig. 3a). Immunoblot analysis indicated that each expressed segment reacted with polyclonal YHV rabbit antiserum (Fig. 3b). However, staining of the C-terminal segment was significantly stronger, suggesting a relatively greater concentration of immunoreactive epitopes in this half of the protein. In immunoblots using mouse monoclonal antibodies Y19, Y20 and YII4, each reacted only with the C-terminal segment (Fig. 1, Fig. 3). Immunoreactive bands migrating below the 32 and 35 kDa bands were detected with both polyclonal antiserum and monoclonal antibodies and appear to be breakdown products of the expressed fusion proteins.
Fig. 3.
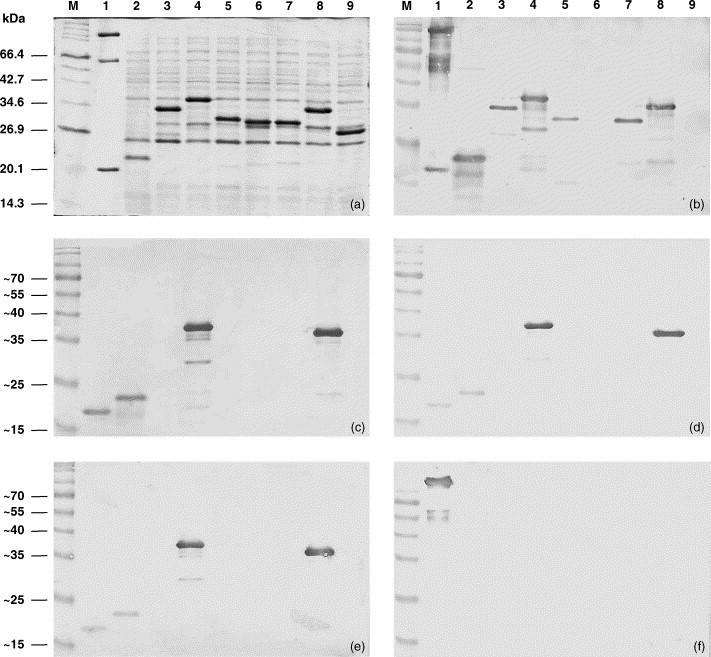
Full-length recombinant YHV His6-ORF2 protein and GST-fusion proteins containing partial segments of the ORF2 protein expressed in E. coli and analysed by SDS-PAGE (panel a) and immunoblot using polyclonal rabbit antiserum to YHV (panel b), monoclonal antibody Y19 (panel c), monoclonal antibody Y20 (panel d), monoclonal antibody YII4 (panel e), and gp116 monoclonal antibody V3-2B (panel f). Molecular weight markers (lane M); purified virus (lane 1); and E. coli transformed with plasmids pET17b-FL (lane 2), pGEX-5X-N (lane 3), pGEX-5X-C (lane 4), pGEX-5X-NN (lane 5), pGEX-5X-CN (lane 6), pGEX-5X-NC (lane 7), pGEX-5X-CC (lane 8) and vector plasmid pGEX-5X-1 (lane 9).
For more precise location of linear epitopes, E. coli BL21 cells transformed with recombinant plasmids pGEX-5X-NN, pGEX-5X-CN, pGEX-5X-NC and pGEX-5X-CC were induced with IPTG and analyzed by SDS-PAGE and immunoblotting. Following induction, GST-fusion proteins containing four linear quadrants of the N protein, i.e. NN (M1–A36), CN (V37–E73), NC (Q74–K109) and CC (R110–S146), were expressed at similar levels and appeared in gels as 29.5, 29, 29 and 32 kDa bands, respectively (Fig. 3a). The relatively slower migration of the recombinant protein containing the CC quadrant was consistent with slower migration of the GST-fusion protein containing the C-terminal half of the N protein. Immunoblot analysis using YHV polyclonal rabbit antiserum indicated that the NN, NC and CC quadrants contained immunoreactive epitopes (Fig. 1, Fig. 3). The strongest reaction was with the CC quadrant (R110–S146) which, like the longer C-terminal half, appeared to generate a series of smaller immunoreative breakdown products. The CN quadrant (V37–E73) did not react with the polyclonal rabbit antiserum. Immunoblot analysis using mouse monoclonal antibodies Y19, Y20 and YII4 indicated that each reacted only with the CC quadrant (Fig. 1, Fig. 3).
Further mapping of linear epitopes within the CC quadrant was conducted using three terminally truncated clones of the YHV nucleoprotein. E. coli BL21 cells transformed with recombinant plasmids pET17b-FL-Δ9, pET17b-FL-Δ18 and pET17b-FL-Δ27 were induced with IPTG and analyzed by SDS-PAGE and immunoblotting. Following induction, recombinant nucleoproteins truncated at the C-terminus by 9, 18 and 27 amino acids were expressed at similar levels and appeared in gels as 16, 14.5, and 13.5 kDa bands, respectively (Fig. 4a). The significant reduction in M r (5 kDa) following removal of the C-terminal 9 amino acids confirms that this short highly acidic domain is the primary cause of the difference between the observed M r and deduced molecular weight of the full-length nucleoprotein. Immunoblot analysis indicated each truncated nucleoprotein segment reacted with YHV polyclonal antiserum (Fig. 1, Fig. 4). In the Δ27-truncated protein, a second minor 14.5 kDa band detected by SDS-PAGE following induction also reacted strongly with the polyclonal antiserum. The origin of this band is not yet determined but monoclonal antibody reactions (see below) indicate that it also includes the Δ27 truncation. Monoclonal antibodies Y19 and YII4 reacted with each of the Δ9- and Δ18-truncated nucleoproteins but not the Δ27-truncated protein, indicating the epitopes are located either within the deleted E129–E137 segment or span the point of truncation (Fig. 1, Fig. 4). Monoclonal antibody Y20 reacted only with the Δ9-truncated nucleoprotein indicating that the epitope is located either within the deleted P120–A128 segment or spans the point of truncation (Fig. 1, Fig. 4).
Fig. 4.
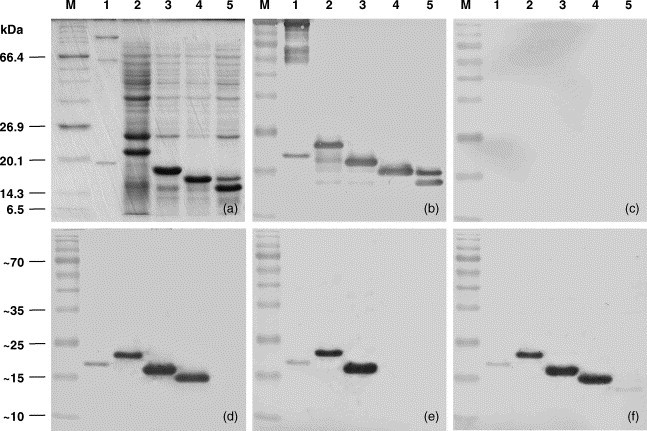
C-terminal-deleted segments of recombinant YHV ORF2 protein expressed in E. coli and analysed by SDS-PAGE (panel a) and immunoblot using polyclonal rabbit antiserum to YHV (panel b), normal rabbit pre-bled serum (panel c) and mouse monoclonal antibodies Y19 (panel d), Y20 (panel e) and YII4 (panel f). Molecular weight markers (lane M); purified virus (lane 1); and E. coli transformed with plasmids pET17b-FL (lane 2), pET17b-FL-Δ9 (lane 3), pET17b-FL-Δ18 (lane 4), and pET17b-FL-Δ27 (lane 5).
3.4. Purification of recombinant YHV nucleocapsid protein
E. coli BL21 cells transformed with recombinant plasmid pET17b-FL containing full-length YHV ORF2 gene were cultured to mid-log phase and induced with IPTG for 4 h. Following induction, His6-ORF2 protein was purified from the cell lysate by Ni+-NTA affinity chromatography. The expressed nucleoprotein remained primarily in the insoluble fraction which was relatively free of other cellular proteins. The soluble fraction contained useful quantities of nucleoprotein which was purified to near homogeneity following elution from the Ni+-NTA column (Fig. 5 ). The concentration of the expressed recombinant nucleoprotein in the whole cell lysate was approximately 1.15 mg/ml and the overall efficiency of purification was approximately 30%.
Fig. 5.

Purification of recombinant YHV His6-ORF2 protein by Ni+-NTA affinity chromatography analysed by SDS-PAGE (panel a) and immunoblot using YHV monoclonal antibody Y19 (panel b). Molecular weight markers (lane M); purified virus (lane 1); whole cell lysate of E. coli BL21 transformed with plasmid pET17b-FL (lane 2); insoluble fraction of the cell lysate (lane 3); soluble supernatant fraction of the cell lysate (lane 4); and column fractions eluted with LB containing 250 mM imidazole (lanes 5–9). Arrowheads indicate the purified His6-tagged recombinant nucleoprotein.
4. Discussion
Yellow head virus is an invertebrate nidovirus that is classified in the genus Okavirus of the family Roniviridae (Walker et al., 2004). Key criteria for the taxonomic placement of YHV and closely related Gill-associated virus (GAV) are the existence of a (+) ssRNA genome containing two overlapping long open-reading-frames (ORF1a and ORF1b) that are aligned during translation by a −1 ribosomal frame-shift, and a characteristic ‘SDD’ motif in the active site of the RNA-dependent RNA polymerase (Cowley et al., 2000, Sittidilokratna et al., 2002). It has also been reported that the GAV genome is transcribed as a nested set of 3′-coterminal sub-genomic mRNAs (Cowley et al., 2002). However, unlike the known vertebrate nidoviruses (coronaviruses, toroviruses and arteriviruses), the GAV nucleoprotein gene (ORF2) is located upstream, rather than downstream, of the glycoprotein gene (Cowley et al., 2004). In this paper, we confirm that the YHV nucleocapsid protein (p20) is also encoded in ORF2 which immediately precedes the gene encoding the transmembrane glycoproteins gp116 and gp64 (i.e. ORF3) (Jitrapakdee et al., 2003). The function of the YHV ORF2 gene is evident from the deduced amino acid sequence, which shares a high level identity (84%) with the GAV nucleoprotein. It is also demonstrated by the reaction of the E. coli-expressed recombinant ORF2 protein with YHV monoclonal antibody Y19 which is known to be specific for virion structural protein p20 (Sithigorngul et al., 2002), the only known polypeptide component of YHV nucleocapsids (Soowannayan et al., 2003). The unusual location of the YHV and GAV nucleoprotein genes may be a consequence of genetic recombination in an ancestral nidovirus. High-frequency genetic recombination is known to occur commonly in (+) ssRNA viruses, including nidoviruses (Lai et al., 1985, van der Most and Spaan, 1995, Snijder et al., 1991, Lai, 1992), and it has been proposed that the diverse structural and morphological characteristics of nidoviruses may have occurred by a modular evolutionary process that results in exchange by recombination of complete genes or gene sets (Snijder and Horzinek, 1993, Snijder and Horzinek, 1995, Snijder and Spaan, 1995).
There is a significant difference between the molecular weight of the YHV nucleocapsid protein predicted from the deduced amino acid sequence (M.W. = 16.6 kDa) and the observed electrophoretic mobility of the virion nucleoprotein by SDS-PAGE (M r ∼20 kDa). The deduced sequence indicates several unusual structural features that may contribute to aberrant migration in gels including a relatively high proportion of proline and glycine residues and a highly polar arrangement of charged residues. The discrepancy between predicted size and electrophoretic mobility is also evident in the His6-tagged full-length recombinant protein expressed in E. coli (Fig. 2). It was also observed that the expressed GST-fusion proteins containing the C-terminal half (Q74–S146) and the CC quadrant (R110–S146) of the nucleoprotein were retarded relative to the migration of fusion proteins containing other segments of similar size (Fig. 3). The 5 kDa reduction in M r following removal of the C-terminal 9 amino acids indicates that, despite the overall predominance of basic residues (pI = 9.9), this short highly acidic domain is primarily responsible for the relatively slow electrophoretic migration of the nucleoprotein.
Immunoblot analysis using polyclonal rabbit antiserum raised against nonionic detergent-disrupted, purified YHV identified reactive linear epitopes in all quadrants of the nucleoprotein except for the CN quadrant (V37–E73). This region has a low antigenicity index (Hopp and Woods, 1981) and a low predicted surface probability (Janin, 1979) and may well be buried in the core of the folded nucleoprotein. The most intense reactions occurred with the acidic CC quadrant (R110–S146), which also contained the epitopes for three available mouse monoclonal antibodies. Monoclonal antibodies Y19 and YII4 each mapped to linear sites either within the deleted E129–E137 segment or the upstream region spanning the point of truncation. Y19 and YII4 were derived from different cell fusions. Each reacts with both YHV and GAV and so must target highly conserved sequences comprising the 6–7 amino acids of a linear B cell epitope. This identifies the sequence IVPDPSL spanning the point of truncation as the only possible epitope for Y19 and YII4 (Fig. 1b). Monoclonal antibody Y20 is specific for YHV and so must target a 6–7 amino acid sequence that is not identical in GAV either within the deleted E129–E137 segment or the upstream region spanning the point of truncation (Fig. 1b). This identifies a maximum span of 15 amino acids from D123 to E137 (DIENLAEGIHAMSLE) which contains a core of common sequence (N126–M134). Therefore, the Y20 epitope must be located in one of the variable regions at either end of this 15 amino acid sequence.
His6-tagged recombinant YHV N protein was expressed in E. coli at high levels and was easily purified in quantity from the bacterial cell lysate by Ni+-NTA-affinity chromatography. The purified product will assist future structural studies of the YHV nucleoprotein and may be applied for the development of immunodiagnostic reagents. It has been reported previously that structural proteins VP19 and VP28 of white spot syndrome virus expressed in E. coli can induce a protective response following oral vaccination of shrimp (Witteveldt et al., 2004). While the nature of the protective mechanism is not known, it is possible that structural proteins of other shrimp viruses could also induce protective immunity. High-level expression and purification of recombinant YHV nucleoprotein will facilitate similar studies for this second major pathogen of farmed shrimp.
Acknowledgements
The authors wish to thank Dr Paisarn Sithigorngul for providing mouse monoclonal antibodies. We also thank Dr Jeff Cowley for useful discussions and Mr Lee Cadogan for assistance with preparation of rabbit antiserum.
References
- Chantanachookin C., Boonyaratpalin S., Kasornchandra J. Histology and untrastructure reveal a new granulosis-like virus in Penaeus monodon affected by yellow-head disease. Dis. Aquat. Org. 1993;17:145–157. [Google Scholar]
- Cowley J.A., Cadogan L.C., Spann K.M. The gene encoding the nucleocapsid protein of gill-associated nidovirus of Penaeus monodon prawns is located upstream of the glycoprotein gene. J. Virol. 2004;78:8935–8941. doi: 10.1128/JVI.78.16.8935-8941.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M. Gill-associated virusPenaeus monodon prawns: an invertebrate nidovirus with ORF1a and ORF1b gene related arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic RNAs that do not possess 5′-leader sequences. J. Gen. Virol. 2002;83:927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. The complete sequence of Gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 1977-1987;147 doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel T.W., Boonyaratpalin S., Withyachumnarnkul B. Progress in research on yellow-head virus and white-spot virus in Thailand. In: Flegel T.W., MacRae I.H., editors. Diseases in Asian Aquaculture III. Fish Health Section, Asian Fisheries Society; Manila: 1997. pp. 285–295. [Google Scholar]
- Hopp T.P., Woods K.R. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. U.S.A. 1981;86:152–156. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nat. Lond. 1979;277:491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S., Unajak S., Sittidilokratna N. Identification and analysis of gp116 and gp64 structural glycoproteins of yellow head nidovirus of Penaeus monodon shrimp. J. Gen. Virol. 2003;84:863–873. doi: 10.1099/vir.0.18811-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat. Lond. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Baric R.S., Makino S. Recombination between nonsegmented RNA genomes of murine coronaviruses. J. Virol. 1985;56:449–456. doi: 10.1128/jvi.56.2.449-456.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsuwan C. Tamsetakit Co., Ltd.; Bangkok: 1991. Handbook for Cultivation of Black Tiger Prawns. (in Thai) [Google Scholar]
- Nadala E.C.B., Tapay L.M., Loh P.C. Yellow head virus: a rhabdovirus-like pathogen of penaeid shrimp. Dis. Aquat. Org. 1997;31:141–146. [Google Scholar]
- Sithigorngul P., Chauychuwong P., Sithigorngul W. Development of a monoclonal antibody specific to Yellow head virus (YHV) from Penaeus monodon. Dis. Aquat. Org. 2000;492:27–34. doi: 10.3354/dao042027. [DOI] [PubMed] [Google Scholar]
- Sithigorngul P., Rukpratanporn S., Longyant S. Monoclonal antibodies specific to Yellow head virus (YHV) of Penaeus monodon. Dis. Aquat. Org. 2002;49:71–76. doi: 10.3354/dao049071. [DOI] [PubMed] [Google Scholar]
- Sittidilokratna N., Hodgson R.A.J., Cowley J.A. Complete ORF1b-gene sequence indicates Yellow head virus is an invertebrate nidovirus. Dis. Aquat. Org. 2002;50:87–93. doi: 10.3354/dao050087. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., den Boon J.A., Horzinek M.C. Comparison of the genome organization of toro- and coronaviruses: both divergence from a common ancestor and RNA recombination have played a role in Berne virus evolution. Virology. 1991;180:448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 1993;74:2305–2316. doi: 10.1099/0022-1317-74-11-2305. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C. The molecular biology of toroviruses. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 238–291. [Google Scholar]
- Snijder E.J., Spaan W.J.M. The coronaviruslike superfamily. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 239–255. [Google Scholar]
- Soowannayan C., Flegel T.W., Sithigorngul P. Detection and differentiation of yellow head complex viruses using monoclonal antibodies. Dis. Aquat. Org. 2003;57:193–200. doi: 10.3354/dao057193. [DOI] [PubMed] [Google Scholar]
- van der Most R.G., Spaan W.J.M. Coronavirus replication, transcription and RNA recombination. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 11–31. [Google Scholar]
- Walker P.J., Cowley J.A., Spann K.M. Yellow head complex viruses: transmission cycles and topographical distribution in the Asia-Pacific region. In: Browdy C.L., Jory D.E., editors. The New Wave: Proceedings of the Special Session on Sustainable Shrimp Culture, Aquaculture 2001. The World Aquaculture Society; Baton Rouge: 2001. pp. 227–237. [Google Scholar]
- Walker P.J., Bonami J.R., Boonsaeng V. Roniviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., editors. Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press; London: 2004. pp. 973–977. [Google Scholar]
- Witteveldt J., Cifuentes C.C., Vlak J.M. Protection of Penaeus monodon against infection with white spot syndrome virus by oral vaccination. J. Virol. 2004;78:2057–2061. doi: 10.1128/JVI.78.4.2057-2061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongteerasupaya C., Vickers J., Sriurairatana S. A non-occluded, systemic baculovirus that occurs in cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis. Aquat. Org. 1995;21:66–77. [Google Scholar]
- Ziebuhr J., Bayer S., Cowley J.A. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 2003;77:1415–1426. doi: 10.1128/JVI.77.2.1415-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


