Highlights
-
•
Hepatitis B core antigen (HBcAg) protein with PEDV epitopes can assemble into virus like particles.
-
•
Epitope placement within HBcAg can affect the immunogenicity of a vaccine.
-
•
The YSNIGVCK antigen from PEDV has a strong correlation with virus neutralization.
Keywords: HBcAg, PEDV, Vaccine, VLP, Purification
Abbreviations: PEDV, Porcine Epidemic Diarrhea Virus;; EFP, ELISA Fusion Protein;; Ab, Antibody;; mAb, monoclonal antibody;; HBcAg, Hepatitis B virus core antigen;; VLP, Virus-like Particle;; MIR, Major Immunodominant Region;; VN, Virus Neutralization;; GST, Glutathione-S-Transferase;; AEX, Anion Exchange;; IMAC, Immobilized Metal Affinity Chromatography;; IPTG, Isopropyl β-D-1-thiogalactopyranoside;; ORF, Open Reading Frame;; S, Spike;; E, Envelope;; M, Membrane;; N, Nucleocapsid;; MLV, Modified Live Virus;; KV, Killed Virus;; aa, Amino Acid
Abstract
Porcine epidemic diarrhea virus (PEDV) is a member of the Alphacoronaviridae genus within the Coronaviridae family. It is the causative agent of porcine epidemic diarrhea, a disease that can have mortality rates as high as 100% in suckling piglets. PEDV causes severe economic loss, and has been in existence for decades. A panzootic starting in 2010 renewed interest in the development of a universal vaccine toward PEDV. This report details several design changes made to a Hepatitis B virus core antigen (HBcAg)-based recombinant vaccine strategy, and their effect in vivo. Initially, several multi-antigen vaccine candidates were able to elicit antibodies specific to three out of four B-cell epitopes inserted into the chimeric proteins. However, a lack of virus neutralization led to a redesign of the vaccines. The focus of the newly redesigned vaccines was to elicit a strong immune response to the YSNIGVCK amino acid motif from PEDV. Genetically modified new vaccine candidates were able to elicit a strong antibody (Ab) response to the YSNIGVCK epitope, which correlated with an increased ability to neutralize the CO strain of PEDV. Additionally, the location of the inserted PEDV epitopes within the vector protein was shown to affect the immune recognition toward the native HBcAg during vaccination.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is an enteric virus that causes severe diarrhea, dehydration, and depression [1], [2]. PEDV is a species within the Alphacoronavirus genus (previously the type I Coronaviridae) in the Coronaviridae family of the Nidovirales order [3], [4]. Instances of PEDV began in the early 1970s, but outbreaks had been relatively tame, small, and sporadic until a panzootic occurred beginning in 2010. A mutation in the virus led to an increased virulence in some strains. This most recent outbreak caused severe economic losses worldwide as well as the introduction of the disease into North America and the United States in 2013 [5], [6], [7], [8], [9].
Transmission of PEDV occurs primarily through the oral-nasal route. Incubation time for PEDV varies with age and strain, and the original strain, CV777, can take up to 8 days before viral shedding is detected [10]. Newborn piglets (<7 days) develop symptoms to PEDV including fecal shedding within 24 h of infection and die at a high rate within 3–4 days, causing significant economic loss to the swine industry [11], [12]. The successful replication of PEDV in intestinal epithelial cells of young piglets causes mal-absorptive diarrhea induced by decreased efficacy of brush border membrane-bound digestive enzymes and leaky junctions in the small intestine [13], [14]. The symptoms are severe enough in these cases to result in a death rate of nearly 100% upon exposure due to dehydration. This is not true for adult pigs, however, which may be due to their delayed infection and more mature microvilli.
The typical genome for all Coronaviridae contains a 5′ and a 3′ untranslated region (UTR), and contains a range of open reading frames (ORFs) [15]. PEDV has an approximately 28 kb genome with ORFs arranged 5′ to 3′ in the order of: ORF1a, ORF1b, spike (S) protein, envelope (E) protein, membrane (M) glycoprotein, ORF3, and nucleocapsid (N) protein. For most coronaviruses, the S protein undergoes post-translational modifications including cleavage into an S1 and an S2 domain [16]. This is not true for PEDV, but there is a conventional separation of the protein into these domains around amino acids (aa) 781/782. Recent research in recombinant vaccination against PEDV has predominantly focused on the S protein, with particular attention paid to the S1 domain [17], [18], [19], [20], [21]. There are two known, small, and linear B-cell epitopes within the S1 domain corresponding to the aa sequences 748YSNIGVCK755, and 764SQYGQVKI771 from the original CV777 strain [22], [23], [24]. An additional small B-cell epitope was discovered within the S2 domain corresponding to the aa sequence 1368GPRLQPY1374 [22]. A fourth short, linear B-cell epitope is located in the M protein corresponding to the sequence 195WAFYVR200 [25].
Hepatitis B virus core antigen (HBcAg) is a popular protein expressed in E. coli for use in vaccine development. The resultant proteins are able to assemble into virus-like particles (VLPs) in vitro after purification [26] . Used in a chimeric form, foreign antigens from other viruses can be inserted into the protein genetically for efficient immune system presentation from the VLP platform. HBcAg is typically divided into two separate domains for chimeric protein design. The change from the N-terminal to the C-terminal domain occurs around Isoleucine-80. Insertion of foreign epitopes near this residue will have a higher immunogenic response compared to an N-terminal or C-terminal insertion, and this site was identified as the major immunodominant region (MIR) [27]. Previous work by this lab used HBcAg as a vector protein in a novel PEDV vaccination strategy. During the original experiment, five treatment vaccines were designed including four candidates with each of the four (748YSNIGVCK755, 764SQYGQVKI771, 1368GPRLQPY1374, 195WAFYVR200) linear B-cell epitopes inserted individually into the MIR. A fifth candidate had all four epitopes together in a linear peptide sequence inserted into the MIR of the HBcAg “backbone” protein [28]. The resultant epitope antibody (Ab) titers as measured by ELISA were not strongly correlated with virus neutralization with the exception of the 748YSNIGVCK755 motif. It was therefore concluded that an immune response to this particular antigen from PEDV might be more critical than others for virus neutralization. The inclusion of the YSNIGVCK epitope in future vaccine designs could therefore be critical to their efficacy.
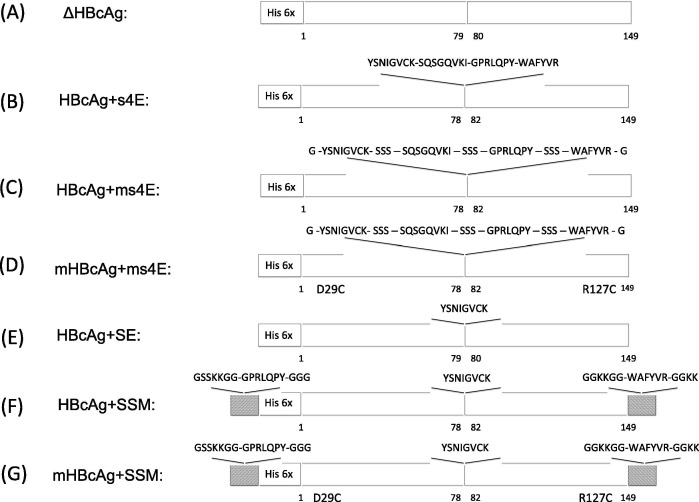
This report focuses on several design changes to previous HBcAg-based PEDV vaccines in an effort to improve the exposure of the inserted PEDV epitopes to the immune system and increase the overall immune response. The elimination of three residues from the MIR of the HBcAg protein, 79PIS81, was reported to increase the immune response to the inserted epitope [29]. This modification was performed for 5 vaccine candidates (Fig. 1 B–D and Fig. 1F–G). The addition of flexible, water-soluble residues between foreign epitope insertions at the MIR was also tested in two vaccine candidates to increase the solubility and presentation of the included antigens (Fig. 1C and D) [30]. The substitution of two residues, D29C and R127C, in the HBcAg vector protein of two vaccine candidates (Fig. 1D and G) was tested for increased efficacy. Assembly of HBcAg-based VLPs with these substitutions was previously shown by Lu et al to add a network of disulfide covalent bonds between pentamers and hexamers leading to an increased structural stability (Native VLP shown in Fig. S1A and C, additional disulfide bonds shown in red in Fig. S1B and D) [31]. Finally, the locations of the epitopes 1368GPRLQPY1374 and 195WAFYVR200 were changed so as to only include the 748YSNIGVCK755 motif in the MIR of the assembled VLPs. All vaccine design changes were performed and tested in vivo in a first animal trial (Fig. 1A–D), and a second animal trial (Fig. 1A, E–G).
Fig. 1.
Schematic diagrams of each vaccine tested. Foreign epitope sequences are indicated for the MIR, the N-terminus, and the C-terminus of the HBcAg protein as well as their location within the vector protein. Additional mutations are reported at their approximate location. (A) The truncated HBcAg protein with an N-terminal Histidine tag; (B–D) treatment vaccines used during the first animal trial; (E–G) treatment vaccines used during the second animal trial.
2. Materials and methods
2.1. Plasmid construction
Plasmid construction was performed using the pET-28a (+) plasmid (Novagen, Madison, WI). Briefly, genes coding for the optimized sequences (IDT, Coralville, IA) for each protein were amplified to contain an overlapping sequence corresponding to their placement in the vector plasmid. The genes were then used to amplify the pET-28a plasmid, which then contained the gene of interest. Each circular plasmid was transformed into BL21 (DE3) E. Coli T7 express® (NEB) cells using the heat shock method. Each construct was verified by Sanger sequencing after transformation for the correct sequence and codon usage.
2.2. Expression
Overnight cultures of BL21 (DE3) E. coli cells that were grown in LB media supplemented with 30 μg/mL kanamycin were used to inoculate 1 L batches of 2 × YT media supplemented with the same antibiotic at a rate of 0.2% v/v. The E. coli cultures were incubated at 28 °C until the OD600 reached 0.7 AU. Protein production was induced with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG), and cells were incubated overnight. The resulting cells were centrifuged and stored at −20 °C until purification.
2.3. Purification
Protein purification was performed as previously described [28]. Briefly, cell pellets were re-suspended and sonicated in PBS. All proteins aside from ΔHBcAg were located in the inclusion bodies. Proteins were denatured in 6 M urea and loaded onto an anion exchange chromatography (AEX) column utilizing DEAE Sepharose FF resin (GE Healthcare, Marlborough, MA). Eluents were then pH adjusted and loaded onto an immobilized metal affinity chromatography (IMAC) column. Following IMAC purification, step-wise dialysis was performed in PBS at pH 7.8 with additives. These additives were, in order: 0.9% Sarkosyl, 0.45% Sarkosyl, 0.01% Sarkosyl, 0% Sarkosyl (PBS only). Each vaccine candidate was then diafiltered further into PBS buffer using Spin X UF tubes with a molecular weight cut off of 100 kDa (Corning, Corning, NY).
2.4. Formulation
Concentrated vaccines in PBS were sterile filtered through 30 mm, 0.10 μm syringe filters (Celltreat, Pepperell, MA) followed by adjuvant addition and dilution with sterile PBS such that each dosage of 0.5 mL contained 20 μg of VLPs. Final vaccines for the first animal trial contained 20 μg protein in PBS at pH 7.8 with 100 μg per dose of chitosan (Invivogen) [32]. Candidates formulated for the second experiment had a total of 20 μg protein in PBS at pH 7.8 with 100 μg Alhydrogel (Invivogen). Vaccine candidates were then held for < 8 h at 4 °C until ready for use [33]. Vaccine dosages were chosen to be at or below previous studies [29], [34], [35].
2.5. Confirmation of assembly
A zetasizer nano (Malvern Instruments, Malvern UK) instrument was used according to manufacturers instructions, where particle size distributions were measured in PBS for initial confirmation of VLP assembly. Formvar coated copper grids were treated with protein samples for a few seconds until the grid was blotted with filter paper and allowed to dry. The samples were then stained with 2% phosphotungstic acid at pH 7, again for a few seconds before the stain was blotted with filter paper and allowed to dry [36]. Images were taken using a JEM-1400 (JEOL, Peabody, MA) microscope at a maximum of 300,000x.
2.6. Protein visualization
Protein samples were prepared for analysis by adding water, NuPage Reducing Agent (10×) (Invitrogen, Carlsbad, CA), and NuPage LDS Buffer (4×) (Invitrogen) for a total of 10 μL per well. Non-reduced samples were prepared similarly, substituting 1 μL of water for the reducing agent. Each sample was put into a NuPage™ 4–12% Bis-Tris PAGE gel (Invitrogen) along with a PrecisionPlus™ protein ladder (Biorad, Hercules, CA). Gels were run in MES buffer (Invitrogen). After electrophoresis, gels were rinsed three times with 18 MΩ water for 5 min each. Gels were then treated with SimplyBlue™ Safestain (Invitrogen) for at least one hour, and then rinsed with water overnight. Images of stained gels were taken using a Chemidoc Touch Imager (BioRad). Densitometry measurements were performed using ImageLab software (BioRad). Densitometry readings of >96% were considered to be pure enough to proceed with animal trials.
2.7. Western blot
Western blots were performed using the same protocol as reduced PAGE samples, but with a Precision Plus WesternC protein ladder (Biorad). Gels were loaded with 2 μL of cell lysate per well. Each Gel was blotted onto a nitrocellulose membrane using a Transblot® Turbo™ instrument (Biorad) according to manufacturers instructions. Membranes were blocked for at least one hour using 5% non-fat dry milk in TBST. A 3 × wash was performed between each step consisting of three five-minute rinses in TBST. Each membrane was treated with a primary mouse anti-histag antibody (Thermo) followed by a secondary HRP conjugated goat anti-mouse antibody (Millipore, Billerica, MA). Detection was done using a Chemidoc Touch instrument (Biorad) with Clarity® ECL substrate (Biorad).
2.8. Endotoxin testing
Endotoxin levels were tested after removal of detergent and chaotropic agents. Tests were performed using a Pierce® LAL Chromogenic Endotoxin Quantitation Kit (Thermo). The kit was used according to manufacturer’s instructions, with a endotoxin cut off level of <5 EU/mL.
2.9. Animals
All animal experiments were performed in accordance with guidelines and approved by the Virginia Tech Institutional Animal Care and Use Committee (IACUC). Groups of 8 germ free balb/c mice from Charles River were used for in vivo experiments. 7–8 week old mice were allowed to acclimate for 13 days prior to a Day -1 blood draw via submandibular bleed. Vaccine candidates and two negative controls (ΔHBcAg and PBS Only) were administered intraperitoneally on day 0, day 14, and day 28. Blood samples were collected on days 13 and 27 via the cheek punch method, and on day 42 by exsanguination. Collected blood samples were incubated at 4 °C for 2 h and centrifuged. Aliquots of supernatants were made, if required, and frozen at −20 °C until tested.
2.10. Enzyme linked immunosorbent assay (ELISA)
Ab titers to the inserted foreign epitopes were measured by a previously described process utilizing custom designed and purified ELISA fusion proteins (EFPs) [28]. All proteins used in coating were diluted to 10 μg/mL in coating buffer (50 mM Na2CO3, 50 mM NaHCO3, pH 9.6). Plates were coated at 37 °C for > 2 h, washed, and blocked overnight at 4 °C. Serum samples from Day 42, serially diluted in blocking buffer with a starting dilution of 1:25 were applied for 1 h at room temperature (RT). Plates were next incubated with goat anti-mouse IgG HRP (Millipore, Billerica, MA). Visualization was done with TMB One Component Microwell Substrate (SouthernBiotech) and read at 450 nm using a Biotek Synergy HTX plate reader (Winooski, VT). Titers were calculated as the reciprocal of the dilution at which test sera measurements were statistically similar to PBS negative control samples (n = 3) located on each plate. The Acinetobacter phage-derived VLP, AP205, was expressed and purified from the same plasmid (pET-28a) as the PEDV vaccine candidates using the same process. Different fractions were collected during AEX and the proteins were not assembled. ELISA plates coated with VLP proteins were performed at the same dilutions as the EFP coated plates, with the same process.
2.11. Whole virus (WV) antibody ELISA
The WV ELISA process used was adapted from Thomas et al. [37]. Microlon 600 flat bottom plates were coated with isolated CO strain (USDA APHIS, Ames, IA) of PEDV at a concentration of 104.5 TCID50 diluted 1:50 in sterile PBS at 4 °C for 6 h. Plates were then washed 5 × in sterile TBS. Plates were blocked in 1% BSA at 4 °C overnight, followed by a wash. Murine serum samples were diluted 1:25 in TBS and each well was filled with 100 μL of diluted serum. Serum samples were allowed to incubate for 1 h at RT, and were then washed 5 × with sterile TBS. Each well was next treated with a 100 μL volume of 1:10,000 diluted goat anti-mouse IgG HRP conjugate (Millipore) for 1 h at RT. After 5 × washes in TBS, the wells were detected with 100 μL of TMB One Component Microwell Substrate (SouthernBiotech). The reaction was stopped using 0.05% sulfuric acid. A Biotek Synergy HTX plate reader was used to read plates at 450 nm.
2.12. PEDV virus neutralization (VN) test
PEDV VN titers were measured using a focus forming assay (FFA) as described previously and executed by the Animal Disease Research and Diagnostic Laboratory in the Veterinary and Biomedical Sciences Department at South Dakota State University [38]. Briefly, serum samples were heat-treated and diluted 1:10 in minimal essential medium (MEM) with 1.5 μg/mL tosyl phenylalanyl chloromethyl ketone (TPCK) trypsin. A serial, 2-fold dilution was then performed in 96-well plates with the MEM/TPCK-trypsin diluent. Cell culture adapted PEDV was then added at 100 foci forming units (FFU)/100 μL and incubated for 1 h at 37 °C. The resulting mixture was then added to confluent monolayers of Vero-76 cells and incubated for 2 h at 37 °C. These new wells were then washed with MEM/TPCK-trypsin and allowed to incubate for 20–24 h. Plates were fixed with acetone and stained with fluorescein isothiocyanate (FITC) conjugated mAb SD6-29 for visualization. End point titers were determined as the lowest concentration resulting in a 90% or greater reduction in fluorescence relative to controls.
2.13. Statistics
JMP® Pro 12 software (SAS, Cary, NC) was used in all statistical analysis. All samples were run in a nested experimental design using administration, peptide/protein, and group, respectively. Individual ANOVA contrasts were performed to determine statistical significance between individual groups.
3. Results
3.1. Epitope antibody generation
All vaccines used for in vivo testing were successfully purified (Fig. S2, further discussion in supplemental material) and were able to form VLPs as indicated by dynamic light scattering (data not shown) and by transmission electron microscope (TEM) images (Fig. 2 A–H). After the completion of the first animal trial (Fig. 3 ), there was an immune response seen to the SQSGQVKI, GPRLQPY, and WAFYVR epitopes as seen in Fig. 3A–D for two of the three test vaccine groups compared to the ΔHBcAg control (HBcAg + s4E and mHBcAg + ms4E). In each case, the epitope-specific Ab titer increased in a dose-dependent manner over the course of the experiment (data not shown). It is important to note that the responses seen to the YSNIGVCK motif were minimal or non-existent (Fig. 3A). The lack of response to the YSNIGVCK motif may indicate that there is an interaction taking place between residues in the MIR of all test candidates that reduces or eliminates the presentation of the YSNIGVCK epitope [39].
Fig. 2.
TEM verification of VLP assembly. TEM images were taken of each vaccine candidate prior to each animal trial. Concentration and buffer vary (18 MΩ H2O, or PBS) with each sample for visual confirmation. Larger images were taken at the maximum magnification. Smaller images are close-ups manipulated after the images were taken for greater detail.
Fig. 3.
Epitope antibody titers from the first animal trial. Ab titers from mouse sera after completion of the first animal trial as measured by ELISA corresponding to plates coated with EFPs are shown. Ab titers specific to (A) EFP-YSNIGVCK, (B) EFP-SQSGQVKI, (C), EFP-GPRLQPY and (D) EFP-WAFYVR were measured. Each treatment group’s mean titer is compared to the ΔHBcAg negative control by ANOVA analysis. Standard error bars are shown for each treatment group (* = P ≤ 0.05, ** = P ≤ 0.01, *** = P ≤ 0.001).
During the second animal trial (Fig. 4 ), alum was included in the formulation of three of the vaccine candidates in an effort to test its efficacy as an adjuvant. Fig. 4A indicates that the addition of Alum to the formulation was detrimental to vaccine efficacy as measured by the immune response to the YSNIGVCK epitope (comparison of the HBcAg + SE candidate vaccine with and without alum added). This phenomenon appears to hold true for the HBcAg + SSM candidate as well. Interestingly, the addition of alum did not appear to disrupt the response to the mHBcAg + SSM vaccine, which was identical to the HBcAg + SSM vaccine with the exception of the D29C and R127C mutations. These results indicate the addition of Cysteine residues enabled the mHBcAg + SSM vaccine candidate to invoke an immune response even in the presence of alum, possibly due to the added network of disulfide bonds. However, inefficient antigen presentation in the MIR of the HBcAg + SSM candidate due to peptide interactions cannot be ruled out as a contributing factor to the low Ab response level. The mHBcAg + SSM candidate was also able to elicit an immune response to the N-terminal GPRLQPY and C-terminal WAFYVR epitopes as seen in Fig. 4B and C. Similar to the first animal trial, Ab titers for each treatment increased in a dose dependent manner throughout the experiment (data not shown).
Fig. 4.
Epitope antibody titers from the second animal trial. Ab titers from mouse sera after completion of the second animal trial as measured by ELISA corresponding to plates coated with EFPs are shown. Ab titers specific to (A) EFP-YSNIGVCK, (B) EFP-GPRLQPY, and (C) EFP-WAFYVR were measured. Each treatment group’s mean titer is compared to the ΔHBcAg negative control by ANOVA analysis. Standard error bars are shown for each treatment group (* = P ≤ 0.05, ** = P ≤ 0.01, *** = P ≤ 0.001).
3.2. Vector protein antibody generation
There is a great concern in the recombinant production of chimeric protein-based vaccines that pre-existing Abs to the vector protein may cause the elimination of the vaccine prior to antigen presentation. The Ab titers specific to the ΔHBcAg protein itself were therefore tested in conjunction with the individual epitope inserts. Results from Fig. 5 A indicate that the Ab titers to ΔHBcAg were statistically similar across all treatments during the first animal trial, indicating that each vaccine induced an immune response, though not necessarily to the inserted epitopes (Fig. 3). During the second animal trial (Fig. 5B), however, the data suggest that the two vaccine candidates with an epitope inserted N-terminal to the Histidine tag (HBcAg + SSM and mHBcAg + SSM) had significantly lower Ab responses to ΔHBcAg compared to the other treatments. To further elucidate the immune response to the vector protein, Ab titers to an unrelated vector protein containing the same N-terminal Histidine tag and cleavage site were tested (AP205, as described in the materials and methods section). Fig. 5C and D indicate that all vaccines elicited a significantly higher Ab titer to the Histidine tag than those vaccines with a foreign epitope insertion N-terminal to the Histidine tag (HBcAg + SSM and mHBcAg + SSM), following the same pattern as the Ab titers to the whole ΔHBcAg protein. Together these data demonstrate that the histidine tag located at the N-terminus of the vaccine candidates may play a large role in Ab recognition to each protein, as has been seen previously by others, and the placement of an epitope N-terminal to the tag may reduce its immune interaction [40].
Fig. 5.
Antibody titers to the vector protein and Histidine tag. Ab titers from mouse sera after completion of both animal trials as measured by ELISA corresponding to plates coated with His6xΔHBcAg (A and B) or His6xAP205 (C and D) are shown. Sera were tested for the presence of Abs after the first animal trial (A and C) as well as the second animal trial (B and D). Each treatment group’s mean titer is compared to the ΔHBcAg negative control by ANOVA analysis. Standard error bars are shown for each treatment group (* = P ≤ 0.05, ** = P ≤ 0.01, *** = P ≤ 0.001).
3.3. Virus neutralization (VN)
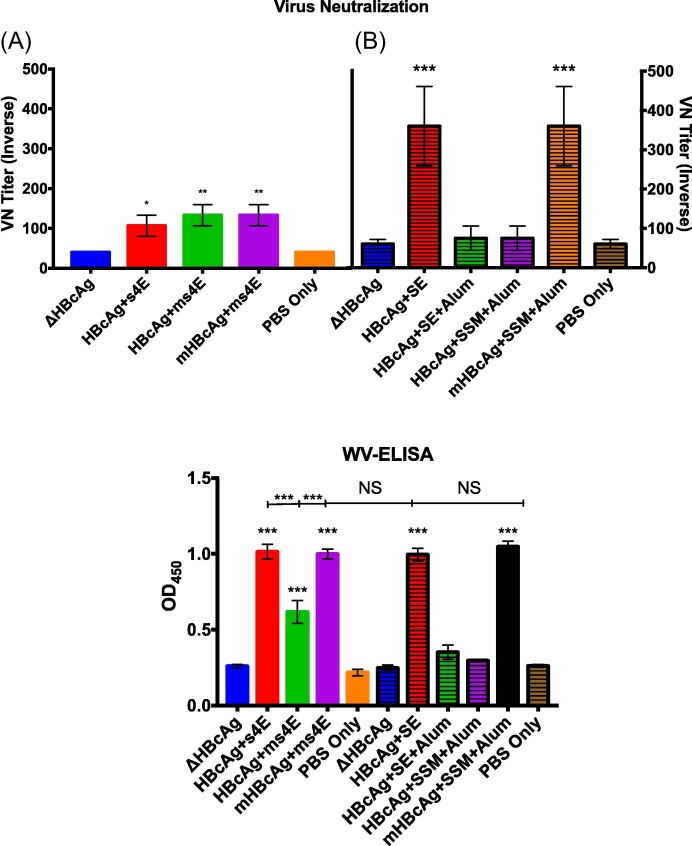
To further test the efficacy of the vaccine candidates, the response of the murine serum to the virus itself was determined by foci forming assay (FFA) utilizing the CO strain of PEDV. Fig. 6 A shows the VN titers from the first animal trial. Although there is statistical significance between the three test groups (HBcAg + 4E, HBcAg + ms4E, and mHBcAg + ms4E) and the negative controls (ΔHBcAg, and PBS Only), the numbers remain relatively low. Serum samples from the second animal trial (Fig. 6B) demonstrated a stronger ability to neutralize virus for two of the treatment groups only (HBcAg + SE and mHBcAg + SSM). When combined with peptide Ab titers, the data demonstrate a strong correlation between high VN titer and high Ab titer to the YSNIGVCK epitope (Figs. 6B and 4A) compared to the GPRLQPY epitope (Figs. 6A and 3C).
Fig. 6.
VN and WV ELISA. A foci forming assay (FFA) and a whole virus ELISA were used to determine the serum Ab titers specific to the CO strain of PEDV. Sera collected at Day 42 for each treatment group from (A) the first animal trial and (B) the second animal trial were tested for PEDV neutralization in vitro. These same serum samples were subsequently tested for their ability to bind to PEDV without neutralization measurement (C). Each treatment group’s mean titer is compared to both the ΔHBcAg and PBS Only negative controls by ANOVA analysis. Standard error bars are shown for each treatment group (* = P ≤ 0.05, ** = P ≤ 0.01, *** = P ≤ 0.001).
3.4. Whole virus (WV) ELISA
To determine if a low binding affinity of Abs generated through recombinant vaccination to the native proteins from virions could contribute to VN discrepancies, a WV ELISA assay was performed on all serum samples (Fig. 6C). For each treatment, the WV ELISA results appear to correlate well with individual peptide ELISA results independent of the specificity of the Abs generated. The candidates with high Ab titers to B-cell epitopes from PEDV (HBcAg + 4E, HBcAg + ms4E, and mHBcAg + ms4E from Fig. 3, HBcAg + SE, and mHBcAg + SSM + Alum from Fig. 4) had significantly higher absorption signals than the ΔHBcAg, and PBS Only treatments. These data indicate that the peptides presented by a vector protein did not prevent them from eliciting Abs that could bind to the virus during the virus neutralization assay, and that there may be a true difference in the ability of the YSNIGVCK to neutralize PEDV compared to GPRLQPY.
4. Discussion
The original goal of this project was to improve upon vaccine designs made and tested previously for PEDV by design modification to increase antigen presentation and virus neutralization on a per dose basis [28]. The elimination of the 79PIS81 residues and the addition of Serine residues between foreign epitopes tested during the first animal trial revealed a major flaw where either or both modifications prevented the presentation of the YSNIGVCK epitope. It is hypothesized that this phenomenon is due to residue interactions at the MIR of the VLP, as each modification is predicted to increase the flexibility of the region [29]. This newly added flexibility might therefore have removed a small α-helix native to the vector protein, allowing the new interactions to take place. These treatments were able to generate Abs to the other B-cell epitopes and to the vector protein, indicating that assembly of the VLPs with some antigen presentation did indeed take place. VN titers to these vaccine candidates were significant but low, indicating that high Ab titers to the SQSGQVKI, GPRLQPY, and WAFYVR epitopes are not necessarily associated with virus neutralization.
With the preliminary data in mind, the focus of the second experiment was two-fold: to test the use of alum as an adjuvant, and attempt two redesigns to increase the immune response to the YSNIGVCK epitope while maintaining the immune response to the GPRLQPY and WAFYVR B-cell epitopes. Although neutralization is a key measurement, there may be a benefit to including multiple conserved antigens in a vaccine for future mutations in some strains. The HBcAg + SE protein was used as a positive control during the second animal trial in two different formulations. The addition of alum to the formulation of the HBcAg + SE treatment group appeared to be detrimental to its immune response as indicated by a lack of YSNIGVCK-specific Abs to the adjuvanted vaccine (Fig. 4A). Interestingly, the HBcAg + SE + Alum treatment group was capable of inducing a response to ΔHBcAg (Fig. 5B), possibly through the recognition of the Histidine tag (Fig. 5D). Together, the data indicate that the HBcAg + SE VLPs were assembled with successful YSNIGVCK antigen presentation prior to formulation with alum, but the antigen presentation was reduced upon formulation leading to the Ab recognition of vector protein alone. The use of alum in an IP vaccine delivery may indeed reduce the immune response as seen by Chang et al. [41]. Cubas et al have also indicated that IP vaccine delivery of VLPs may not be the best route for drainage into the lymph nodes, which might reduce the exposure of the foreign antigens included in the VLP designs [42]. However, the complete elimination of an immune response to the foreign antigen with a similar immune response to the vector protein indicates that there may be additional factors present within this study. Future in vivo work may therefore require a more detailed analysis of the administration route as a possible variable in vaccine improvement.
There was also a significant immune response difference seen between the two redesigned vaccine candidates (HBcAg + SSM and mHBcAg + SSM) during the second animal trial. The mHBcAg + SSM vaccine, with the Cysteine substitutions, generated Abs specific to all three PEDV epitopes included (Figs. 4A–3C) and was able to neutralize virus (Fig. 6B), while the non-substituted version elicited an immune response to the vector protein alone (Fig. 5B). The sole difference between the HBcAg + SSM and mHBcAg + SSM candidate designs was in the vector protein located away from the MIR. Non-reduced PAGE analysis (Fig. S2B) along with TEM images (Fig. 2G) have indicated that there were no issues with assembly of the HBcAg + SSM VLPs. The difference in immune response therefore seems to indicate that the Cysteine residues have improved antigen presentation in the mHBcAg + SSM vaccine, possibly due to an increase in VLP stabilization as reported by Lu et al. [31]. We hypothesize this increased stabilization gives the mHBcAg + SSM vaccine a higher half-life in vivo, leading to a prolonged antigen presentation. However, this hypothesis was not investigated further.
The placement of the GPRLQPY foreign epitope N-terminal to the Histidine tag in two vaccine candidates (HBcAg + SSM and mHBcAg + SSM) is correlated with a reduced immune reaction to ΔHBcAg when compared to all of the other treatments tested (Fig. 5A and B). Ab response to the histidine tag cannot be separated from that of ΔHBcAg in this case, as the Histidine tag was used for purification of the ΔHBcAg coat proteins used in the assay. However, ELISA values for an unrelated protein containing the same N-terminal sequence showed a similar pattern amongst all treatment groups (Fig. 5C and D), indicating that the placement of the foreign epitope upstream may reduce the Ab recognition of the Histidine tag. These results may make physiological sense. If an extended N-terminus on the ΔHBcAg protein forms spikes upon VLP assembly, then it is possible that antigens located within that region (including Histidine tags) would have an opportunity to interact with immune cells [43]. The N-terminal placement of a more desirable epitope within this region may therefore cause a preferential interaction between the inserted foreign epitope and the immune system compared to that of the tag. It seems unlikely that the other changes (removal of 79PIS81, C-terminal addition of the WAFYVR epitope) would influence the detection of the Histidine tag, but they cannot be ruled out as possibilities from the presented results.
Fig. 6A and B indicates that a high Ab response to the YSNIGVCK epitope is of higher significance in vaccine design for virus neutralization compared to the other B-cell epitopes used in this study. Although several design changes were made between the two animal trials outlined in this report, the individual epitope Ab concentrations were measured using the same assay. Furthermore, the significance of the YSNIGVCK motif has been implicated in previous reports by this lab as well as others [24], [28]. The inclusion of B-cell epitopes other than YSNIGVCK in vaccines for PEDV elicited Abs to multiple peptide sequences without any increase in VN titers. This result would make sense if virus neutralization were more dependent upon the concentration of Abs specific to the YSNIGVCK peptide sequence, meaning the addition of other epitope specific Abs have diminished returns. Since apoptosis of piglet intestinal epithelial cells is of primary concern, virus neutralization should be of the utmost importance.
One possible explanation for the discrepancy in VN titers could be a fundamental difference in the GPRLQPY amino acid presentation in the context of the recombinant VLP compared to that of the native PEDV virus. If a difference were to exist, then it would be expected that Abs generated by the E. coli expressed proteins would have reduced or negligible binding to the PEDV virions. However, WV ELISA results (Fig. 6C) demonstrate a strong correlation between epitope Ab titers and WV titers independent of the antigen sequence. Thus it seems as though the preferential neutralization of the YSNIGVCK epitope is a true phenomenon. VN results in this report are based on systemic IgG levels in a murine model. The results may therefore not correlate directly with prevention of viral infection in the mesenteric epithelia of suckling piglets without further development. However, the results herein help demonstrate an improved efficacy of a vaccine in generating neutralizing Abs toward PEDV. Future studies building on these results should therefore include strategies to enhance lactogenic immunity from the pregnant sow in the form of secretory IgAs specific to the YSNIGVCK antigen.
Acknowledgments
Acknowledgement
We thank the generous financial support by Smithfield Foods and Murphy-Brown LLC and the insightful discussion with their scientists, especially Dr. Terry Coffey, throughout the project period.
We also wish to thank Aaron Singrey, Eric Nelson, and the rest of the team at the Animal Disease Research and Diagnostic Laboratory in the Veterinary and Biomedical Sciences Department at South Dakota State University.
Conflict of interest
None.
Funding
This work was supported by Smithfield Foods and Murphy-Brown LLC.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.vaccine.2018.06.015.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec. 1977;100:243. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- 2.Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callebaut P., Debouck P., Pensaert M. Enzyme-linked immunosorbent assay for the detection of the coronavirus-like agent and its antibodies in pigs with porcine epidemic diarrhea. Vet Microbiol. 1982;7:295–306. doi: 10.1016/0378-1135(82)90009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pensaert M., Callebaut P., Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet Q. 1986;8:257–261. doi: 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4 doi: 10.1128/mBio.00737-13. e00737–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, et al. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. Journal of veterinary diagnostic investigation : official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 2013;25:649–54. [DOI] [PubMed]
- 7.Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diep N.V., Norimine J., Sueyoshi M., Lan N.T., Yamaguchi R. Novel porcine epidemic diarrhea virus (PEDV) variants with large deletions in the spike (S) gene coexist with PEDV strains possessing an intact S gene in domestic pigs in Japan: A new disease situation. PloS One. 2017;12:e0170126. doi: 10.1371/journal.pone.0170126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Arriba M., Carvajal A., Pozo J., Rubio P. Isotype-specific antibody-secreting cells in systemic and mucosal associated lymphoid tissues and antibody responses in serum of conventional pigs inoculated with PEDV. Vet Immunol Immunopathology. 2002;84:1–16. doi: 10.1016/S0165-2427(01)00386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata I., Tsuda T., Mori M., Ono M., Sueyoshi M., Uruno K. Isolation of porcine epidemic diarrhea virus in porcine cell cultures and experimental infection of pigs of different ages. Vet Microbiol. 2000;72:173–182. doi: 10.1016/S0378-1135(99)00199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paarlberg PL. Updated estimated economic welfare impacts of porcine epidemic diarrhea virus (PEDV). Purdue University, Department of Agricultural Economics, Working Papers. 2014;14:1–38.
- 13.Jung K., Ahn K., Chae C. Decreased activity of brush border membrane-bound digestive enzymes in small intestines from pigs experimentally infected with porcine epidemic diarrhea virus. Res Vet Sci. 2006;81:310–315. doi: 10.1016/j.rvsc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Jung K., Eyerly B., Annamalai T., Lu Z., Saif L.J. Structural alteration of tight and adherens junctions in villous and crypt epithelium of the small and large intestine of conventional nursing piglets infected with porcine epidemic diarrhea virus. Vet Microbiol. 2015;177:373–378. doi: 10.1016/j.vetmic.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brian D.A., Baric R.S. Coronavirus genome structure and replication. In: Enjuanes L., editor. Coronavirus replication and reverse genetics. Berlin, Heidelberg; Springer, Berlin Heidelberg: 2005. pp. 1–30. [Google Scholar]
- 16.Duarte M., Laude H. Sequence of the spike protein of the porcine epidemic diarrhoea virus. J General Virol. 1994;75(Pt 5):1195–1200. doi: 10.1099/0022-1317-75-5-1195. [DOI] [PubMed] [Google Scholar]
- 17.Subramaniam S., Cao D., Tian D., Cao Q.M., Overend C., Yugo D.M. Efficient priming of CD4 T cells by Langerin-expressing dendritic cells targeted with porcine epidemic diarrhea virus spike protein domains in pigs. Virus Res. 2017;227:212–219. doi: 10.1016/j.virusres.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hain K.S., Joshi L.R., Okda F., Nelson J., Singrey A., Lawson S. Immunogenicity of a recombinant parapoxvirus expressing the spike protein of Porcine epidemic diarrhea virus. J General Virol. 2016;97:2719–2731. doi: 10.1099/jgv.0.000586. [DOI] [PubMed] [Google Scholar]
- 19.Makadiya N., Brownlie R., van den Hurk J., Berube N., Allan B., Gerdts V. S1 domain of the porcine epidemic diarrhea virus spike protein as a vaccine antigen. Virol J. 2016;13:57. doi: 10.1186/s12985-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh J., Lee K.-W., Choi H.-W., Lee C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- 22.Cruz D.J., Kim C.J., Shin H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun D., Feng L., Shi H., Chen J., Cui X., Chen H. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okda F.A., Lawson S., Singrey A., Nelson J., Hain K.S., Joshi L.R. The S2 glycoprotein subunit of porcine epidemic diarrhea virus contains immunodominant neutralizing epitopes. Virology. 2017;509:185–194. doi: 10.1016/j.virol.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Chen J., Shi H., Chen X., Shi D., Feng L. Identification of a conserved linear B-cell epitope in the M protein of porcine epidemic diarrhea virus. Virol J. 2012;9:225. doi: 10.1186/1743-422X-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murthy A.M., Ni Y., Meng X., Zhang C. Production and evaluation of virus-like particles displaying immunogenic epitopes of porcine reproductive and respiratory syndrome virus (PRRSV) Int J Mol Sci. 2015;16:8382–8396. doi: 10.3390/ijms16048382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schodel F., Moriarty A.M., Peterson D.L., Zheng J.A., Hughes J.L., Will H. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J Virol. 1992;66:106–114. doi: 10.1128/jvi.66.1.106-114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillam F., Zhang J., Zhang C. Hepatitis B core antigen based novel vaccine against porcine epidemic diarrhea virus. J Virol Methods. 2017 doi: 10.1016/j.jviromet.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Yin Y., Zhang S., Cai C., Zhang J., Dong D., Guo Q. Deletion modification enhances anthrax specific immunity and protective efficacy of a hepatitis B core particle-based anthrax epitope vaccine. Immunobiology. 2014;219:97–103. doi: 10.1016/j.imbio.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Schodel F., Peterson D., Hughes J., Wirtz R., Milich D. Hybrid hepatitis B virus core antigen as a vaccine carrier moiety: I. presentation of foreign epitopes. J Biotechnol. 1996;44:91–96. doi: 10.1016/0168-1656(95)00118-2. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y., Chan W., Ko B.Y., VanLang C.C., Swartz J.R. Assessing sequence plasticity of a virus-like nanoparticle by evolution toward a versatile scaffold for vaccines and drug delivery. PNAS. 2015;112:12360–12365. doi: 10.1073/pnas.1510533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNeela E.A., O'Connor D., Jabbal-Gill I., Illum L., Davis S.S., Pizza M. A mucosal vaccine against diphtheria: formulation of cross reacting material (CRM(197)) of diphtheria toxin with chitosan enhances local and systemic antibody and Th2 responses following nasal delivery. Vaccine. 2000;19:1188–1198. doi: 10.1016/s0264-410x(00)00309-1. [DOI] [PubMed] [Google Scholar]
- 33.Tan W.S., Dyson M.R., Murray K. Hepatitis B virus core antigen: enhancement of its production in Escherichia coli, and interaction of the core particles with the viral surface antigen. Biol Chem. 2003;384:363–371. doi: 10.1515/BC.2003.042. [DOI] [PubMed] [Google Scholar]
- 34.Klamp T., Schumacher J., Huber G., Kuhne C., Meissner U., Selmi A. Highly specific auto-antibodies against claudin-18 isoform 2 induced by a chimeric HBcAg virus-like particle vaccine kill tumor cells and inhibit the growth of lung metastases. Cancer Res. 2011;71:516–527. doi: 10.1158/0008-5472.CAN-10-2292. [DOI] [PubMed] [Google Scholar]
- 35.Duan X.Y., Han D.G., Zhang M.X., Wang J.S. Generation of fusion protein EGFRvIII-HBcAg and its anti-tumor effect in vivo. J Exp Clin Cancer Res: CR. 2009;28:133. doi: 10.1186/1756-9966-28-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barreto-Vieira DF, Barth OM. Negative and positive staining in transmission electron microscopy for virus diagnosis; 2015.
- 37.Thomas J.T., Chen Q., Gauger P.C., Giménez-Lirola L.G., Sinha A., Harmon K.M. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naive conventional neonatal and weaned pigs. PloS One. 2015;10:e0139266. doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okda F., Liu X., Singrey A., Clement T., Nelson J., Christopher-Hennings J. Development of an indirect ELISA, blocking ELISA, fluorescent microsphere immunoassay and fluorescent focus neutralization assay for serologic evaluation of exposure to North American strains of Porcine Epidemic Diarrhea Virus. BMC Vet Res. 2015;11:180. doi: 10.1186/s12917-015-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karpenko L.I., Ivanisenko V.A., Pika I.A., Chikaev N.A., Eroshkin A.M., Veremeiko T.A. Insertion of foreign epitopes in HBcAg: how to make the chimeric particle assemble. Amino Acids. 2000;18:329–337. doi: 10.1007/s007260070072. [DOI] [PubMed] [Google Scholar]
- 40.Randolph T.W. The two faces of His-tag: Immune response versus ease of protein purification. Biotechnol J. 2012;7:18–19. doi: 10.1002/biot.201100459. [DOI] [PubMed] [Google Scholar]
- 41.Chang H., Li X., Teng Y., Liang Y., Peng B., Fang F. Comparison of adjuvant efficacy of chitosan and aluminum hydroxide for intraperitoneally administered inactivated influenza H5N1 vaccine. DNA Cell Biol. 2010;29:563–568. doi: 10.1089/dna.2009.0977. [DOI] [PubMed] [Google Scholar]
- 42.Cubas R., Zhang S., Kwon S., Sevick-Muraca E.M., Li M., Chen C. Virus-like particle (VLP) lymphatic trafficking and immune response generation after immunization by different routes. J Immunother. 2009;32:118–128. doi: 10.1097/CJI.0b013e31818f13c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGonigle R., Yap W.B., Ong S.T., Gatherer D., Bakker S.E., Tan W.S. An N-terminal extension to the hepatitis B virus core protein forms a poorly ordered trimeric spike in assembled virus-like particles. J Struct Biol. 2015;189:73–80. doi: 10.1016/j.jsb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.