Abstract
It is possible to visualize rapidly viral particles by electron microscopy (EM) in patient samples and in cell cultures, and characterize the particles on the basis of their size and morphology. In many instances, EM has contributed to the diagnosis of specific infectious agents. Four different types of viruses with different characteristics of particle size, capsid structure, the presence or absence of an envelope, genomic content and stability outside the host were screened and diagnosed by EM at the level of family/genus. The results were confirmed at the species level by elution of the sample material from the grids used for EM examination and nucleic acid amplification. This approach could be valuable in situations where the immediate diagnosis is unclear, or when new infectious agents appear.
Keywords: Virus, Electron microscopy, Nucleic acid amplification
1. Introduction
Electron microscopy (EM) can be used to visualize virus particles directly at high magnifications. The first investigations of the morphology and later the ultrastructure of viruses infecting humans and animals became possible, due to the invention of the EM technique in the early 1930s (Agar, 1996), and the first micrographs showing the size and morphology of poxviruses were published in 1938 (Von Borries et al., 1938). With the development of the negative contrast staining method by Brenner and Horne (1959), viral particles could be investigated at the ultrastructural level.
During the following decades EM was used together with other techniques such as immunoassays, and later also with nucleic acid amplification techniques, to characterize a variety of viruses (reviewed in Biel and Gelderblom, 1999). This progress made it possible to establish viral classification based upon the morphology of virus particles (Tyrell and Almeida, 1967). EM was used in many laboratories for routine diagnosis of human viruses causing viral gastroenteritis such as the unculturable noroviruses, and astrovirus and rotavirus (Beards, 1988, Clark and McKendrick, 2004).
Today, EM is used routinely as a diagnostic method in cases of skin associated viral infections (Hawranek et al., 2003, Schimmer et al., 2004) and to some extent for the diagnosis of viral gastroenteritis (Frankenhauser et al., 2002, Suzuki et al., 2005). EM is indispensable in situations where the etiological agent causing outbreaks, epidemics or pandemics is difficult to identify. Recently, there have been two examples, namely the SARS pandemic in 2003 (Ksiazek et al., 2003) and the outbreak of human monkeypox in the USA in 2003, an infection not seen previously outside the African continent (Reed et al., 2004). In both of these situations, EM contributed to the identification of the viral agent. After 11th September 2001, EM has also been adopted in many countries as a rapid screening method to be used for the examination of both environmental and human samples in the case of bioterrorism (Gelderblom, 2003, Miller, 2004).
In the present study a strategy was developed whereby it is possible to screen by transmission EM for viruses in various types of patient samples, and then confirm and characterize the findings further at the species level by using nucleic acid amplification techniques.
2. Materials and methods
2.1. Experimental design
Four different viruses were investigated for detection by EM and subsequent nucleic acid amplification by the method outlined in Fig. 1 . The physical and chemical properties of these viruses are shown in Table 1 . The initial experiments were carried out using viruses propagated in cell cultures. Thereafter, samples containing the viruses were also included in order to test the application of the method to different types of diagnostic samples.
Fig. 1.
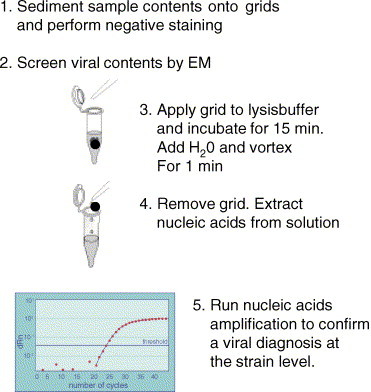
Flow-diagram showing a method for elution of viral nucleic acids from copper grids after EM analysis.
Table 1.
Physical and chemical properties of viruses tested by EM and PCR techniques
| Virus | Adenovirus | CMV | Influenza A | Norovirus |
| Family | Adenoviridae | Herpesviridae | Orthomyxoviridae | Caliciviridae |
| Genome | Linear, double-stranded DNA | Linear, double-stranded DNA | Segmented, single-stranded RNA | Linear, single-stranded RNA |
| Envelope | None | Yes | Yes | None |
| Particle size (nM) | 70–80 | 150–200 | 80–90 | 27–40 |
| Stability outside host | High | Low | Medium | High |
2.2. Propagation of viruses in cell cultures
The viruses propagated in cell culture for use in the experiments were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). Adenoviruses (ATCC VR-5) were propagated in VERO cells using Eagles MEM culture medium with 2% fetal calf serum, influenza A viruses (ATTC VR-97) were propagated in McDarbin canine kidney cells using Eagles MEM serumfree culture medium. When the cells had grown to confluence in 75 cm2 culture flasks (Nunc), viruses were added and allowed to grow until a complete cytophatic effect was attained. The culture flasks with the cell cultures containing the medium were thawed and frozen twice before harvesting the viruses. CMV (VR-977) were propagated in confluent human fibroblasts using Eagles MEM with 1% fetal calf serum. The supernatant was harvested and used directly as a viral preparation when a complete cytopathic effect was seen.
2.3. Patient samples
The patient samples had been submitted to the Department of Virology at the Statens Serum Institut and consisted of one eyeswab positive for adenovirus, one tracheal secretion positive for influenza A virus, one urine sample positive for CMV and one stool sample positive for norovirus. The real time PCR values were obtained from the routine diagnostic test. All patient samples had low cycle threshold (Ct) values, indicating that the samples had high viral titres. The Ct value is the value that is obtained, when the fluorescent signal from the PCR product exceeds that of the background.
2.4. Fixation and preparation of viral samples for EM
Before EM, all samples were inactivated with paraformaldehyde (PFA). Frozen 4% PFA in phosphate-buffered saline (pH 7.4) was heated to 65 °C in closed vials. When the PFA was dissolved completely, it was cooled on ice for 2 min and added to the samples at a final concentration of 2% in a total sample volume of 180 μl. The samples were transferred to airfuge vials and centrifuged at 90.000 rpm onto formvar coated carbon reinforced 400 mesh copper grids (Veco-grid's, AxLab) for 30 min in an airfuge using a A-100/30 rotor (Beckman Coulter). The grids were stained negatively with a 1.5% (pH 6.8) potassium phosphotungstic acid (PTA) solution for 30 s and allowed to dry in a fume-hood. The EM examination was conducted with a transmission electron microscope (Morgagni 268D, FEI Company) for an average of 30 min/specimen. Between 8 and 10 “windows” of the 400 mesh grid were scanned and photographs were taken using a Megaview III digital camera (Soft Imaging System).
2.5. Elution of viral nucleic acids from the EM grids
Viral DNA or RNA was eluted from the viral particles adhering to the copper grids (Fig. 1), by placing the grid into a vial and adding 300 μl of lysis buffer (Roche). After 10 min, 200 μl of sterile water was added, and the grid was vortex’ed for 1 min. The grid was then removed from the vial and the nucleic acids were purified for amplification by extraction on a MagNapure instrument using a total nucleic acids extraction kit (Roche Diagnostics). The nucleic acids were stored in elution buffer at −20 °C until use.
2.6. Real-time PCR and RT-PCR
Adenovirus nucleic acid was detected with fluorescent probes using a Lightcycler (Roche) instrument as described by Heim et al. (2003) with minor modifications. Influenza A virus and CMV nucleic acids were detected with fluorescent probes using a MX3000 detection device (Stratagene) with real-time RT-PCR (influenza A) or PCR (CMV) assays for each virus with minor modifications as compared to published real-time assays (Spackman et al., 2002, Boechk et al., 2004). Norovirus RNA was RT-PCR amplified and detected on an ethidium bromide stained 2% agarose gel as described by Vinje et al. (1997).
3. Results
3.1. Testing for viral genomes derived from viruses adhered to grids for subsequent nucleic acid amplification
Initially, it was determined whether it was possible to elute virus constituents from negatively stained grids and then detect the viral nucleic acids by nucleic acid amplification. The results are shown in Fig. 2 . One sample each of cell propagated adenovirus and influenza A virus, respectively, was extracted directly using MagnaPure extraction and then subjected to nucleic acid amplification (unprocessed sample). In addition, two samples (1x + EM) were subjected to EM examination prior to MagnaPure extraction and nucleic acid amplification as shown in Fig. 1. A quantitative real-time assay was used for adenovirus (Fig. 2A), using the standard curve to calculate the average loss of viral copies per ml for the processed (1x + EM) sample relative to the unprocessed sample, according to the formula (Ctintercept − Ctsample/slope) × inv. log. The difference was estimated to be a 38-fold loss of sensitivity i.e. from 2.3 × 108 copies/ml (Ct = 17.49) to 6 × 106 copies/ml (Ct = 23.20). This corresponded to 5.71 PCR cycles. From the Ct value of the highest dilution (100x + EM, Ct = 32.34) it could then be calculated, that 6.84 × 105 copies/ml of adenovirus could be detected by EM and this corresponded to 6.84 × 105/38 = 1.8 × 104 copies/ml as detected by PCR. Although influenza A virus (Fig. 2B) was detected using a non-quantitative assay, the loss in PCR cycles (Ctsample − Ctunprocessed sample = 17.79 − 12.15) could be measured to be 5.64 which was in good agreement with the result estimated with the adenovirus assay.
Fig. 2.
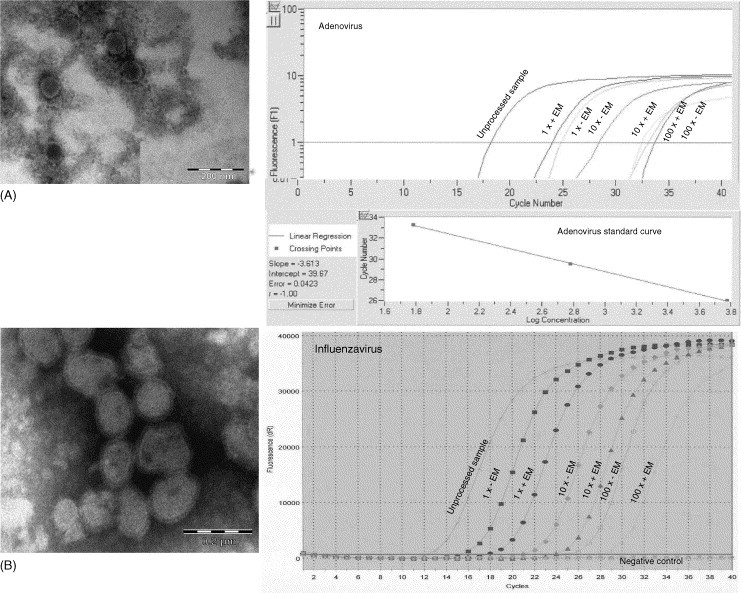
Detection of adenovirus and influenza A virus from cell culture by EM (left) and nucleic acid amplification (right). Magnifications (left): (A) Adenovirus, 89.000×; (B) Influenza A virus, 89.000×. The cell cultures were serially diluted 10-fold and treated as in Fig. 1, except that (+EM) samples were exposed to the EM beam, while (−EM) samples were unexposed. The unprocessed samples were untreated and extracted directly using MagnaPure extraction. The standard curve for the adenovirus quantitative real-time PCR is also shown on the figure.
3.2. Testing of differences in viral nucleic acid concentration between grids with viral preparations with and without EM
Next, the difference in the amount of viral copies between grids that had been processed by the method outlined in Fig. 1 and grids that had also been processed by the method except for exposure to the EM beam was investigated [(+EM) and (−EM)]. As depicted in Fig. 2, over a 100-fold dilution range, viruses were detected as particles by EM and also as viral genomes by nucleic acids amplification in all samples tested and no differences were detected between any of the samples for each dilution.
3.3. Amplification of viral nucleic acids from patient samples after elution from grids
Finally, the same procedure was examined with stored patient samples that had been diagnosed previously as adenovirus, influenza A, CMV or norovirus positive in the PCR diagnostic laboratory. For this purpose patient samples of different origin with a known high viral load were chosen, allowing detection by EM. The patient samples that were used are listed in Table 2 .
Table 2.
Virus containing sample material tested by EM and PCR techniques
| Virus | Sample material |
|---|---|
| Adenovirus | Eyeswab |
| CMV | Urine |
| Influenza A | Tracheal-secretion |
| Norovirus | Faeces |
3.4. Adenovirus
The results are shown in Fig. 3A. Adenovirus particles were detected in the sample by EM and subsequently, after elution, also the viral genomes by nucleic acid amplification. As a positive control, a standard of 105 copies/ml was included in the RT-PCR assay. The negative control is not shown in the figure, as this curve is not available with the lightcycler assay due to the lack of a reference dye.
Fig. 3.
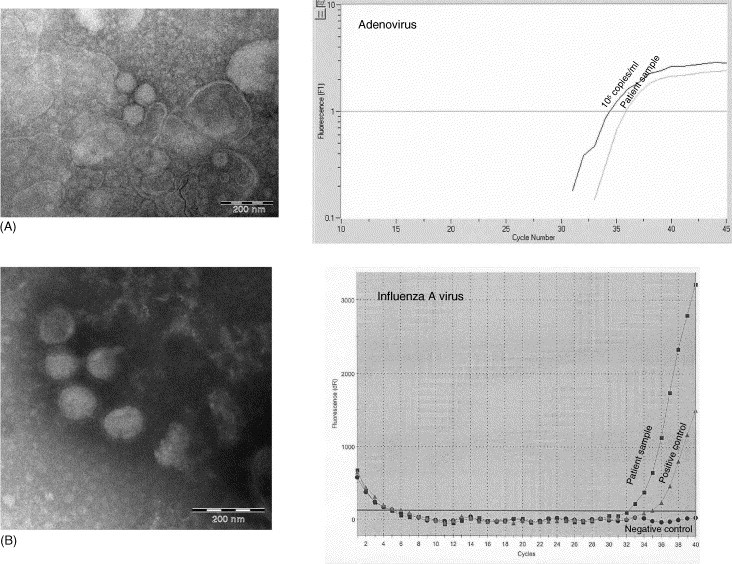
EM pictures of adenovirus (A) and influenza A virus (B) in patient samples used in this study and the EM sample material (right) subjected to real-time PCR (adenovirus) or real-time RT-PCR (influenza A virus). EM magnifications: (A) adenovirus, 89.000×; (B) influenza A virus, 89.000×.
3.5. Influenza A virus
The results are shown in Fig. 3B. A positive control used in the routine diagnostic assay was included in the real-time RT-PCR together with a non-template negative control.
3.6. Cytomegalovirus
In Fig. 4A, an EM picture of CMV particles derived from a cell culture sample is included for comparison to the EM picture of the patient sample, since only solitary CMV particles were found in the patient sample. The real-time PCR was quantitative, and the positive control was a sample containing 3.5 × 105 copies/ml which was included together with a non-template negative control.
Fig. 4.
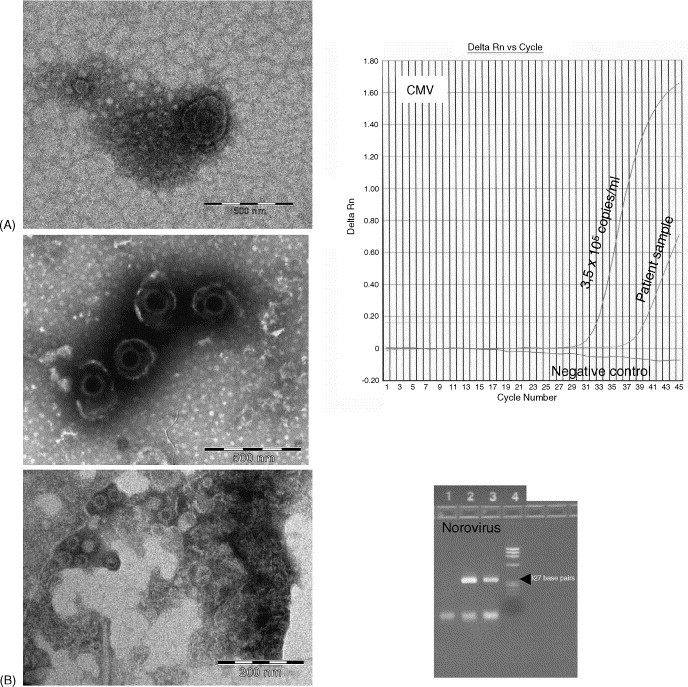
Left: the upper picture of CMV (A) and the norovirus picture (B) originate from the patient samples that were used in the study. The lower CMV picture is from a cell culture. Right: the EM sample material subjected to real-time PCR (CMV) or RT-PCR and subsequent gel electrophoresis (norovirus): Lane 1, negative control; lane 2, positive control; lane 3, patient sample; lane 4, ΦX174/Hae III marker. The norovirus-specific bands are 327 base pairs (indicated by an arrow). EM magnifications: CMV, 44.000×; norovirus, 89.000×.
3.7. Norovirus
The results are shown in Fig. 4B. Small rounded 35 nm particles were visualised in the EM sample. In the RT-PCR assay which was non-quantitative, a positive patient sample and a non-template sample were included as controls.
4. Discussion
In the present investigation, viruses differing in respect of both physical and chemical properties were diagnosed by EM and, after elution from the EM grids diagnosed at the species level by nucleic acid amplification.
There was a 38-fold decrease in viral copies, when untreated cell culture derived viral samples were measured by real-time PCR as compared to samples that had been subjected to EM examination. There were no major differences in the number of viral copies that could be detected on grids which had been exposed to the EM beam and the grids that had not been exposed to the EM beam over a 100-fold dilution range, indicating that this step does not contribute to the 38-fold decrease in viral copies. The decrease in viral copies could be due to a percentage of the viruses remaining in the supernatant after centrifugation onto the grids or that part of the viruses were not visible by EM after centrifugation onto the grid, due to the viruses adhering to the copper mesh or due to some degradation of nucleic acids during the sample preparation. As estimated for adenovirus it could, however, be compensated for by the higher sensitivity of the PCR technique and therefore has no negative impact on the results.
In a recent study by Biel et al. (2004) the detection of polyomavirus in urine samples from bone marrow transplant patients was done in parallel with EM and PCR. Within a range of probabilities of detection by EM it was found that there was a 50% probability of identifying viruses in samples containing 106 genome equivalents/ml, and the lower limit for EM detection was estimated to be >103 genome equivalents/ml. Our results showed, that adenovirus particles could be detected in cell culture corresponding to 6.84 × 105 genome equivalents/ml. The lower limit of detection by EM when using the approach described in the present study awaits further investigation.
The morphology of viruses belonging to the same family is unique for each particular family. For instance, coronaviruses with their club-like peplomers, or influenza viruses with their cogwheel-like appearance are recognised easily.
The EM technique has been used occasionally in parallel with other virological methods to identify the causative viral agent in patients during outbreaks of zoonotic origin and during seasonal and out-of-season epidemics and pandemics. For instance, in one case report, reovirus-like particles were found by EM in rhesus monkey kidney cells inoculated with cerebrospinal fluid from a young patient, and further investigations confirmed the diagnosis reovirus serotype 2 (Hermann et al., 2004). The same approach was used to diagnose Chandipura virus causing an outbreak of acute encephalitis with a high fatality rate in children from India (Rao et al., 2004). The etiological agent involved in a zoonotic outbreak in a rural area of Brasil causing cowpox-like symptoms in a dairy herd, and an associated human infection, was resolved partly with the aid of EM and was found to be a vaccinia-like virus, which was named Aracatuba virus (Trinidade et al., 2003). The peak of rotavirus infections in children during the winter in Japan was found to have changed to early spring by confirming the rotavirus diagnosis in parallel with EM and the latex agglutination method (Suzuki et al., 2005). In recent years, noroviruses have become a major problem in hospitals worldwide, causing outbreaks of viral gastroenteritis (reviewed by Clark and McKendrick, 2004). The high diversity of this group of viruses leads sometimes to the failure of detection by conventional routine diagnostic RT-PCR assays. The use of EM as an additional method can in this instance be essential (Gallimore et al., 2004, Vipond et al., 2004). Finally, the recent pandemic caused by SARS was also resolved partly by EM (Ksiazek et al., 2003).
A morphological diagnosis can be made by EM, including enrichment (see Gentile and Gelderblom, 2005), within 1 h after receiving a specimen and will provide a diagnosis. In addition, the “open view” of EM allows for the simultaneous detection of more than one agent in multiply infections, which is also sometimes the case in patients with gastroenteritis (Hazelton and Gelderblom, 2003).
In summary, a method was developed whereby it is possible to diagnose viruses in cell culture and in patient samples by EM pre-diagnosis and subsequently, by using the same sample material eluted from a grid, establish the viral diagnosis at the strain level using nucleic acid amplification. The method is versatile and can be used for different types of sample material and all kinds of agents.
Acknowledgements
We thank Helle Jensen and Sofie Midgley for critically reviewing the manuscript, Elizabeth Engels and Annette Nielsen for technical assistance and the diagnostic laboratory for helping with the nucleic acids amplification analyses.
References
- Agar A.W. The story of European electron microscopes. In: Mulvey T., editor. Advances in Imaging and Electron Physics, 1st ed. Academic Press; London: 1996. pp. 415–584. [Google Scholar]
- Beards G.M. Laboratory diagnosis of viral gastroenteritis. Eur. J. Clin. Microbiol. Infect. Dis. 1988;7:11–13. doi: 10.1007/BF01962164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel S.S., Gelderblom H.G. Diagnostic electron microscopy is still a timely and rewarding method. J. Clin. Virol. 1999;13:105–119. doi: 10.1016/S1386-6532(99)00027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel S.S., Nitsche A., Kurth A., Siegert W., Özel M., Gelderblom H.R. Detection of human polyomaviruses in urine from bone marrow transplant patients: comparison of electron microscopy with PCR. Clin. Chem. 2004;50:306–312. doi: 10.1373/clinchem.2003.024539. [DOI] [PubMed] [Google Scholar]
- Boechk M., Huang M., Ferrenberg J., Stevens-Ayers T., Stensland L., Nichols W.G., Corey L. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 2004;42:1142–1148. doi: 10.1128/JCM.42.3.1142-1148.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., Horne R.W. A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta. 1959;34:103–110. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- Clark B., McKendrick M. A review of viral gastroenteritis. Curr. Opin. Infect. Dis. 2004;17:461–469. doi: 10.1097/00001432-200410000-00011. [DOI] [PubMed] [Google Scholar]
- Frankenhauser R.L., Monroe S.S., Noel J.S., Humphrey C.D., Bresee J.S., Parashar U.D., Ando T., Glass R.I. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 2002;186:1–7. doi: 10.1086/341085. [DOI] [PubMed] [Google Scholar]
- Gallimore C.I., Green J., Richards A.F., Cotterill H., Curry A., Brown D.W., Gray J.J. Methods for the detection and characterisation of noroviruses associated with outbreaks of gastroenteritis: outbreaks occurring in the north-west of England during two norovirus seasons. J. Med. Virol. 2004;73:280–288. doi: 10.1002/jmv.20088. [DOI] [PubMed] [Google Scholar]
- Gelderblom H.R. Elektronenmikroskopie im methodenspektrum der bioterrorismus-diagnostik. Bundesgesundheitsbl. Gesundheitsforsch. Gesundheitsshutz. 2003;46:984–988. [Google Scholar]
- Gentile M., Gelderblom H.R. Rapid viral diagnosis: role of electron microscopy. New Microbiol. 2005;28:1–12. [PubMed] [Google Scholar]
- Hawranek T., Tritscher M., Muss W.H., Jecel J., Nowotny N., Kolodziejek J., Emberger M., Schaeppi H., Hintner H. Feline orthopoxvirus infection transmitted from cat to human. J. Am. Acad. Dermatol. 2003;49:513–518. doi: 10.1067/s0190-9622(03)00762-x. [DOI] [PubMed] [Google Scholar]
- Hazelton P.R., Gelderblom H.R. Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg. Infect. Dis. 2003;9:294–303. doi: 10.3201/eid0903.020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A., Ebnet C., Harste G., Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003;70:228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- Hermann L., Enbree J., Hazelton P., Wells B., Coombs K. Reovirus type 2 isolated from cerebrospinal fluid. Pediatr. Dis. Infect. Dis. J. 2004;23:373–375. doi: 10.1097/00006454-200404000-00026. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Miller S.E. Surveillance of bioterrorism agents: considerations for EM laboratories. Microsc. Today. 2004;12:56–59. [Google Scholar]
- Rao B.I., Basu A., Wairagkar N.S., Gore M.M., Arankalle V.A., Thakare J.P., Jadi R.S., Rao K.A., Mishra A.C. A large outbreak of acute encephalitis with high fertility rate in children in Andhra Pradesh, India, in 2003, associated with Chandipura virus. Lancet. 2003;364:869–874. doi: 10.1016/S0140-6736(04)16982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K.D., Melski J.W., Graham M.B., Regnery R.L., Sotir M.J., Wegner M.V., Kazmierczak J.J., Stratman E.J., Li Y., Fairley J.A., Swain G.R., Olson V.A., Sargent E.K., Kehl S.C., Frace M.A., Kline R., Foldy S.L., Davis J.P., Damon I.K. Detection of monkeypox in humans in the western hemisphere. N. Engl. J. Med. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- Schimmer B., Sprenger H.G., Wismans P.J., van Genderen P.J. Three patients with orf (ecthyma contagiosum) Ned. Tijdschr. Geneeskd. 2004;148:788–791. [PubMed] [Google Scholar]
- Spackman E., Senne D.A., Myers T.J., Bulanga L.L., Garber L.P., Perdue M.L., Lohman K., Daum L.T., Suarez D.L. Development of a real-time reverse transcription PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Sakai T., Tanabe N., Okabe N. Peak rotavirus activity shifted from winter to early spring in Japan. Pediatr. Infect. Dis. J. 2005;24:257–260. doi: 10.1097/01.inf.0000154327.00232.4d. [DOI] [PubMed] [Google Scholar]
- Trinidade G.S., Fonseca F.G., Marques J.T., Nogueira M.L., Mendes L.C., Borges A.S., Peiro J.R., Pituco E.M., Bonjardim C.A., Ferreira P.C., Kroon E.G. Aracatuba virus: a vaccinialike virus associated with infection in humans and cattle. Emerg. Infect. Dis. 2003;9:155–160. doi: 10.3201/eid0902.020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrell D.A.J., Almeida J. Direct electron-microscopy of organ cultures for the detection and characterisation of viruses. Arch. Ges. Virusforsch. 1967;22:417–425. doi: 10.1007/BF01242962. [DOI] [PubMed] [Google Scholar]
- Vinje J., Altena S.A., Koopmans M.P. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 1997;176:1374–1378. doi: 10.1086/517325. [DOI] [PubMed] [Google Scholar]
- Vipond I.B., Caul E.O., Hirst D., Carmen B., Curry A., Lopman B.A., Pead P., Pickett M.A., Lambden P.R., Clarke I.N. National epidemic of Lordsdale norovirus in the UK. J. Clin. Virol. 2004;30:243–247. doi: 10.1016/j.jcv.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Von Borries B., Ruska E., Ruska H. Bakterien und virus in übermikroskopischer aufname. Klin. Wochenschr. 1938;17:921–925. [Google Scholar]


