Abstract
Background
Human rhinoviruses (HRVs) are some of the earliest identified and most commonly detected viruses associated with acute respiratory tract infections (ARTIs) and yet the molecular epidemiology and genomic variation of individual serotypes remains undefined.
Objectives
To molecularly characterise a novel HRV and determine its prevalence and clinical impact on a predominantly paediatric population.
Study design
Nucleotide sequencing was employed to determine the complete HRV-QPM coding sequence. Two novel real-time RT-PCR diagnostic assays were designed and employed to retrospectively screen a well-defined population of 1244 specimen extracts to identify the prevalence of HRV-QPM during 2003.
Results
Phylogenetic studies of complete coding sequences defined HRV-QPM as a novel member the genus Rhinovirus residing within the previously described HRV-A2 sub-lineage. Investigation of the relatively short VP1 sequence suggest that the virus is resistant to Pleconaril, setting it apart from the HRV A species. Sixteen additional HRV-QPM strains were detected (1.4% of specimens) often as the sole micro-organism present among infants with suspected bronchiolitis. HRV-QPM was also detected in Europe during 2006, and a closely related virus circulated in the United States during 2004.
Conclusions
We present the molecular characterisation and preliminary clinical impact of a newly identified HRV along with sequences representing additional new HRVs.
Keywords: Rhinovirus, ARTI, LRTI, Newly identified virus, Respiratory infection, Phylogeny, Characterisation, RT-PCR, Epidemiology
1. Introduction
Acute respiratory tract infections (ARTIs) are a leading cause of human morbidity worldwide and are most frequently viral in origin. Many newly identified viruses have recently been associated with ARTI, including human metapneumovirus (HMPV; van den Hoogen et al., 2001), human coronaviruses (HCoV) NL63 (van der Hoek et al., 2004) and HKU1 (Woo et al., 2005), human bocavirus (HBoV; Allander et al., 2005) and the KI and WU polyomaviruses (Allander et al., 2007, Gaynor et al., 2007).
Human rhinoviruses (HRVs) are the most common infectious agents associated with ARTIs, comprising the largest genus of human pathogens, Rhinovirus, in the family Picornaviridae. Despite a significant and growing body of evidence to implicate HRVs in more serious clinical outcomes (Gern and Busse, 1999, Hayden, 2004), the presence and role of more than 100 genetically and serologically distinct HRVs in illness is often unexplored. Additionally, the viruses themselves are not considered to be individual entities with distinct circulation patterns as other respiratory viruses are. Studies have produced unusual HRV-like sequences or made cursory reports of untypeable or variant HRV strains in the United Kingdom (Mori and Clewley, 1994), Switzerland (Deffernez et al., 2004), Finland (Savolainen et al., 2002), Belgium (Loens et al., 2006), Australia (Arden et al., 2006) and the United States (Lamson et al., 2006). Many of these strains may represent novel HRV serotypes.
During an investigation of picornavirus PCR products we identified a cluster of more than 30 nucleotide sequences demonstrating <20% identity with any sequence on GenBank (Arden et al., 2006). We proposed that these belonged to a novel genetic sub-lineage, HRV-A2. We further investigated one of these putative viruses and herein present the complete polyprotein coding sequence of a novel HRV, HRV-QPM, which was first detected in an infant with bronchiolitis.
2. Materials and methods
2.1. Specimen population
Clinical specimens (n = 1244) were predominately nasopharyngeal aspirates (NPA; 92%) collected from patients (52.9% male) aged between 1 day and 80 years (mean age 9.2 years, median age 1.3 years) who had presented to Queensland hospitals or general practitioners with symptoms of ARTI during 2003. NPAs and purified nucleic acid extracts were stored at −70 °C. Extracts were tested by RT-PCR (OneStep, QIAGEN®) for HMPV, HRVs and enteroviruses (HEVs), all four non-SARS HCoVs, HBoV, respiratory syncytial virus (HRSV), influenza A and B viruses, parainfluenzaviruses and human adenoviruses (Arden et al., 2006).
2.2. Reverse-transcription and PCR
Real-time RT-PCR employed 0.2–0.4 pmol of oligonucleotide. Amplicons were purified when necessary and both strands sequenced (Big Dye v3.1, Applied Biosystems). Real-time RT-PCR was performed on the Rotor-Gene™ 3000 (Corbett Research).
First strand cDNA synthesis (Transcriptor RT, Roche) using (T)17ATA or gene specific primers was followed by second strand synthesis and amplification (Platinum® Pfx, Invitrogen). Oligonucleotide sequences are available upon request.
2.3. Cell culture
Human cell lines (HeLa-Ohio, A549, MRC-5 and WI-38) were inoculated with patient respiratory secretions stored at −70 °C. Inoculated cells were rolled at 33 °C, watched daily for cytopathic effect and HRV-QPM RNA levels were monitored by real-time RT-PCR.
2.4. Phylogenetic diversity
Nucleotide or amino acid sequences were aligned with the program MUSCLE (Edgar, 2004) and, where necessary, backtranslated with the program Protogene (Moretti et al., 2006). Maximum likelihood trees were constructed using a GTR + I + Γ4 model of nucleotide substitution in PAUP* (Swofford, 2003), with outgroups as indicated. Trees were visualized with TreeEdit (http://evolve.zoo.ox.ac.uk/software.html). The nucleotide distance matrix for neighbour-joining trees was generated using the Kimura two-parameter estimation in MEGA 3.1 (Kumar et al., 2004). Nodal confidence values indicate the results of bootstrap resampling (n = 1000).
2.5. Determination of the complete HRV-QPM coding sequence
Initial HRV-QPM sequences were used to design new PCR primers permitting determination of the continuous coding sequence from overlapping fragments, bracketed by partial untranslated regions (UTRs). Additional primers from HRV (Loens et al., 2006) and HEV (Caro et al., 2001, Oberste et al., 2004, Simmonds and Welch, 2006) studies supplemented the process. To confirm that our primary sequence belonged to a distinct virus and was not the result of mixed template in the patient's specimen, a second round of RT-PCR and sequencing of overlapping fragments was performed using newly designed HRV-QPM-specific primers targeting RNA from the original specimen extract. Discrepant sequence results were confirmed by a third round of RT-PCR and sequencing.
3. Results
3.1. Detection of HRV-QPM in Brisbane, Australia
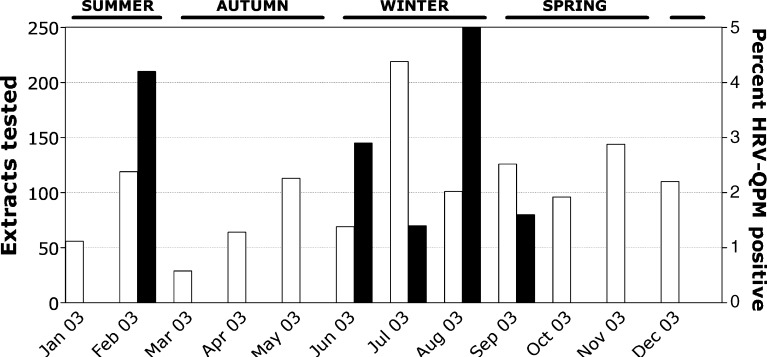
To determine whether HRV-QPM was a distinct pathogen endemically circulating in Brisbane, 1244 specimen extracts spanning all of 2003 were examined (Fig. 1 ). HRV-QPM 1B RNA was detected in 1.4% (n = 17) of extracts by real-time RT-PCR and verified using a confirmatory assay targeting the 1D region. HRV-QPM prevalence peaked in winter but was also detected in spring and summer. No other micro-organisms were detected in 64.7% (n = 11) of positives (Table 1 ). There were six co-detections with: HCoV-NL63, HCoV-NL63 and HBoV, HBoV, HMPV or HCoV-229E.
Fig. 1.
The monthly distribution of tested respiratory specimens (open bars). The HRV-QPM detections (filled bars) are presented as a percentage of the specimens tested each month. Seasons are indicated.
Table 1.
Patients positive for HRV-QPM
| Diagnosis upon presentation and HRV-QPM strain | Age | Other micro-organisms |
|---|---|---|
| Bronchiolitis (n = 5; 35.7%*) | ||
| HRV-QPM001a | 10 months | –b |
| HRV-QPM012 | 5 months | – |
| HRV-QPM010 | 10 months | – |
| HRV-QPM004 | 27 days | – |
| HRV-QPM006 | 7 months | – |
| Febrile convulsion (n = 1; 7.1%) | ||
| HRV-QPM011 | 2 years 6 months | – |
| Asthma (n = 2; 14.3%) | ||
| HRV-QPM002 | 3 years 7 months | – |
| HRV-QPM007 | 4 years 7 months | – |
| Viral URTI (n = 1; 7.1%) | ||
| HRV-QPM008 | 3 years 7 months | HBoV, NL63 |
| Persistent/hacking/whooping cough (n = 3; 21.4%) | ||
| HRV-QPM007 | 9 months | – |
| HRV-QPM013 | 1 year 4 months | HBoV |
| HRV-QPM016 | 11 months | HMPV |
| Underlying condition (n = 1; 7.1%) | ||
| HRV-QPM009 | 29 years 9 months | 229E |
| Exacerbation of COPD (n = 1; 7.1%) | ||
| HRV-QPM005 | 67 years 11 months | – |
| No notes (n = 3) | ||
| HRV-QPM017 | 11 months | NL63 |
| HRV-QPM003 | 2 years 8 months | – |
| HRV-QPM015 | 1 month | NL63 |
*Percentage of those with clinical notes available.
HRV-QPM strain detected.
Negative after all microbial investigations.
Overall respiratory picornaviruses were the most frequently detected micro-organisms at 41.0% of microbial detections (n = 346) followed by HBoV (12.0%, n = 101), HRSV (8.4%, n = 71) and HMPV (7.5%, n = 63). No detections were made in 44.7% (n = 556) of extracts. A small study of consecutively selected NPA extracts obtained during the winters of 2002, 2004 and 2006 (n = 160 per year) yielded no additional HRV-QPM detections.
Isolation of HRV-QPM was unsuccessful due to the age of the inoculum or perhaps because the HRV-A2 viruses are particularly fastidious.
3.2. Clinical impact of HRV-QPM infection
Clinical information from the 10-month-old male index case (yielding HRV-QPM strain 001) revealed bronchiolitis following a 5-day history of rhinorrhoea, cough and respiratory distress. There were no fevers vomiting or diarrhoea noted. The infant had an audible wheeze with scattered crackles in both lung fields, right otitis media and pharyngitis. The chest X-ray showed bilateral patchy changes, worse in the right mid zone, consistent with a non-specific viral lower respiratory infection (LRTI). The infant responded to nebulised ventolin and oral prednisolone and was discharged after a 48 h admission.
Clinical details from the other HRV-QPM positive cases demonstrated the following presenting symptoms and signs: rhinorrhoea, fever, cough and wheeze, with diagnoses including asthma and bronchiolitis, consistent with ARTI. Eight (47.1%) of 17 positive patients presented with indications of LRTI, most commonly presenting as bronchiolitis (5/8), wheezing (2/8) and one case suspected of pertussis (PCR negative) each in the absence of another micro-organism (Table 1). The only two adult cases had underlying conditions.
3.3. In silico characterisation of the HRV-QPM genome
Analysis of the deduced nucleotide sequence suggested HRV-QPM was a putative new member of the genus Rhinovirus. The polyprotein coding sequence spanned 6429 nt; shorter than all other reported HRVs and HEVs (Lukashev et al., 2005, Oberste et al., 2004; GenBank #EF186077). Predicted protease cleavage sites, similar to other picornaviruses, were also identified with the unconfirmed division of the polyprotein into typical picornavirus structural (VP1–4) and non-structural proteins (2A–C and 3A–D; NetPicoRNA World Wide Web server, Blom et al., 1996, Skern et al., 1985, Stanway et al., 1984). A highly conserved translation initiation site (MGAQVS) shared with other HRVs and HEVs was also identified.
A striking feature of the HRV-QPM genome was its deviation from all other sequenced members of the HRV-A species. Members of HRV-A1 (represented by the complete coding sequences of serotypes 1B, 2, 16, 39 and 89) shared 71.6–85.5% predicted amino acid identity while HRV-QPM, the sole HRV-A2 coding sequence to date, shared 50.8–51.9% identity with HRV-A1 and 46.8% with HRV-B (serotype 14). Given the extensive recombination observed in other picornaviruses (Lukashev et al., 2005, Simmonds and Welch, 2006), we next investigated the possible role of recombination in the emergence of HRV-QPM. Phylogenies of multiple, distinct HRV polyprotein regions gave no indication of recombination involving HRV-QPM (data not shown), similar to recent findings (Simmonds and Welch, 2006). The paucity of full-genome HRV sequences, however, is a limitation in these analyses.
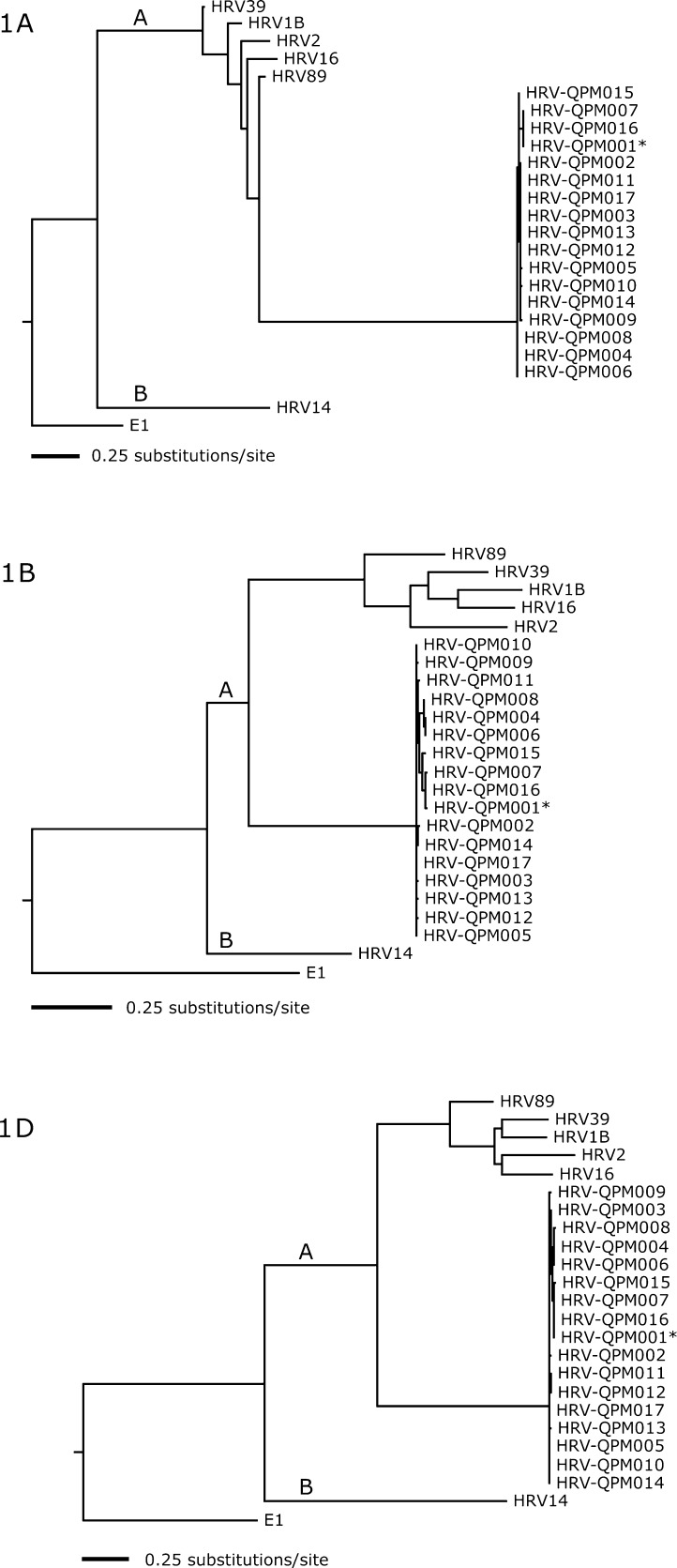
A number of further investigations were undertaken to confirm that HRV-QPM was neither a strain of an existing HRV serotype nor an artefact of PCR amplification or sequencing. The complete 1A, 1B and 1D regions (putatively encoding cleavage products VP4, VP2 and VP1) of all 17 HRV-QPM strains (001–017) were sequenced and compared to representative HRVs and HEVs (Fig. 2 ). Strains exhibited >96% nucleotide identity. These analyses confirmed that HRV-QPM exists as a novel genetic sub-lineage within the HRV-A species.
Fig. 2.
Nucleotide-based maximum likelihood phylogenies of the complete HRV structural genes 1A, 1B, and 1D (Genbank #EF512632–EF512682). The trees depict relationships among HRV-QPM strains, HRV species B serotype 14, and HRV species A serotypes 89, 39, 16, 1B, and 2 (accession numbers NC_001490, NC_001617, AY751783, L24917, D00239, X02316). The trees are rooted with human echovirus 1 (E1), a distantly related cluster B HEV, and branch lengths are drawn to scale. The two HRV species are indicated.
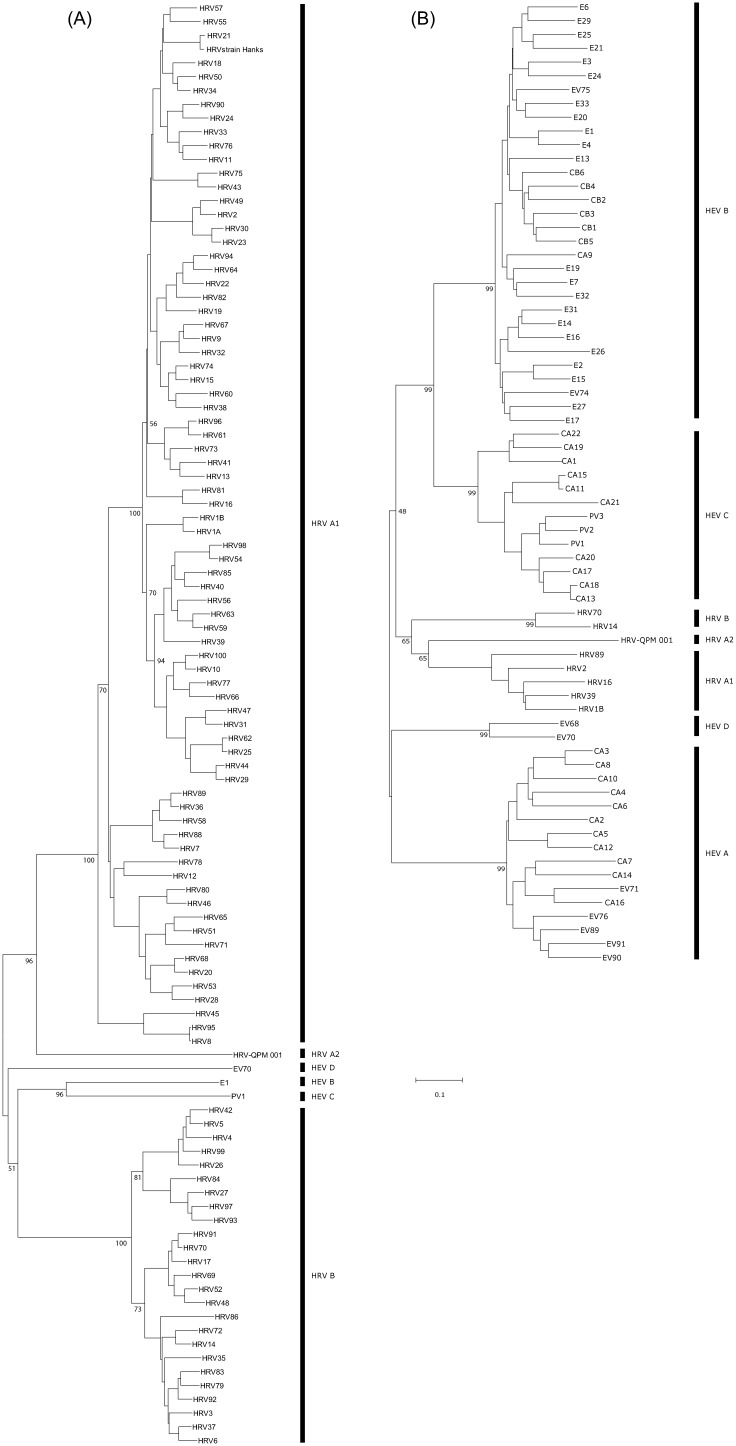
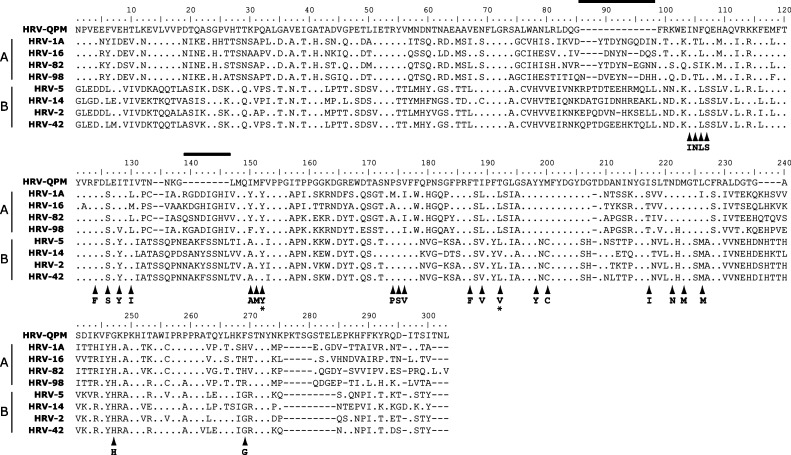
VP1 is the predominant viral surface protein and the common target both for serotyping and genotyping studies (Oberste et al., 1999) and the development of antiviral agents (Ledford et al., 2004, Ledford et al., 2005). Complete VP1 sequences from all known HRV and HEV serotypes were available for comparison to the prototype strain of HRV-QPM, 001. These data further confirmed that HRV-QPM was similar but distinct from other known HEVs and HRVs (Fig. 3a and b and Table 2 ) most likely representing a novel serotype within the HRV-A species. Importantly, the putative HRV-QPM VP1 is 6–22 amino acids shorter than that of any other HRV serotype because of several deletions. Additional in silico studies predicted that infection by HRV-QPM is not likely to respond to treatment with the capsid binding antiviral, Pleconaril. To date all other members of the HRV-A species have proven susceptible to this antiviral compound. Two amino acids reportedly conserved among all HRV-B viruses (Ledford et al., 2004), were altered in the HRV-QPM VP1 sequence (Fig. 4 ). Using the amino acid numbering system from our study, the HRV-QPM-specific changes were Tyr152 to Phe and a hydrophilic Thr in place of a hydrophobic Val191 or Leu191. Changes at position 152 and 191 are implicated in complete resistance to the antiviral among naturally occurring HRV-B species (Ledford et al., 2005).
Fig. 3.
Unrooted neighbour-joining trees of the complete VP1 amino acid sequences from representatives of (A), all four HEV clusters (includes examples from each HRV species; PV: poliovirus; CAV: coxsackievirus A; CBV: coxsackievirus B; EV: enterovirus; E: echovirus; Oberste et al., 1999) and (B), known HRV serotypes (includes examples from each HEV cluster). HRV-QPM is included in both trees.
Table 2.
Predicted VP1 amino acid identity between HRV-QPM and HEV or HRV serotypes
| Mina (%) | Max (%) | |
|---|---|---|
| HRV-QPM vs. HEVs | 26.9 | 36.0 |
| HRV-QPM vs. HRVs | 32.4 | 45.4 |
| HRV vs. HEV | 27.6 | 42.1 |
| HRV vs. HRV | 34.0 | 98.5 |
| HEV vs. HEV | 28.2 | 97.2 |
Data were obtained from the full VP1 sequences presented in Fig. 3.
Minimum to maximum amino acid identity shared between HRV-QPM and the established HRV and HEV serotypes.
Fig. 4.
Alignment of predicted VP1 amino acid sequences HRV-QPM and HRV-A (HRV-1A, 16, 82 and 98) and HRV-B (HRV-5, 14,2 and 42) species. Numbering based on the HRV-QPM sequence (EF186077). The 25 residues of the pleconaril binding pocket are arrowed (majority HRV-B amino acid sequence listed below each arrow, Ledford et al., 2005). Two regions, indicated by overhead lines, appeared to be absent in HRV-QPM when compared with all other HRV VP1 sequences on GenBank (representative data shown here). Two residues predicted by Ledford to convey complete antiviral resistance to natural HRV-B strains (e.g. in HRV-5 and 14) are indicated by stars. Residues identical to HRV-QPM are indicated by dots. The absence of a residue is indicated by a dash.
3.4. Similarity to other uncharacterised HRV sequences
A review of sequences located on GenBank and in the literature reveals that variants and incompletely characterised HRVs have been noted over many years. Most PCR-based HRV studies have targeted the 5′-UTR region for nucleotide sequencing of these variants but this region's use as a discriminator of serotype is hindered by the paucity of complete UTR sequences available both from known serotypes and HRV variants. The sequences encoding VP4/VP2 and VP1 have proven more reliable for genotyping and are comparable to serotyping (Laine et al., 2005, Ledford et al., 2004). A recent study found sequences in Belgium (sampled in 1998–1999) which appear to be related to HRV-QPM in the VP4 region (Loens et al., 2006).
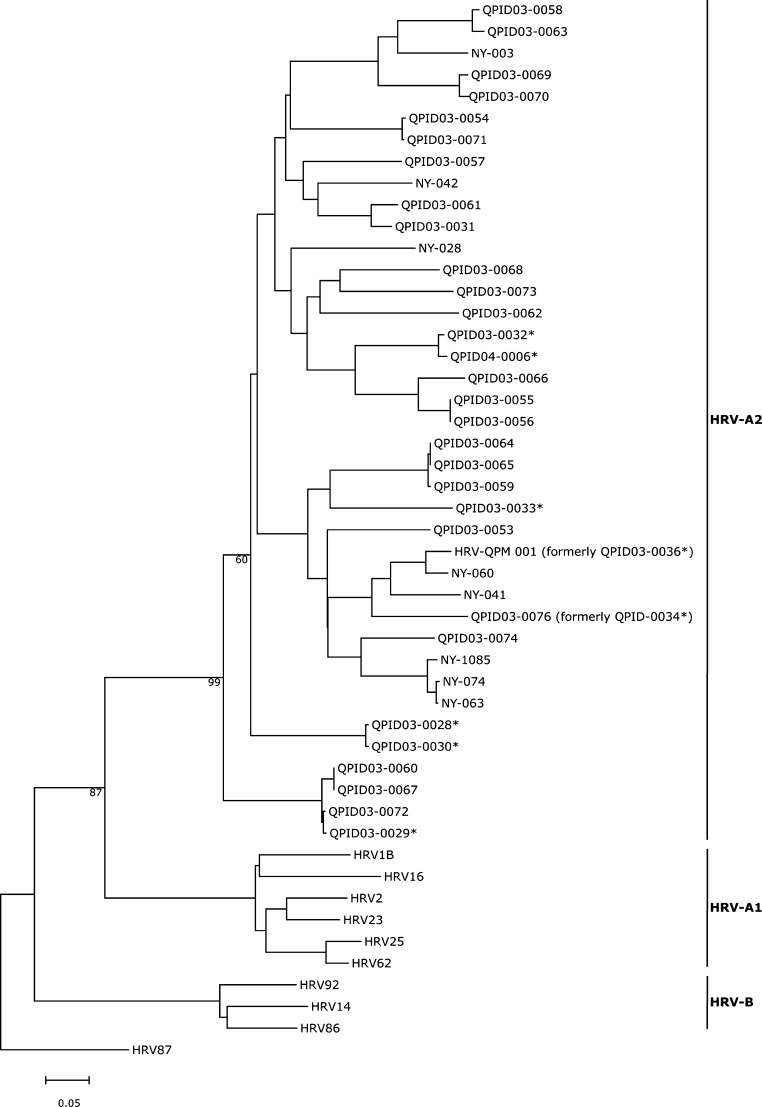
In collaboration with other investigators in Western Europe, we searched small numbers of specimens for HRV-QPM and were able to identify the virus in one country, with a similar prevalence, during its peak 2006 respiratory virus season (data not shown). We were unable to find HRV-QPM in the Netherlands (specimens from 2002/2003 and 2004/2005) in New South Wales, Australia (2006) or in New Zealand (2001). However, a very similar virus was circulating in New York during 2004 (Fig. 5 ). Together, these results suggest that HRV-QPM is not a geographically isolated HRV but one circulating globally.
Fig. 5.
Relationships among HRV-A2 strains, as depicted by a maximum likelihood phylogeny of VP4/VP2 nucleotide sequences. Sequences described here (QPID strains and HRV-QPM prototype are compared to similar strains detected in the state of New York (as reported by Lamson et al., 2006) and HRV-A1 and B serotypes virus. HRV-87 is included as an outgroup. Accession numbers: HRV-2, AB079139; HRV-14, K02121; HRV-16, L24917; HRV-23, AF343597; HRV-25, AF343617; HRV-62, AF343618; HRV-86, AF343648; HRV-92, AY040238; HRV-87/HEV-68, AY040243; NY-074, DQ875932; NY-028, DQ875931; NY-003, DQ875929; NY-060, DQ875928; NY-042, DQ875926; NY-1085, DQ875925; NY-063, DQ875924; NY-041, DQ875921; HRV-QPM, EF186077. Previously identified but uncharacterised strains of the HRV-A2 sublineage (*; Arden et al., 2006) are indicated.
4. Discussion
In this study, we have described the detection and distinguishing features of a newly identified HRV together with a summary of its clinical impact on a predominantly paediatric population. HRV-QPM was identified by intensively investigating 1 of more than 30 picornavirus-like sequences obtained during a previous study. In the present study we tested a well characterised set of respiratory specimens, mainly from children, and showed the virus to be present in 1.4% of them, with monthly detection frequencies as high as 5.0% in August and 4.2% in February. HRV-QPM was associated with a variety of clinical syndromes involving mild to severe illness, including a strong linkage with bronchiolitis in the absence of other detectable pathogens. Our findings add to recent data about the largely unrecognised diversity of HRV and their importance in clinical illness, in particular, LRTI.
We found that approximately half of all HRV-QPM detections were from patients with LRT symptoms in whom HRV-QPM was the sole micro-organism detected, suggesting that this HRV at least is associated with more serious illness. Detailed prospective clinical studies will be required to compare the prevalence of HRV-QPM in other ill populations and in suitably matched control groups without symptoms.
Detections of HRV-QPM in Europe and a closely related virus in New York suggest that HRV-QPM is not a geographically isolated HRV but a global respiratory pathogen. A small, preliminary study of local specimen extracts collected during other years did not find any additional HRV-QPM strains. This is the first molecular epidemiology study to address the annual prevalence of an individual HRV and these findings may be suggestive of the nature of all HRV serotypes. Immunity resulting from infection may protect a particular community from a certain serotype until a suitably large naïve community, either due to increasing numbers of non-immune infants and young children or adults with waning immunity, is available to again facilitate that serotype's unfettered infection and transmission. This period may encompass several years. It will be important to carry out future ‘HRV-hunting’ studies over many respiratory seasons to maximise the number of new viruses detected, overcome the restricting effects of herd immunity and better identify the length of time between cycles of infection for each HRV.
Since their discovery in the 1950s, infection by HRVs has been synonymous with the ‘common cold’ and, despite reports of association with more serious clinical outcomes (Gern and Busse, 1999, Hayden, 2004, Kaiser et al., 2006), these viruses are still considered to be of minor clinical significance by many. Consequently, the genus Rhinovirus has been poorly characterised by modern standards; only six unique genomes from over 100 HRV serotypes currently reside on GenBank. The paucity of sequence data undoubtedly parallels our understanding of individual serotypes. In a time when molecular diagnostics are considered the gold standard, a lack of sequence reduces our detection capabilities and therefore limits our understanding of the seasonal distribution and recurrence of each serotype. In turn, this has perpetuated the concept that the genus is a collection of “just rhinoviruses” rather than a large number of individual viruses each worthy of detailed characterisation.
Detailed studies of HRV-QPM and related HRV-A2s may now determine if this novel group represents a recently emerged global subset of HRVs that are more frequently associated with clinically severe respiratory outcomes or a previously unidentified viral group that has been circulating for many years. These and future findings will hopefully invigorate research into the genomics and epidemiology of these long ignored and clinically important respiratory viruses.
Acknowledgments
This work was supported by NHMRC Project grant 455905 and the Royal Children's Hospital Foundation grants 912-011 and 922-202 sponsored by the Woolworth's Fresh Futures Programme. We are grateful to Dr. Lia van der Hoek and Dr. Eric C.J. Claas (The Netherlands), Dr. William Rawlinson (NSW, Australia) and Dr. Lance Jennings (New Zealand) for local HRV-QPM testing. We thank Queensland Heath Pathology Service for the provision of clinical specimens.
References
- Allander T., Andreasson K., Gupta S., Bjerkner A., Bogdanovic G., Persson M.A. Identification of a third human polyomavirus. J Virol. 2007 doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Hansen J., Blaas D., Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro V., Guillot S., Delpeyroux F., Crainic R. Molecular strategy for ‘serotyping’ of human enteroviruses. J Gen Virol. 2001;82:79–91. doi: 10.1099/0022-1317-82-1-79. [DOI] [PubMed] [Google Scholar]
- Deffernez C., Wunderli W., Thomas Y., Yerly S., Perrin L., Kaiser L. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J Clin Microbiol. 2004;42:3212–3218. doi: 10.1128/JCM.42.7.3212-3218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor AM, Nissen MD, Whiley DM, et al. 2007. Identification of a Novel Human Polyomavirus from Patients with Acute Respiratory Tract Infections. PLoS Pathogens, in press. [DOI] [PMC free article] [PubMed]
- Gern J.E., Busse W.W. Association of rhinovirus infections with asthma. Clin Microbiol Rev. 1999;12:9–18. doi: 10.1128/cmr.12.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden F.G. Rhinovirus and the lower respiratory tract. Rev Med Virol. 2004;14:17–31. doi: 10.1002/rmv.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser L., Aubert J.-D., Pache J.-C., Deffernez C., Rochat T., Garbino J. Chronic rhinoviral infection in lung transplant recipients. Am J Respir Crit Care Med. 2006;174:1392–1399. doi: 10.1164/rccm.200604-489OC. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetic analysis and sequence alignment. Briefings Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laine P., Savolainen C., Blomqvist S., Hovi T. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J Gen Virol. 2005;86:697–706. doi: 10.1099/vir.0.80445-0. [DOI] [PubMed] [Google Scholar]
- Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York state during 2004–2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford R.M., Collett M.S., Pevear D.C. Insights into the genetic basis for natural phenotypic resistance of human rhinoviruses to pleconaril. Antiviral Res. 2005;68:135–138. doi: 10.1016/j.antiviral.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Ledford R.M., Patel N.R., Demenczuk T.M., Watanyar A., Herbertz T., Collett M.S. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 2004;78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loens K., Goossens H., de Laat C., Foolen H., Oudshoorn P., Pattyn S. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse transcription-PCR, in children with acute respiratory infections during a winter season. J Clin Microbiol. 2006;44:166–171. doi: 10.1128/JCM.44.1.166-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev A.N., Lashkevich V.A., Ivanova O.E., Koroleva G.A., Hinkkanen A.E., Ilonen J. Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J Gen Virol. 2005;86:3281–3290. doi: 10.1099/vir.0.81264-0. [DOI] [PubMed] [Google Scholar]
- Moretti S, Reinier F, Poirot O, Armougom F, Audic S, Keduas V, Notredame C. PROTOGENE: turning amino acid alignments into bona fide CDS nucleotide alignments. Nucl Acids Res 2006;34 (Web Server issue):W600–3. [DOI] [PMC free article] [PubMed]
- Mori C., Clewley J.P. Polymerase chain reaction and sequencing for typing rhinovirus RNA. J Med Virol. 1994;44:323–329. doi: 10.1002/jmv.1890440403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M.S., Maher K., Kilpatrick D.R., Pallansch L.A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M.S., Maher K., Schnurr D., Flemister M.R., Lovchik J.C., Peters H. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol. 2004;85:2577–2584. doi: 10.1099/vir.0.79925-0. [DOI] [PubMed] [Google Scholar]
- Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Welch J. Frequency and dynamics of recombination within different species of human enteroviruses. J Virol. 2006;80:483–493. doi: 10.1128/JVI.80.1.483-493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skern T., Sommergruber W., Blaas D., Gruendler P., Fraundorfer F., Pieler C. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985;13:2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway G., Hughes P.J., Mountford R.C., Minor P.D., Almond J.W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12:7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, Mass: 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A.M. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004 doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C.-M., Chan K.-H., Tsoi H.-W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]