Abstract
Porcine deltacoronavirus (PDCoV) have recently emerged in several swine producing countries. Our survey found that in addition to porcine epidemic diarrhoea virus (PEDV), PDCoV has also been a causative enteric pathogen of diarrhoeic outbreaks occurring at swine farms around Japan since late 2013. Phylogenetic analysis using the complete genomes of PDCoVs detected in Japan in 2014 demonstrated that the PDCoVs from Japan may be closely related to the PDCoVs from the U.S. and Korea during 2013 to 2014 but not the PDCoVs from China and Hong Kong during 2004 to 2016 and from Thailand, Vietnam and Laos during 2015 to 2016. To investigate the pathogenicity of a representative Japanese PDCoV, we performed an experimental infection using hysterectomy-produced colostrum-deprived piglets. The PDCoV-inoculated piglets showed acute, watery diarrhoea, but all recovered and survived. In addition, all piglets inoculated with the Japanese PDCoV exhibited virus shedding at high level in faeces and viremia corresponding to their clinical symptoms. In the PDCoV-inoculated group, viruses were mainly detected from jejunums to colons by a quantitative PDCoV-specific PCR and microscopic observation. These findings would provide useful information for establishing a diagnostic methodology for distinguishing diarrhoea caused by PDCoV from that caused by other enteric pathogens, such as PEDV.
Keywords: Porcine deltacoronavirus, Complete genome, Phylogenetic analysis, Pathogenicity, Hysterectomy-produced colostrum-deprived piglets
Highlights
-
•
Our survey finds that PDCoV is a causative agent of outbreaks of diarrhoea in pigs in Japan since late 2013.
-
•
Our phylogenetic analysis reveals that PDCoVs from Japan are closely related to the recent PDCoVs from the U.S. and Korea.
-
•
Our study indicates that PDCoV-inoculated piglets showed acute, watery diarrhoea, but all recovered and survived.
-
•
Our study demonstrates that virus shedding in faeces and sera, and viral distribution in the PDCoV-inoculated piglets.
1. Introduction
Coronaviruses belonging to the Coronavirinae subfamily are traditionally divided into three genera, Alphacoronavirus, Betacoronavirus, and Gammacoronavirus, based on antigenic relationships (Woo et al., 2010). To these traditional classifications, another novel genus, Deltacoronavirus, was recently added, which is found in diverse host species, including some mammalian and avian species, as described by the International Committee for Taxonomy of Viruses (Dong et al., 2007; Chan et al., 2013). In a large-scale molecular surveillance study performed in Hong Kong, additional deltacoronaviruses were identified in many avian species and swine (Woo et al., 2012). Thereafter, porcine/swine deltacoronavirus (PDCoV/SDCV) was first detected in pigs with diarrhoea in Ohio in 2014, and it has since also been identified in other states of the United States (U.S.) (Marthaler et al., 2014a, Marthaler et al., 2014b; Wang et al., 2014a, Wang et al., 2014b). PDCoVs have also recently been detected in Asian countries such as Korea, China, Thailand and Laos (Lee and Lee, 2014; Dong et al., 2015; Janetanakit et al., 2016; Lorsirigool et al., 2016; Zhang, 2016). PDCoV is an enveloped, positive-sense, single-stranded RNA virus, and its genome contains the following elements in the order: 5'untranslated region (UTR), open reading frame 1a/1b (ORF1a/1b), spike (S), envelope (E), membrane (M), nonstructural protein 6 (NS6), nucleocapsid (N), nonstructural protein 7 (NS7), and 3'UTR (Woo et al., 2012).
From October 2013 to September 2015, over 1000 outbreaks of PED have occurred in Japan spanning almost all prefectures, as reported by the Ministry of Agriculture, Forestry and Fisheries (http://www.maff.go.jp). In contrast, we found that diarrhoeic faecal samples from several swine farms have often been negative for PED virus (PEDV), transmissible gastroenteritis virus (TGEV), other enteric viruses (rotaviruses and enteroviruses) since 2014. Some previous studies have reported that PEDV and PDCoV could be simultaneously and frequently detected in faecal samples from pigs with diarrhoea in the regions of the U.S. in which PEDV was epidemic (Marthaler et al., 2014a, Marthaler et al., 2014b; Li et al., 2014). In addition, three research groups demonstrated that PDCoV is enteropathogenic in pigs in experimental infections (Chen et al., 2015; Jung et al., 2015, Jung et al., 2016; Ma et al., 2015). Therefore, these findings suggest the possibility that PDCoV is one of the major pathogens, in addition to PEDV, causing outbreaks of diarrhoea in multiple swine farms in Japan.
In the present study, we performed an active survey of PDCoV with a specific PCR for a number of faecal samples from diarrhoeic pigs from 2013 to 2014, in order to confirm the emergence of PDCoV in Japan. Additionally, we determined the complete genome sequences of seven Japanese PDCoV strains using next-generation sequencing and used these to characterize the genetic relationships between Japanese PDCoV strains and published PDCoV strains from other countries via comparative and phylogenetic analyses. Furthermore, we investigated its pathogenicity in hysterectomy-produced colostrum-deprived neonates experimentally inoculated with YMG/JPN/2014, one of the Japanese PDCoV strains.
2. Materials and methods
2.1. Samples for survey of PDCoV
In order to investigate association between PDCoV and diarrhoea in pigs in Japan, we conducted survey of PDCoV using 477 faecal samples, negative for PEDV-, TGEV-, and other enteric viruses (rotavirus and enterovirus)- specific PCR, collected from piglets or pigs with diarrhoea as a main clinical sign at multiple swine farms in 17 prefectures throughout Japan during November 2013 to August 2014. The faecal samples were prepared as 10% suspensions diluted in phosphate-buffered saline and were subjected to centrifugation at 3000 ×g for 10 min at 4 °C to remove debris. Viral RNA was extracted from 10% faecal suspensions using QIAamp viral RNA mini kit (Qiagen, Venlo, Limburg, Netherlands) according to the manufacturer's instructions. Detection of virus in faecal samples was performed by real-time RT-PCR including viral standards with known titres for quantification. The real-time RT-PCR using Takara One Step PrimeScript RT-PCR kit (Takara Bio, Inc., Shiga, Japan), PDCoV M gene-based primers (forward, ATCGACCACATGGCTCCAA; reverse, CAGCTCTTGCCCATGTAGCTT) and FAM-labelled probe (CACACCAGTCGTTAAGCATGGCAAGCT) was run on an ABI 7500 (Thermo, Carlsbad, CA, USA) with the following conditions: reverse transcription, 10 min at 48 °C, Taq activation, 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 45 s at 60 °C (Marthaler et al., 2014b).
2.2. Whole-genome sequencing and phylogenetic analysis
Complementary DNA (cDNA) was synthesized from 7 out of 72 samples positive for PDCoV-specific PCR using the PrimeScript High Fidelity RT-PCR Kit (Takara Bio, Inc., Shiga, Japan) with a random 6-mer primer. Nearly complete genomes, consisting of eight overlapping amplicons (approximately 4 kb in length), were generated from the cDNAs using a set of primers originally designed with reference to PDCoV strains reported previously (Table S1). Eight amplicons were pooled in equal amounts, and analysed using next-generation sequencing technology (Ion Torrent PGM; Life Technologies, Carlsbad, CA, USA). The consensus genome sequences of the seven Japanese PDCoV strains were determined with reference to the complete genomes of the 17 U.S., two Hong Kong, and one Korea PDCoV strains assigned previously in GenBank. Phylogenetic analysis based on the complete genomes was performed using the maximum-likelihood method with the general time reversible nucleotide substitution model and 1000 bootstrap replicates implemented in the MEGA6 program (Tamura et al., 2013).
2.3. Viral isolation
ST (swine testis) cells (ECACC, #92040221) were maintained in Eagle's minimum essential medium supplemented with 10% foetal bovine serum (FBS), 2 mM l-glutamine, 1% non-essential amino acids, and 1 mM sodium pyruvate (defined as growth medium). Confluent ST cells in six wells were inoculated with 200 μl of 10% intestinal suspension diluted with an equal volume of growth medium without FBS containing 5 μg/ml trypsin (defined as trypsin medium). The viral isolation was attempted from a 10% intestinal suspension from a PDCoV-infected pig with severe diarrhoea detected in Yamagata prefecture in December 2014. After 1 h incubation at 37 °C with 5% CO2, the inocula were removed, and cells were washed three times with growth medium without FBS, and added to 3 ml of trypsin medium. Until a cytopathic effect (CPE), characterized by rounded cells, was observed, passage of the isolated virus in ST cells was conducted at 5-day intervals. This viral stock was serially 10-fold diluted with trypsin medium and inoculated into ST cells grown in 96-well plates at 100 μl per well in eight wells per dilution. The plate was incubated at 37 °C with 5% CO2 for 7 days. Viral CPEs were monitored daily, and viral titres were determined according to the method described by Reed and Muench (1938), and expressed as TCID50/ml.
2.4. Animal study design
Hysterectomy-produced colostrum-deprived newborn piglets, which are highly susceptible to infectious agents, were obtained from two specific pathogen-free sows, according to the guidelines for the proper conduct of animal experiment at National Institute of Animal Health. Eleven piglets (3-days-old) were randomly separated into two groups: group 1, comprising piglets inoculated with the Japanese PDCoV isolate, YMG/JPN/2014 (n = 8), and group 2 (negative-control group), comprising piglets inoculated with Eagle's minimum essential medium (n = 3). The individuals from groups 1 were inoculated orally with 2 × 106TCID50/head.
Faecal samples were collected from each piglet every day from 0 day post-inoculation (DPI) to 8 DPI and every 3–4 days after 8 DPI. Serum samples were collected from each piglet every 2 days from 0 DPI to 8 DPI and every 3–4 days after 8DPI. Three and one piglets from groups 1 and 2 were euthanized for pathological examination at 4 and 7 DPI, respectively. The remaining piglets from each group were monitored for clinical signs and virus shedding until 27 DPI. The small and large intestines and other organs (heart, lung, kidney, liver, spleen, tonsil, trachea, muscle, stomach, and mesenteric lymph nodes) were sampled from the PDCoV-inoculated and control piglets, in order to examine the viral distribution in those tissues.
Viral RNA was extracted from 10% faecal suspensions, sera, and 10% tissue homogenates collected from animals according to the method described above. Virus shedding in faeces, sera, and various tissues were determined by the real-time RT-PCR targeting the PDCoV M gene as described above.
2.5. Preparation of PDCoV-specific rabbit antiserum
We obtained the U.S. PDCoV isolate, Michigan/8977/2014 strain from National Veterinary Services Laboratories at the USDA (Ames, IA, USA), to generate rabbit antiserum against the PDCoV validated in a previous study (Ma et al., 2015). The obtained U.S. PDCoV isolate was propagated in ST cells in our laboratory as described above. Culture fluid of the isolated virus on ST cells was centrifuged at 5600g for 20 min at 4 °C to remove debris. Thereafter, semi-purified virus was obtained at the interface of the discontinuous gradient of 20 and 50% (wt/vol) sucrose in phosphate-buffered saline after centrifugation at 100,700g for 2 h at 4 °C. One specific pathogen-free, JW/CSK (Japan SLC, Inc., Shizuoka, Japan) were subcutaneously immunized three times at 2 weeks intervals with semi-purified virus mixed 1:1 with adjuvant liquid (TiterMax Gold; Sigma-Aldrich, Munich, Germany). The rabbit serum was collected at 2 weeks after the last immunization.
The rabbit antiserum against PDCoV were evaluated its specificity on ST cells inoculated the U.S. PDCoV isolate by an indirect immunofluorescence assay (IFA) using FITC-labelled goat anti-rabbit IgG antibody (Bethyl Laboratories, Inc. Montgomery, TX, USA). Moreover, the cross-reactions with PDCoV-specific rabbit antiserum were evaluated by IFA on both PEDV-infected Vero cells and TGEV-infected CPK cells inoculated the TGEV isolate, but the antiserum did not react with the PEDV and TGEV.
2.6. Immunohistochemistry (IHC)
Small and large intestines and other organs (heart, lung, kidney, liver, spleen, tonsil, trachea, muscle, stomach, and mesenteric lymph nodes) from the PDCoV-inoculated and control piglets at 4 DPI, 7 DPI and 27 DPI were fixed in 10% formalin, paraffin-embedded, sectioned, and mounted on glass slides. The tissues were stained with PDCoV-specific rabbit antiserum as a primary antibody (see Section 2.5. Preparation of PDCoV-specific rabbit antiserum), and with diaminobenzidine as a second labelled antibody. Antigen detection by IHC staining was semi-quantitatively assessed ratio of enterocytes with positive staining signal to enterocytes of villous and crypt epithelium as follows as reference with a previous report (Chen et al., 2015): –, no staining; +, approximately 1–9%; ++, approximately 10–19%; +++, approximately 20–49%; ++++, approximately 50–100%.
3. Results
3.1. Survey of PDCoV in Japan
In the survey of PDCoV using 477 faecal samples, 72 samples (15.1%) were positive according to PDCoV specific real-time RT-PCR. Detection rates of PDCoV positive samples by age group are summarized in Table 1 . Among sows, almost half (48.3%) were positive according to PDCoV-specific PCR. Fattening pigs (over 120 days old) as well as weaned pigs (21–60 days old) showed the next highest positive rates at 10.5% and 10.3%, respectively. In addition, our survey showed that PDCoV was present in Japan since February 2014.
Table 1.
Detection of porcine deltacoronavirus in faecal samples negative for other enteric viruses-specific PCR collected around Japan from 2013 to 2014.
| Groups | Number of Samples | Number of positive samples | Rate of positive samples [%] |
|---|---|---|---|
| Newborn pigs (<21 days old) | 191 | 15 | 7.3 |
| Weaned pigs (21 to 60 days old) | 53 | 4 | 10.3 |
| Nursing pigs (61 to 120 days old) | 51 | 1 | 2.0 |
| Fattening pigs (120 days old <) | 95 | 10 | 10.5 |
| Sows | 87 | 42 | 48.3 |
| Total | 477 | 72 | 15.1 |
3.2. Whole-genome sequencing and phylogenetic analysis
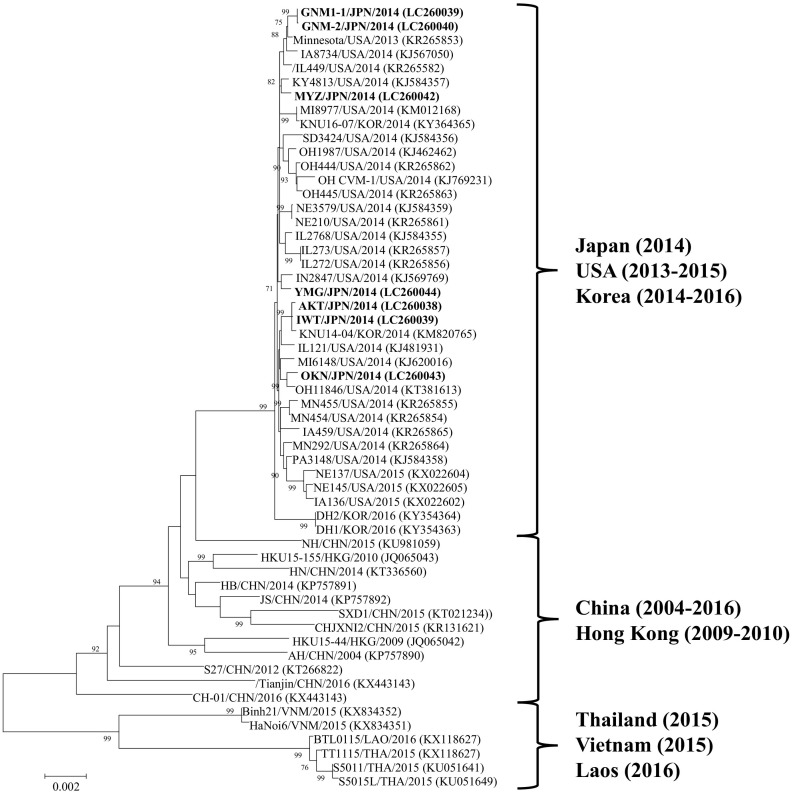
The complete genomes of seven Japanese PDCoV strains were analysed using next-generation sequencing technology on an Ion PGM platform. The data were assembled based on the known complete genomes of PDCoV strains from other countries. The genome lengths, GenBank accession numbers, and collection information for the seven Japanese PDCoV strains are summarized in Table 2 . Phylogenetic analysis based on the complete genomes showed that the seven PDCoV strains detected in Japan in 2014 clustered with those detected in the U.S. and Korea from 2013 to 2016, which could be distinguished from those detected in China including the Hong Kong from 2004 to 2016, and in Thailand, Laos, and Vietnam from 2015 to 2016 (Fig. 1 ). In addition, comparative genomic analysis of PDCoV strains from Japan and other countries revealed that the complete genomes of the Japanese PDCoV strains were almost identical to those of the recent U.S. and Korean PDCoV strains (99.7–100% identity), which were comparatively distinctive from those of the Chinese PDCoV strains including the Hong Kong PDCoV strains (98.6–99.3% identity) and those of the Thai, Vietnamese and Lao PDCoV strains (97.4–97.8% identity) (Table 3 ).
Table 2.
Summary of sample collection and genomic features of the seven porcine deltacoronavirus strains used in the present study.
| Strains | Collection prefecture | Collection date | Length (nucleotides) | GenBank accession number |
|---|---|---|---|---|
| AKT/JPN/2014 | Akita | May 2014 | 25,362 | LC260038 |
| GNM-1/JPN/2014 | Gunma | May 2014 | 25,362 | LC260039 |
| GNM-2/JPN/2014 | Gunma | May 2014 | 25,362 | LC260040 |
| IWT/JPN/2014 | Iwate | May 2014 | 25,362 | LC260041 |
| MYZ/JPN/2014 | Miyazaki | Mar 2014 | 25,362 | LC260042 |
| OKN/JPN/2014 | Okinawa | Aug 2014 | 25,362 | LC260043 |
| YMG/JPN/2014 | Yamagata | Dec 2014 | 25,362 | LC260044 |
Fig. 1.
Phylogenetic tree based on complete genomes of seven PDCoV strains from Japan and global PDCoV strains from other countries. The tree was constructed using the maximum-likelihood method with the general time reversible nucleotide model implemented in MEGGA 6.0. The number at each node indicates groups with >70% bootstrap support using 1000 replicates. Strains abbreviations, country, years of detection, and GenBank accession numbers in parenthesis are indicated. The seven Japanese PDCoV strains used in the present study are shown in bold. The scale bar indicates nucleotide substitutions per site.
Table 3.
Nucleotide identities [%] among porcine deltacoronavirus strains from Japan and other countries used in the present study.
| Strain | Full genome | ORF1 | S | E | M | NS6 | N | NS7 |
|---|---|---|---|---|---|---|---|---|
| Japan vs. US, Korea (n = 31) | 99.7–100 | 99.7–100 | 99.5–100 | 99.6–100 | 99.5–100 | 99.3–100 | 99.1–100 | 98.8–100 |
| Japan vs. China, Hong Kong (n = 12) | 98.6–99.3 | 98.6–99.4 | 98.1–99.1 | 98.8–99.6 | 98.8–99.5 | 98.2–100 | 98.4–99.4 | 98.7–99.7 |
| Japan vs. Thailand, Laos, Vietnam (n = 6) | 97.4–97.8 | 97.5–97.7 | 96.1–96.8 | 99.2–99.6 | 98.0–99.1 | 98.2–98.9 | 97.1–98.8 | 97.1–98.7 |
| Japan vs. Japan (n = 7) | 99.8–100 | 99.9–100 | 99.7–100 | 100 | 99.8–100 | 99.6–100 | 99.6–100 | 99.5–100 |
3.3. Viral isolation
The isolated virus was efficiently propagated in ST cells at the third passage. The isolate, YMG/JPN/2014, was evaluated to be PDCoV by real-time RT-PCR targeting the PDCoV M gene and by an immunofluorescent assay using PDCoV-specific rabbit antiserum. The isolate was additionally propagated four times in ST cells, and its titre was 106TCID50/ml when used in the experimental study.
3.4. Clinical signs and virus shedding
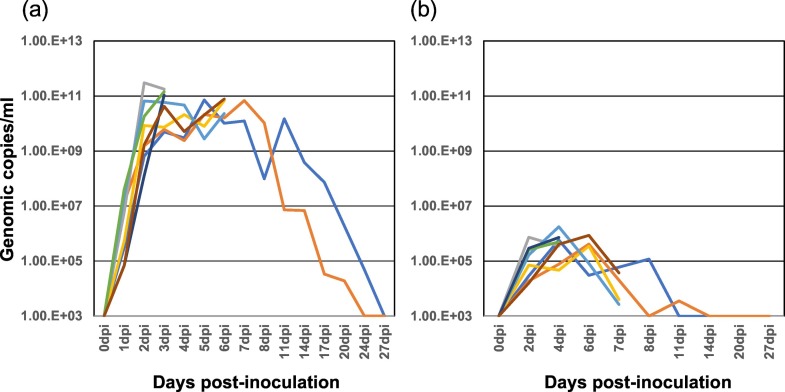
PDCoV-inoculated newborn piglets developed acute, watery diarrhoea, anorexia, rough hair, and vigorous prostration during 2–11 DPI, and then all recovered and survived throughout the observation period. PDCoV RNA shedding detected in faecal and serum samples from the PDCoV-inoculated piglets are shown in Fig. 2 . All piglets inoculated with the Japanese PDCoV isolate exhibited peak viral shedding of 1010–1011 copies/ml in faeces at 2–6 DPI, and shed viruses in faeces at high levels for approximately a week. Afterwards, viral shedding gradually decreased and reached a level undetectable by PCR at 27 DPI. Viral RNAs in the ranges of 105–106 copies/ml were detected in the sera of PDCoV-inoculated piglets at 2–6 DPI.
Fig. 2.
Viral shedding in faeces (a) and sera (b) from PDCoV-inoculated piglets. Individual viral shedding (shown in different colours) was detected by quantitative PDCoV M specific real-time RT-PCR at each time point.
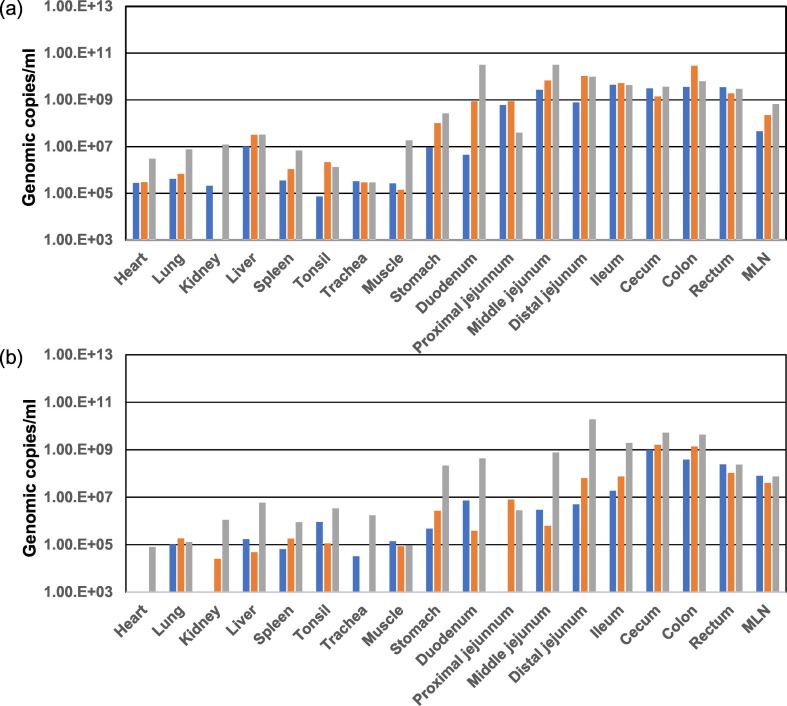
Viral distributions in various tissues from the PDCoV-inoculated group were examined at 4 and 7 DPI by using PDCoV-specific real-time RT-PCR (Fig. 3 ). In the PDCoV-inoculated group, viruses were mainly detected in intestinal tissues from the proximal jejunums to rectums, and in mesenteric lymph nodes (MLNs) in the ranges of 108–1010 copies/ml at 4 DPI. In addition, PDCoV-specific PCR also detected viral RNA copies of over 107 from the livers, from 2/3 of stomach and duodenum samples, and from 1/3 of muscle samples from PDCoV-inoculated piglets at 4 DPI. At 7 DPI, viruses were mainly distributed in the large intestines, from the caecum to rectum, at high levels (108–109 copies/ml). In addition, viral genomic copies of >107 were detected from the MLN and from 2/3 of distal jejunum and ileum samples. All faecal, sera, and tissue samples from piglets in the negative-control group were negative according to PDCoV-specific PCR.
Fig. 3.
Viral shedding in various tissues of PDCoV-inoculated piglets at 4 DPI (a) and 7DPI (b). Individual viral shedding (shown in different colours) was detected by quantitative PDCoV M specific real-time RT-PCR.
3.5. IHC
IHC staining detected PDCoV antigens in the cytoplasm of villus and crypt enterocytes from the proximal jejunum to colon in samples from the PDCoV-inoculated group using PDCoV-specific rabbit antiserum at 4 DPI necropsy (Table 4 ). The average IHC scores of the PDCoV-inoculated piglets were low in the proximal jejunum and caecum to colon, high in the middle jejunum to ileum, and negative in the duodenum and rectum. On 7 DPI, viruses were mainly distributed in large intestines, from ceca to colons at low level. No positive staining was identified in any tissues from the PDCoV-inoculated group at 27 DPI. IHC staining was not detected on all examined tissues from the negative-control piglets.
Table 4.
Average immunohistochemical scores in various intestinal tissues from PDCoV-inoculated and negative-control piglets at 4, 7 and 27 days post-infection.
| Group | Days post-inoculation | IHC positive piglets (average IHC scores) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Duodenum | Proximal jejunum | Middle jejunum | Distal jejunum | Ileum | Cecum | Colon | Rectum | ||
| PDCoV-inoculated | 4 DPI | 0/3 | 2/3 (0.7) | 3/3 (3.0) | 3/3 (1.7) | 3/3 (1.7) | 3/3 (1.3) | 3/3 (1.0) | 0/3 |
| 7 DPI | 0/3 | 0/3 | 0/3 | 1/3 (1.0) | 1/3 (0.3) | 2/3 (0.7) | 3/3 (1.0) | 1/3 (0.3) | |
| 27 DPI | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | |
| Negative control | 4 DPI | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| 7 DPI | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | |
| 27 DPI | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | |
4. Discussion
Our survey showed that 72 (15.1%) out of 477 faecal samples that tested negative for PEDV-, TGEV- and other enteric viruses-specific RNA obtained from diarrhoeic pigs in several prefectures in Japan from 2013 to 2014 were positive for PDCoV-specific RNA. This detection rate of PDCoV-positive samples is similar to that (22%) observed in a survey of porcine samples collected in the U.S. in January–February 2014 (Marthaler et al., 2014b). Because we have no detailed information of swine herds which collected diarrhoeic samples, our data cannot deny the possibility that these diarrhoeas might be caused by other pathogens (bacteria and parasites), but suggests that PDCoV would associated with these diarrhoeas. Moreover, our data also suggests that PDCoV can infect into all age groups as similar as PEDV (Saif et al., 2012). Particularly, our survey revealed the highest positive detection rates of PDCoV in sows. Because pregnant pigs are stressful, and decrease self-immunity, sows are susceptible to pathogens around them. In fact, the report for first detection of PDCoV in U.S. exhibited that the U.S. PDCoVs were detected from sows with watery diarrhoea (Wang et al., 2014a, Wang et al., 2014b). In field observation, it reports that sows showed not only severe diarrhoea but also vomiting and anorexia (in our personal communication). These findings suggest that PDCoV might be susceptible to older pig than piglets. However, its hypothesis would need to examine clinical signs and dynamics of viral distribution in older pigs experimentally inoculated with PDCoV in future.
A phylogenetic tree constructed with complete genome sequences demonstrated that seven PDCoV strains detected in Japan grouped with those derived from the U.S. and Korea during 2013 to 2016 and but not with those found in China and Hong Kong during 2004 to 2016 and those found in Thailand, Vietnam, and Laos during 2015 to 2016. Comparative genomic analysis of the PDCoV strains collected in Japan and those collected in other countries from 2004 to 2016 revealed that the complete genomes of the PDCoV strains from Japan were almost identical to those from the U.S. and Korea (99.7–100%). Therefore, our analyses indicated that the PDCoV strains that had emerged in Japan since 2014 were genetically related to those emerging in the U.S. and Korea since 2013 (Marthaler et al., 2014a, Marthaler et al., 2014b; Wang et al., 2014a, Wang et al., 2014b; Lee and Lee, 2014). Furthermore, our findings suggest that the PDCoV outbreaks that have occurred in Japan since 2014 may have been caused by the invasion of recent PDCoV strains from other countries with large pig industries into Japan.
In order to examine the pathogenicity of one of these strains, YMG/JPN/2014, we conducted experimental infections using hysterectomy-produced colostrum-deprived neonatal piglets, which are highly susceptible to causative pathogens. In field observation, this strain developed mild diarrhoea not vomiting and anorexia in few piglets, but the infected piglets recovered from diarrhoea after five days. In contrast, piglets inoculated with the Japanese PDCoV isolate suffered from severe diarrhoea from 2 DPI, but all recovered from diarrhoea at 11 DPI, and survived until 27 DPI. Our results therefore demonstrated that the PDCoV isolate as well as the field strain developed diarrhoea. However, the differences of severity and duration of diarrhoea in piglets in the field and experimental study may have resulted from different conditions such as the inoculated volume or host susceptibility.
In the present study, PDCoV-inoculated piglets exhibited high amounts of faecal viral shedding from 1 DPI, with lower amounts of sera viral shedding than faecal viral shedding from 2 DPI. These results were identical to those in the report by Ma et al. (2015). High levels of PDCoV genomic RNA were identified from intestinal tissues, from the duodenum to rectum, in the PDCoV-inoculated group at 4 DPI necropsy. Lower viral genomic copies were also detected in the livers and MLNs of PDCoV-inoculated piglets using PDCoV-specific real-time RT-PCR at 4 DPI. The presence of virus in these tissues might mean that PDCoV genomic RNA is transferred via the blood stream, as piglets inoculated with PDCoV isolate showed the peak viral shedding in sera at 4 DPI. In addition, the PDCoV-inoculated piglets necropsied at 4 DPI showed positive IHC staining for PDCoV in enterocytes in the proximal jejunum to colon, particularly the middle jejunum to ileum. In contrast, all piglets necropsied at 7 DPI in the PDCoV-inoculated group mainly exhibited viral genomic copies and IHC antigens at high levels in the caecum and colon according to PDCoV-specific PCR and microscopic observation, respectively. Our findings suggest that the PDCoV isolate was widely distributed in the small and large intestines and mainly propagated in the middle jejunum to ileum until 4 DPI, at which point it was transmitted to and propagated in the caecum to colon. These data, especially the viral distribution in the intestines at 7 DPI, differed from those obtained from conventional piglets (5-day-old) inoculated with a PDCoV isolate cultured in ST cells in a previous report (Chen et al., 2015).
In conclusion, we first identified that PDCoV has been emerging in Japan since February 2014. Our analysis demonstrates that the PDCoV might be a causative agent of the outbreaks resulting in diarrhoea in pigs in Japan since late 2013. Because the PDCoVs detected in our study were genetically related with those emerging in other swine-producing countries, such as the U.S. and Korea, since 2013, they may represent an invasion from overseas, like the PEDV invasion into Japan. Our findings provide valuable information on the genetic relationships of PDCoV among countries worldwide, as well as the clinical signs and viral distribution of PDCoV in piglets.
The following is the supplementary data related to this article.
A set of primers used for eight overlapping amplicons, consisting of the nearly entire genome of the porcine deltacoronavirus.
Acknowledgements
This work was supported in part by grants from Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (26111C), and the Ito Foundation (vol.34 and 35).
We thank O. Takisaki, H. Tsuboi, S. Mizukoshi, S. Matsuura, H. Sekine, H. Seo, A. Kimura, and Y. Nakamura for assisting with animal care in NIAH, NARO; K. Honda and K. Hirano for providing clinical information in the field in Yamagata prefecture; S. Itoh, M. Ikezawa, R. Yamaguchi, M. Kobayashi, M. Shimada and B. Xiao for providing technical assistance in NIAH, NARO; K. Kawashima, T. Tsutsui, for providing advice and discussion in NIAH, NARO.
References
- Chan J.F., To K.K., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lesions from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;284:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 2016;22:754–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014;2 doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Chen Q., Harmon K.M., Yoon K., Schwartz K.J., Hoogland M.J., Gauger P.C., Main R.G., Zhang J. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014;2 doi: 10.1128/genomeA.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsirigool A., Saeng-chuto K., Temeeyasen G., Madapong A., Tripipat T., Wegner M., Tuntituvanont A., Intrakamhaeng M., Nilubol D. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch. Virol. 2016;161:2909–2911. doi: 10.1007/s00705-016-2983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine coronaviruses in the United States. MBio. 2015;6 doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Jiang Y., Collins J., Rossow K. Complete genome sequence of strain SDCV/USA/Illinoi121/2014, a porcine deltacoronavirus from United States. Genome Announc. 2014;2 doi: 10.1128/genomeA.00218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. Asimple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Saif L., Pensart M.B., Sestak K., Yeo S.G., Jung K. Coronaviruses. In: Zimmerman J.J., Karriker L., Ramirez A., Schwartz K.J., Stevenson G.W., editors. Diseases of Swine. 10th ed. Wiley-Blackwell; 2012. pp. 501–524. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pig, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Porcine deltacoronavirus HUK15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronavirus as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Porcine deltacoronavirus: overview of infraction dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A set of primers used for eight overlapping amplicons, consisting of the nearly entire genome of the porcine deltacoronavirus.