Abstract
The development of a rapid, specific, and sensitive SYBR Green I-based duplex real-time quantitative PCR assay is described for the simultaneous detection of porcine epidemic diarrhea virus (PEDV) and porcine circovirus type 3 (PCV3). The assay specifically detected PEDV and PCV3, with no fluorescence detected for other non-targeted pig pathogens. The assay showed a good linear relationship, and the limits of detection for this assay were 34.6 copies/μL and 61.2 copies/μL for PEDV and PCV3, respectively. The assay exhibited high repeatability and reproducibility, with intra-assay and inter-assay variation coefficients less than 2.0%. A clinical evaluation using intestinal tissue and fecal samples from piglets suffering from diarrhea at different pig farms in China revealed that the singular infection rates of PEDV and PCV3 were 43.94% (29/66) and 16.67% (11/66), respectively, while the co-infection rate of PCV3 with PEDV was 27.27% (18/66). The results indicate this assay is a rapid and reliable diagnostic tool for PEDV and PCV3 monitoring and surveillance in the field, and provides technical support for the quantitative detection of clinical samples infected or co-infected with PEDV and PCV3.
Keywords: Porcine epidemic diarrhea virus, Porcine circovirus type 3, Duplex, Real-time fluorescence quantitative PCR, Detection
Highlights
-
•
PED outbreaks have been resulted in a huge economic loss in the pig farming industry.
-
•
PCV3 is a novel virus and has been detected in piglets affected with diarrhea.
-
•
A duplex qPCR assay was developed for the simultaneous detection of PEDV and PCV3.
-
•
The LOD for this assay were 34.6 copies/μL and 61.2 copies/μL for PEDV and PCV3, respectively.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is a large, enveloped, single-stranded, positive-sense RNA virus with a genome approximately 28 kb in size, and is taxonomically classified as a member of the genus Alphacoronavirus in the family Coronaviridae [1,2]. PEDV has been recognized in veterinary medicine since the early 1970s in Europe and subsequently identified in many other swine-breeding regions of the world [3]. PEDV is the causative agent of porcine epidemic diarrhea (PED), an acute and highly contagious enteric disease characterized by vomiting, watery diarrhea, dehydration, and high mortality in nursing piglets [4]. PED outbreaks have been reported in many swine-raising countries, including Europe and Asia [[5], [6], [7]]. With the widespread use of inactivated or attenuated bivalent vaccines for PEDV in China, the nationwide spread of PED was controlled until 2010; however, since December 2010, there has been a remarkable increase (>50%) in PED observed on many pig farms in China [8]. A high morbidity rate of between 90% and 100% among neonatal piglets was observed, with a mortality rate of 70%–100% on affected pig farms, resulting in a huge economic loss in the pig farming industry [9,10]. In 2013, PED emerged in swine in the USA [11] and spread rapidly across the country, resulting in the substantial economic losses. And then, one PEDV strain called the S INDEL was identified by sequence analyses in US swine in 2014, which had insertions and deletions (INDEL) in the spike gene and formed a distinct phylogenetic cluster compared to U.S. PEDV prototype strains [12,13], and has also been detected in South Korea, Germany, Belgium, France, Portugal, Japan, Italy and Austria [14,15].
Porcine circoviruses (PCVs), belong to the genus Circovirus within the Circoviridae family, and are non-enveloped, single-stranded circular DNA viruses with the smallest genome (1766–1768 nucleotides) of all animal viruses [16]. Prior to 2016, only two types of PCVs were known to infect pigs around the world: PCV1 and PCV2 [17]. Although PCV1 is non-pathogenic to pigs, PCV2 is pathogenic and has been associated with a number of diseases affecting pigs worldwide [18,19]. In 2016, a novel PCV, called PCV3, was identified in the USA using metagenomic sequencing technology and a genetic evolution analysis [20]. Subsequently, PCV3 was reported to be widely distributed throughtout China in commercial pig farms from 14 provinces [[21], [22], [23], [24]]. Recently, PCV3 was confirmed in Poland, South Korea, Brazil, Italy, and the UK [[25], [26], [27], [28], [29]], indicating that PCV3 is spreading globally. Moreover, previous studies have detected PCV3 in samples obtained from piglets with diarrhea [[30], [31], [32]], suggesting that PCV3 may have a potential association with swine diarrhea.
An accurate laboratory diagnosis of PEDV and PCV3 is essential for epidemiological surveillance and disease management. Among numerous molecular diagnostic methods, real-time quantitative PCR (qPCR) enables the characterization of small quantities of sample with a high level of specificity, sensitivity, and accuracy. Several studies have used a qPCR assay for the detection of PCV3 [20,33,34] and PEDV [35,36], and the recently, a report described multiplex qPCR assay for the differential detection of PCV2 and PCV3 [37]; however, there are currently no reports validating the use of SYBR Green I -based duplex qPCR for the simultaneous detection of PEDV and PCV3 in the field. In this study, a SYBR Green I -based duplex qPCR assay was developed for the simultaneous detection of PEDV and PCV3, and its applicability was evaluated for the detection of PEDV and PCV3 in intestinal tissue and fecal samples of piglets suffering from diarrhea.
2. Materials and methods
2.1. Virus and clinical samples
PCV3-positive and PEDV-positive clinical samples were used to optimize the duplex qPCR conditions. The positive tissue samples were collected from pig farms and identified as positive for PCV3 or PEDV using previously described qPCR assays [33,38] and sequencing. A total of 66 samples (intestinal tissues and fecal) were collected from 66 piglets suffering from diarrhea at different pig farms in Hebei province and Henan province (China) from 2014 to 2018. The tissue samples were homogenized, and diluted 1:10 with phosphate-buffered saline (PBS) in a mortar. The clinical samples were thawed and frozen three times and centrifuged at 12000×g for 5 min. The supernatants were used for nucleic acid extraction immediately or stored at −80 °C until testing.
To test specificity of the duplex qPCR, other porcine viral pathogens, including porcine circovirus type 2 (DF-2), porcine transmissible gastroenteritis virus (HN-HJ2007), porcine reproductive and respiratory syndrome virus (HeN-21), classical swine fever virus (Qixian), porcine pseudorabies virus (HN2012), and porcine parvovirus (HN-K) were used as control strains, and their titers were 105.4 TCID50/mL, 106.8 TCID50/mL, 106.5 TCID50/mL, 105.6 TCID50/mL, 107.4 TCID50/mL, and 108.5 TCID50/mL, respectively.
Total nucleic acids (viral genomic DNA or/and RNA) were extracted from 200 μL of sample (cell cultures infected with each of the control viruses, intestinal tissue and fecal homogenates 10% in PBS) using an Viral DNA/RNA Kit (Beijing Zoman Biotechnology Co., Ltd., China) according to the manufacturer's instructions, and nucleic acids were finally eluted in 30 μL of the RNase free H2O. Viral genomic DNA or/and RNA were immediately used for qPCR or stored at −80 °C until use.
2.2. Primer design
The nucleotide sequences for PEDV (GenBank accession numbers AF353511, KT591944, LC022792, KT199103, KY111278 and MG334554) and PCV3 (GenBank accession numbers KX966193, KX778720, KT869077, KX898030, and KY418606) were collected from GenBank. Two pairs of specific primers were designed based on a highly conserved sequence within the rep region of the PCV3 genome and the ORF1 region of the PEDV genome using the PrimerSelect program in the DNASTAR 5.0 software (DNASTAR Inc.). The PCR primer pairs for each target gene and the size of each amplicon are summarized in Table 1 . The primers were used for the reverse transcription reactions for the PRRSV, TGEV and CSFV controls by Tian's, Kim's and Yang's, respectively [[39], [40], [41]]. The primers were synthesized by Sangon Biotech Shanghai Co, Ltd., China.
Table 1.
Primer sequences for duplex qPCR in this study.
| Virus | Primera | Primer sequence (5′-3′) | Primer location (bp)b | Product size (bp) |
|---|---|---|---|---|
| PEDV | PEDV-F | AAATGGGAAGTCGGCAGA | 15849–15866 | 163 |
| PEDV-R | GTTTTGTTGTGGCGGTAG | 15994–16011 | ||
| PCV3 | PCV3-F | CTACGAGTGTCCTGAAGA | 176–193 | 136 |
| PCV3-R | CCTCCACACTCCACAATA | 294–311 |
F, forward; R, reverse.
Genome position of primer-binding sequences according to the complete genome sequence of PEDV CH-HNKF-16 strain and PCV3 SD2016 strain (GenBank accession no. KY649107.1 and KX966193.1, respectively).
2.3. Construction of standard recombinant plasmids
PCR amplification of PEDV and PCV3 was performed using a PCT-200 Peltier thermal cycler (MJ Research, USA). The PCR protocol was carried out in a 20 μL reaction volume: 10 μL 2×one-step RT-PCR Buffer (Beijing Zoman Biotechnology Co., Ltd., China), 0.4 μL QKRT one-step RT-PCR enzyme mix, 0.5 μL of each primer (0.25 μM), 1pg-1μg DNA/RNA template, RNase-free H2O were mixed to a final volume of 20 μL. The PCR parameters were 45 °C for 30 min, 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 62 °C for 20 s, 72 °C for 20 s, and 72 °C for 10 min.
The positive PCR amplicons corresponding to the PEDV and PCV3 genes were purified using a SanPrep Column DNA Gel Extraction Kit (Sangon Biotech Shanghai Co, Ltd., China) according to the manufacturer's instructions, and cloned into the pMD18-T Vector (Takara) at 16 °C overnight. The resulting recombinant plasmids (pMD-PEDV and pMD-PCV3) were transformed into Escherichia coli DH-5α cells according to the manufacturer's instructions (Takara). The recombinant plasmids were extracted from the bacterial cells using a SanPrep Column Plasmid Mini-Preps Kit (Sangon) and verified using restriction enzyme digestion and PCR. The positive plasmids were submitted to Sangon Biotech Shanghai Co, Ltd., China for sequencing. The sequencing results were confirmed by comparing with the expected sequence of PEDV (GenBank accession number AF353511, KT591944, LC022792, KT199103, KY111278 and MG33455) or the expected sequence of PCV3 (GenBank accession numbers KX966193, KX778720, KT869077, KX898030, and KY418606).
The concentration and purity of the standard plasmids were assessed by measuring the absorbance at 260 nm with a UV spectrophotometer (Thermo Electron, USA) and using the 260/280 nm ratio. The concentratios of the recombinant plasmids were 110.0 ng/μL for PEDV and 189.9 ng/μL for PCV3. The copy numbers of each cloned gene were quantified as previously described: copies/μL = [concentration of plasmid (ng/ul) × (6.022 × 1023)]/[(plasmid length × 660 × 109)] [42], and were 3.46 × 1010 copies/μL for PEDV and 6.12 × 1010 copies/μL for PCV3. Serial 10-fold dilutions of plasmid DNA in Elution buffer were used in the amplification reactions, and stored at −20 °C until further testing was performed.
2.4. One-step duplex RT-qPCR
Before optimization of the duplex qPCR, a single qPCR assay with each PEDV or PCV3 primer was carried out using a LightCycler® 96 Instrument (Roche, Basel, Switzerland). For a single qPCR assay for PEDV, 10 μL 2×HSYBR one-step RT-qPCR Buffer (Beijing Zoman Biotechnology Co., Ltd., China), 0.5 μL (0.25 μM) of each primer, 0.4 μL QK SYBR Enzyme Mix, 1pg-1μg DNA or/and RNA template, and RNase-free H2O were mixed to a final volume of 20 μL. And a single qPCR assay for PCV3 was performed using a commercial qPCR kit (HSYBR one-step RT-qPCR Kit, Zoman Biotechnology, Beijing, China). The 20 μL reaction mixture contained 10 μL of 2×HSYBR one-step RT-qPCR Buffer, 0.5 μL (0.25 μM) of each primer, and 1pg-1μg of DNA or/and RNA template, and the final mixture was adjusted to 20 μL by adding RNase-free H2O. In addition to the concentrations of the two sets of primers were optimized, other reaction components for duplex qPCR were the same as those used for single qPCR for PEDV. The qPCR assay was conducted under the following conditions: 5 min at 45 °C; 5 min at 95 °C, and then 45 cycles of 30 s at 94 °C, 20 s at 60 °C, 20 s at 72 °C. The annealing/extension temperature was optimized, while other amplification conditions for duplex qPCR were the same as those used for single qPCR. The reaction without the template and negative reference samples served as the non-template control.
2.5. Establishment of standard curves
Serial 10-fold dilutions of plasmid DNA in Elution buffer were used in the one-step duplex RT-qPCR amplification reactions, the logarithm of the number of starting templates was plotted along the x-axis, and the cycle threshold (Ct value) was plotted along the y-axis to establish the standard curve.
2.6. Sensitivity and specificity of duplex qPCR
To determine the minimum detectable amount of PEDV and PCV3, the recombinant plasmids, ranging from 106 to 100 copies/μL were 10-fold serially diluted in Elution buffer used as standard positive controls. The sensitivity of the duplex qPCR was compared with conventional PCR.
The specificity of the established duplex qPCR assay was assessed by using the DNA or RNA of above-mentioned six porcine viruses as templates and ddH2O as the negative control, which was subjected to duplex qPCR to detect PEDV and PCV3.
2.7. Reproducibility test for duplex qPCR
The standard samples were selected for the duplex qPCR reaction. Each dilution was subjected to three parallel tests under the same amplification conditions, and the coefficient of variation was calculated to evaluate the repeatability of intra batch detection. The recombinant plasmids pMD-PEDV and pMD-PCV3 were constructed out of three positive samples from different regions. After dilution, duplex qPCR was used to detect the coefficient of variation, and the repeatability of inter batch detection was evaluated.
2.8. Application to clinical samples
Sixty-six clinical samples (the intestinal tissues and fecal) obtained from 66 piglets with diarrhea were tested using the established one-step duplex RT-qPCR method. For clinical samples, single qPCR and duplex qPCR methods were performed as the above described methods using the DNA or RNA from the samples, and compared with the results of previously reported qPCR assays for PEDV [38] and PCV3 [33]. The detection rates of the qPCR methods were compared, and all of the positive clinical samples detected by the duplex qPCR assay were confirmed by DNA sequencing.
3. Results
3.1. Optimization of duplex qPCR
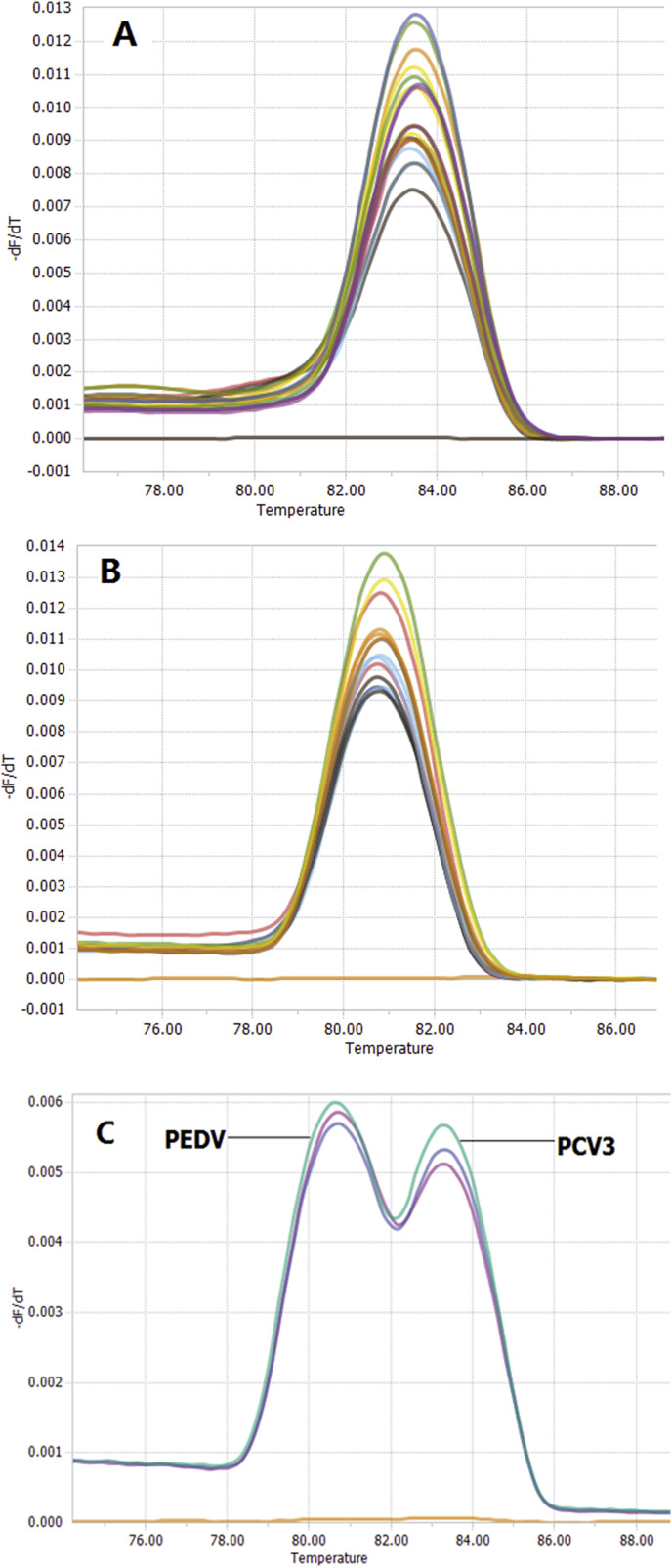
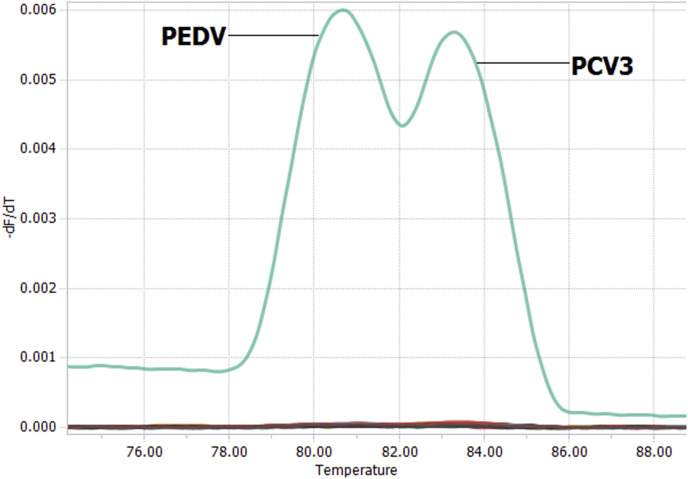
The fluorescent signals were detected for PEDV or PCV3 using single qPCR (Fig. 1 A and B), respectively. For the simultaneous detection of PEDV and PCV3 in a single reaction, the concentration of the primers and the annealing temperature were optimized to contain the highest fluorescence and the lowest threshold cycle. The final optimized primer concentration (0.25 μM of each primer for PEDV and PCV3, respectively) and the annealing temperature (60 °C for PEDV and PCV3, respectively) showed that two fluorescent signals could be detected simultaneously by the duplex qPCR assay. Hence, melting curve analysis revealed that the PEDV melting curve exhibited a Tm of 80.5 °C, whereas the PCV3 curve displayed a Tm of 83.5 °C (Fig. 1C), showing PEDV and PCV3 amplicons can easily be distinguished by specific Tm values due to the different lengths and compositions of two amplicons.
Fig. 1.
Melting curve analysis for the SYBR Green I based qPCR. A and B, single qPCR for PEDV and PCV3; C, duplex qPCR for PEDV and PCV3.
3.2. Sensitivity and specificity of duplex qPCR
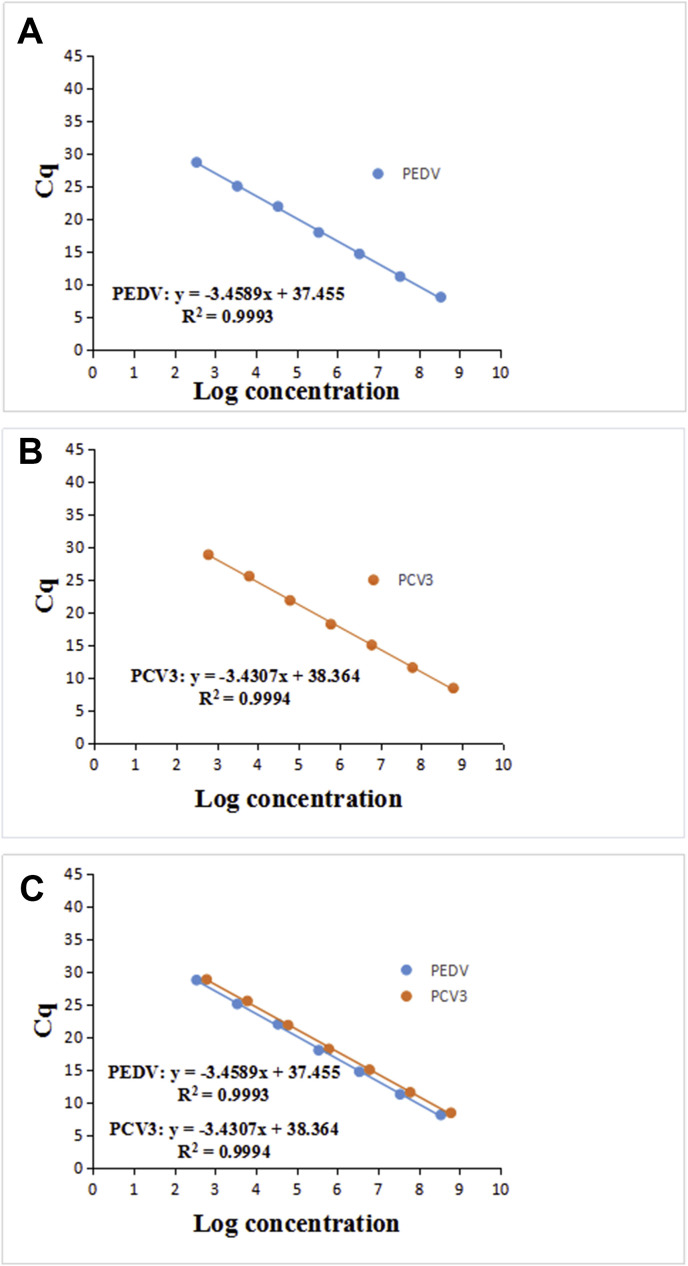
To determine the detection limit and efficiency of the assay, the standard plasmids for PEDV and PCV3 were used as a template, and 10-fold serially diluted with distilled water up to 10−10 (3.46 × 100–3.46 × 109 copies/μL for PEDV and 6.12 × 100–6.12 × 109 copies/μL for PCV3). A standard curve for PEDV and/or PCV3 was generated by plotting the logarithm of the plasmid copy number against the measured Cq values, respectively, and the standard curves had a wide dynamic range of 102–108 copies/μL (Fig. 2 A, Fig. 2B and C), which resulted in the quantification cycle (Cq) values ranging from 6.36 to 34.39 cycles and 6.42 to 33.64 cycles for PEDV and PCV3, respectively, and the standard curves were determined to be y = −3.4589x+37.455 and −3.4307x+38.364 for PEDV and PCV3, with linear correlation (R2) values (square of the correlation coefficient) of 0.99 between the Ct value and the logarithm of the plasmid copy number and reaction efficiency of 94.58% and 95.65%, respectively. When the standard plasmids for PEDV and PCV3 were diluted to 10−9, fluorescence was detected; no reaction was detected at 10−10. The limits of detection (LODs) of this assay were 3.46 × 101 copies/μL and 6.12 × 101 copies/μL for PEDV and PCV3, respectively, which were 100 times more sensitive than those of the conventional PCR (3.46 × 103 copies/μL and 6.12 × 103 copies/μL for PEDV and PCV3, respectively).
Fig. 2.
Standard curves of SYBR Green I -based qPCR generated by plotting the mean Cq values versus 10-fold serial dilutions of the standards over concentrations ranging from 3.46 × 102 to 3.46 × 108 copies/μL for PEDV and 6.12 × 102 to 6.12 × 108 copies/μL for PCV3 in triplicate. The coefficient of determination (R2) and the equation of the regression curve (y) were calculated using a LightCycler® 96 Instrument (Roche). A and B, single qPCR for PEDV and PCV3; C, duplex qPCR for PEDV and PCV3.
The duplex qPCR assay was specific for PEDV and PCV3. In the assay, the fluorescence signals of PEDV and PCV3 were obvious, and specific melting peaks were observed at 80.5 °C and 83.5 °C for PEDV and PCV3, respectively, whereas no specific melting peaks were found for other porcine viruses or the ultrapure water (Fig. 3 ), demonstrating the high specificity of the established duplex qPCR for detection of PEDV and PCV3.
Fig. 3.
Melting curve analysis of the specificity of the duplex SYBR Green I based. qPCR assay. No cross-reactions were detected with porcine circovirus type 2, porcine transmissible gastroenteritis virus, porcine reproductive and respiratory syndrome virus, classical swine fever virus porcine pseudorabies virus, porcine parvovirus and ddH2O.
3.3. Reproducibility of the assay
The reproducibility of the assay was determined by testing three 10-fold serial dilutions of the standard plasmids (3.46 × 104 copies/μL–3.46 × 106 copies/μL for PEDV and 6.12 × 104 copies/μL–6.12 × 106 copies/μL for PCV3) in triplicate using inter- and intra-assay comparisons. For the standard plasmids, the values of the intra-assay co-efficient of variation (CV) ranged from 0.3492% to 0.5583% for PEDV, and the values of the inter-assay CV ranged from 0.1399% to 0.5172% for PCV3 (data not shown). These data indicated that the assay was repeatable and reproducible with low variability.
3.4. Evaluation of duplex qPCR using clinical samples
Among the 66 field samples collected at different time intervals from piglets exhibiting signs of diarrhea, these clinical samples were strongly positive for PEDV and PCV3, with Cq values from 15.04 to 33.43 and from 14.77 to 31.64, respectively. Twenty-nine of 66 samples were positive for PEDV via duplex qPCR, 11 samples were positive for PCV3, and 18 samples were positive for both PEDV and PCV3, which were in accordance with those of the single qPCR assay (Table 2 ). The singular infection rates of PEDV and PCV3 were 43.94% (29/66) and 16.67% (11/66), respectively, and the co-infection rate of PEDV and PCV3 was 27.27% (18/66). The detection rates of PEDV and PCV3 were 71.21% (47/66) and 43.94% (29/66), which are similar to those of Zhou's PEDV qPCR method or Chen's PCV3 qPCR method, respectively. Besides, the singular infection rates of PEDV and PCV3 were 42.42% (28/66) and 15.15% (10/66) using conventional PCR, respectively, and the rate of PEDV and PCV3 co-infection was 24.24% (16/66). The detection rates of PEDV and PCV3 were 66.67% (44/66) and 39.39% (26/66). These results demonstrated that duplex qPCR is applicable for the differential diagnosis of PEDV and PCV3 in field samples, with high sensitivity and specificity.
Table 2.
Comparison of results detected for clinical samples by duplex qPCR and previously reported qPCR assays for PEDV and PCV3.
| Method | Quantity | Results |
Detection rate (%) | |||
|---|---|---|---|---|---|---|
| PEDV | PCV3 | PEDV/PCV3 | Negative | |||
| Duplex qPCR (PEDV/PCV3) | 66 | 29 | 11 | 18 | 8 | 87.88 |
| Single qPCR (PEDV) | 66 | 29 | - | 18 | 19 | 71.21 |
| Single qPCR (PCV3) | 66 | - | 11 | 18 | 37 | 43.94 |
| Zhou's qPCR (PEDV) | 66 | 29 | - | 18 | 19 | 71.21 |
| Chen's qPCR (PCV3) | 66 | - | 11 | 18 | 37 | 43.94 |
| Conventional PCR (PEDV) | 66 | 28 | - | 16 | 22 | 66.67 |
| Conventional PCR (PCV3) | 66 | - | 10 | 16 | 40 | 39.39 |
Note:In this study, the PEDV primers target a 163bp sequence in ORF1. The sensitivity of this assay may be lower than assays targeting other viral genes, such as M and N protein. We compared used a published method (by Zhou et al.) to confirm the positivity of PEDV in clinical samples. However, in the Zhou's method, the primers target PEDV M gene, and the detection rate for PEDV are similar to that of Zhou's PEDV qPCR method. The result suggested that compared with M or N genes, primers designed in ORF1 region may affect the sensitivity of the test, but PEDV primers designed in highly conservative region will not have a significant impact on the positive rate of clinical samples.
4. Discussion
Since the late 1980s, PED has existed throughout China, resulting in huge economic losses to the pig industry [8,43]. As a novel swine virus, PCV3 was reported to be widely circulating throughout the USA, China, South Korea, Poland, Brazil, Italy, and the UK. Previous studies have shown that PCV3 may be linked to diseased pigs with PDNS, reproductive failure, cardiac and multi-systemic inflammation, skin diseases, and porcine respiratory disease complex [20,22,44,45]. However, two additional studies have detected PCV3 in samples obtained from piglets affected with diarrhea. Wang et al. reported that out of 145 archived tissue samples collected from diseased pigs with diarrhea, PMWS, reproductive disorders, respiratory disorders, and PDNS for the detection of PCV3, the PCV3 DNA detection rate was 60.87% (28/46), 20% (12/60), 10% (2/20), 0% (0/16), and 66.67% (2/3), respectively [30]. Moreover, the results showed that the PCV3 detection rate in diarrhea samples was higher than that in the other sample types. Another study analyzed 35 diarrheal weaned pig samples and 35 non-diarrheal weaned pig samples for the presence of PCV3, and found that the rate of PCV3 positivity in the diarrheal samples was 17.14% (6/35), significantly higher than that of the non-diarrhea samples (2.86%, 1/35) [31]. These findings suggest that PCV3 may have a potential association with diarrhea. According to these findings, it is likely that a single PCV3 infection or PEDV and PCV3 co-infection could cause diarrhea in pig populations; however, the virus has not been isolated from these populations and studies investigating pathogenesis using the isolated virus have not been conducted. Thus, a rapid and reliable diagnostic assay is required for the simultaneous detection of PEDV and PCV3 in clinical samples for epidemiological and clinical studies, as well as the development of effective control strategies for PEDV and PCV3 infections.
To date, several approaches have been developed for the laboratory diagnosis of PEDV, including immunohistochemistry [46], enzyme-linked immunosorbent assay (ELISA) [47], RT-PCR [48], reverse transcription loop-mediated isothermal amplification (RT-LAMP) [49], a nanoparticle-assisted PCR assay [50,51], and real-time RT-PCR [35,36]. Similarly, immunohistochemical detection, PCR, and qPCR have all been developed for the detection of PCV3 [20,33,34,37]. However, ELISAs and immunohistochemistry are time-consuming techniques with a relatively low specificity and sensitivity. Thus, they cannot be used to estimate the amount of viral DNA/cDNA copies, which can be an important factor for viral diagnosis via correlations with viremia or clinical signs. On the other hand, qPCR has been widely used as a diagnostic method for its apparent advantages over conventional PCR due to its high sensitivity and specificity, allowing for high-throughput screening and the quantification of viral loads using small volumes. Therefore, the objective of this study was to develop a SYBR Green I-based duplex qPCR assay for the simultaneous detection of PEDV and PCV3.
Previous studies reported amplicons can be distinguished by the Tm of PCR products from melt curves, which is determined by the reduction in relative fluorescence as all dsDNA are denatured to their single-stranded form, and the shape and peak location of the melt curve are functions of the GC/AT ratio, length, and sequence of the fragment [52,53]. In this study, the 163-bp PEDV ORF1 gene or 136-bp PCV3 rep gene was successfully amplified using the single qPCR assay with PEDV- or PCV3-specific primers. Furthermore, the duplex qPCR assay with two sets of PEDV- and PCV3-specific primers simultaneously detected PEDV and PCV3 DNA in a single reaction tube, with similar sensitivity to the corresponding single qPCR assay (Fig. 2). The LOD of the duplex qPCR assay was 34.6 copies/μL for PEDV, which is comparable to those of previously reported qPCR assays [35,54]. The LOD of the duplex qPCR assay was 61.2 copies/μL for PCV3, which is a little higher than that of previously reported Taqman-based qPCR assays [37,55]. These data were 100 times more sensitive than those of conventional PCR. These results indicate the duplex qPCR assay had good sensitivity to detect PEDV and PCV3 in clinical samples.
The efficiency of the duplex qPCR assay was investigated using clinical samples. In this study, the total DNA and RNA were extracted from clinical samples using a commercially available kit (Beijing Zoman Biotechnology Co., Ltd., China). For a sample infected with DNA virus and RNA virus, the conventional nucleic acid extraction method is usually to extract separately, which is time-consuming. In this method, virus genome DNA/RNA nucleic acid extraction kit can be used to extract virus DNA and RNA from samples at the same time, and save time on the basis of cost saving. However, the efficiency of this kit extract RNA and DNA simultaneously may slightly affect the sensitivity of the assay. The nucleic acid content is a little lower than that of separate extraction, but it does not affect the amplification. In this study, we found the sensitivity of PCV3 amplification increased by approximately 1–3 Cq value, whereas the sensitivity of PEDV amplification was not different compared with conventional fluorescence quantitative PCR. Chumakov studies suggest that in the reverse transcriptase stage, reverse transcriptase can lead to the production of primer dimers, which may be the result of DNA-dependent DNA polymerase activity [56]. The reason may be that DNA nucleic acid participates in the first 45 °C 5 min reaction in one-step duplex reaction procedure, which reduces the activity of some DNA. However, the positive rate can be determined by melting peak. Although there was no dimer in two-step RT-PCR, the two-step method must be avoided in clinical virus detection, because it not only increases the workload and prolongs the detection time, but also greatly increases the probability of cross-contamination. Anyhow, the establishment of this method can provide a reference for the establishment of multiple DNA and RNA methods for other pathogens.
Among 66 intestinal tissue and fecal samples, the detection rates of PEDV and PCV3 using duplex qPCR were higher than those of conventional PCR (Table 2), and all samples positive for PEDV or PCV3 in the conventional PCR assay were also positive in the qPCR assay. In contrast, some samples that were negative for PEDV or PCV3 using the conventional PCR assay were found to be positive with the qPCR assay. Furthermore, these samples that were positive for PEDV or PCV3 in the qPCR assay but negative in the conventional PCR assay were confirmed to be positive by DNA sequencing. These results suggest that the conventional PCR assay does not have sufficient sensitivity to detect PEDV or PCV3 in samples with low viral loads, whereas the duplex qPCR assay is better suited to detect low levels of virus and perform a quantitative analysis of the viral load. In addition, the detection rates using duplex qPCR were 71.21% (47/66) for PEDV, which is in accordance with previous studies that reported detection rates of 61.10%–78.49% in China [8]. In this study, the PEDV primers target a 163bp sequence in ORF1. Since coronaviruses generate a set of subgenomic RNAs (coding structural and accessory genes) during the replication in the cells, the sensitivity of this assay may be lower than assays targeting other viral genes, such as M and N protein. However, in this study the positivity of PEDV was confirmed in clinical samples using a published method (by Zhou et al.), and the detection rate for PEDV was similar to that of Zhou's PEDV qPCR method. The result suggested that compared with M or N genes, primers designed in ORF1 region may affect the sensitivity of the test, but PEDV primers designed in highly conservative region will not have a significant impact on the positive rate of clinical samples. Moreover, the detection rate for PCV3 using duplex qPCR was 43.94% (29/66), lower than that reported in diarrheal samples by Wang et al. [30], but higher than that reported in diarrheal weaned pigs by Zhai et al. [31]. This further supports the high sensitivity and specificity of duplex qPCR, and its applicability for the differential diagnosis of PEDV and PCV3 in field samples. This showed that the newly developed duplex qPCR assay for clinical diagnosis has a significant advantage, as PEDV and PCV3 can be amplified simultaneously in a single reaction mixture. Again, it is efficient, simple, quick to detect PEDV and PCV3 nucleic acid, and reduces the cost of diagnosis.
The method has high application value. At present, many suspected cases of DNA virus and RNA virus mixed infection or RNA viruses mixed infection often appear in clinical samples. When it is uncertain which virus is causing the infection, one-step method can be used to diagnose the infection or both simultaneously, so as to achieve the dual purpose of diagnosis and differentiation. In addition, compared with conventional methods, this method is faster and more convenient, and can generally complete the whole detection process, and the results were consistent with those by using a single method, which showed that the stability of the method was better. The model can be extended to the differential diagnosis of other mixed infections with viruses, which has a positive significance in promoting the application of multiple technologies in veterinary medicine.
Since PCV3 was identified as an emerging swine virus in the US, the co-infection of PCV3 with other pathogens (i.e., PCV2 and PRRSV), has been widely reported [31,37,57,58]. In the present study, PCV3 and PEDV nucleic acids were detected in intestinal tissue and fecal samples of piglets suffering from diarrhea using duplex qPCR. These results demonstrated that both singular and co-infections with PEDV and PCV3 exist in the pig population in China. These findings suggest that PCV3 might have a potentially pathogenic association with diarrhea at the tested pig farms; however, there is not enough data to directly confirm that PEDV and PCV3 are relevant in the context of swine diarrhea. Thus, further investigations should be conducted to ascertain the role of PCV3 in pigs affected with diarrhea and monitor the epidemic status of PCV3.
In summary, the present study described the development of a duplex qPCR assay that can sensitively and simultaneously detect the presence of PEDV and PCV3. Therefore, these findings support the use of this technique as a rapid and reliable surveillance method for PEDV and PCV3 infections.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgement
This work was funded by the Henan Province Research Cooperation Projects (152107000003) and Henan Province Education Department Science and Tchnology Specific Projects (18A230007).
References
- 1.Lee J.H., Park J.S., Lee S.W., Hwang S.Y., Young B.E., Choi H.J. Porcine epidemic diarrhea virus infection: inhibition by polysaccharide from Ginkgo biloba exocarp and mode of its action. Virus Res. 2015;195:148–152. doi: 10.1016/j.virusres.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Gene. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin. Exp. Vaccine. Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford K., Lager K.M., Kulshreshtha V., Miller L.C., Faaberg K.S. Status of vaccines for porcine epidemic diarrhea virus in the United States and Canada. Virus Res. 2016;226:108–116. doi: 10.1016/j.virusres.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Stadler J., Zoels S., Fux R., Hanke D., Pohlmann A., Blome S. Emergence of porcine epidemic diarrhea virus in southern Germany. BMC Vet. Res. 2015;11:142. doi: 10.1186/s12917-015-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Liu X., Shi D., Shi H., Zhang X., Feng L. Complete genome sequence of a porcine epidemic diarrhea virus variant. J. Virol. 2012;86:3408. doi: 10.1128/JVI.07150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D.Q., Ge J.W., Qiao X.Y., Jiang Y.P., Liu S.M., Li Y.J. High-level mucosal and systemic immune responses induced by oral administration with Lactobacillus-expressed porcine epidemic diarrhea virus (PEDV) S1 region combined with Lactobacillus-expressed N protein. Appl. Microbiol. Biotechnol. 2012;93:2437–2446. doi: 10.1007/s00253-011-3734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 12.D M., Q W., MR C., KD R., A R., J C. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014.%A Vlasova AN. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leyi W., Beverly B., Yan Z. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pensaert M.B., Martelli P. Porcine epidemic diarrhea: a retrospect from Europe and matters of debate. Virus Res. 2016;226:1–6. doi: 10.1016/j.virusres.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mankertz A., Caliskan R., Hattermann K., Hillenbrand B., Kurzendoerfer P., Mueller B. Molecular biology of Porcine circovirus: analyses of gene expression and viral replication. Vet. Microbiol. 2004;98:81–88. doi: 10.1016/j.vetmic.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Allan G., Krakowka S., Ellis J., Charreyre C. Discovery and evolving history of two genetically related but phenotypically different viruses, porcine circoviruses 1 and 2. Virus Res. 2012;164:4–9. doi: 10.1016/j.virusres.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Opriessnig T., McKeown N.E., Zhou E.M., Meng X.J., Halbur P.G. Genetic and experimental comparison of porcine circovirus type 2 (PCV2) isolates from cases with and without PCV2-associated lesions provides evidence for differences in virulence. J. Gen. Virol. 2006;87:2923–2932. doi: 10.1099/vir.0.82099-0. [DOI] [PubMed] [Google Scholar]
- 19.Opriessnig T., Meng X.J., Halbur P.G. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Invest. 2007;19:591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 20.Palinski R., Pineyro P., Shang P., Yuan F., Guo R., Fang Y. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J. Virol. 2017;91 doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan S., Ku X., Chen F., Wang Y., Yu X., He Q. Complete genome sequence of a novel porcine circovirus type 3 strain, PCV3/CN/Hubei-618/2016, isolated from China. Genome Announc. 2017;5 doi: 10.1128/genomeA.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ku X., Chen F., Li P., Wang Y., Yu X., Fan S. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg. Dis. 2017;64:703–708. doi: 10.1111/tbed.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng S., Wu X., Zhang L., Xin C., Liu Y., Shi J. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg. Dis. 2017;64:1337–1341. doi: 10.1111/tbed.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen H., Liu X., Zhang P., Wang L., Liu Y., Zhang L. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg. Dis. 2018;65:264–266. doi: 10.1111/tbed.12639. [DOI] [PubMed] [Google Scholar]
- 25.Collins P.J., McKillen J., Allan G. Porcine circovirus type 3 in the UK. Vet. Rec. 2017;181:599. doi: 10.1136/vr.j5505. [DOI] [PubMed] [Google Scholar]
- 26.Faccini S., Barbieri I., Gilioli A., Sala G., Gibelli L.R., Moreno A. Detection and genetic characterization of Porcine circovirus type 3 in Italy. Transbound Emerg. Dis. 2017;64:1661–1664. doi: 10.1111/tbed.12714. [DOI] [PubMed] [Google Scholar]
- 27.Kwon T., Yoo S.J., Park C.K., Lyoo Y.S. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet. Microbiol. 2017;207:178–180. doi: 10.1016/j.vetmic.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Stadejek T., Wozniak A., Milek D., Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg. Dis. 2017;64:1350–1353. doi: 10.1111/tbed.12672. [DOI] [PubMed] [Google Scholar]
- 29.Tochetto C., Lima D.A., Varela A.P.M., Loiko M.R., Paim W.P., Scheffer C.M. Full-Genome Sequence of Porcine Circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound Emerg. Dis. 2018;65:5–9. doi: 10.1111/tbed.12735. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Zhang Y., Zhang R., Han Q., Wang J., Liu L. Recombinase polymerase amplification assay for rapid detection of porcine circovirus 3. Mol. Cell. Probes. 2017;36:58–61. doi: 10.1016/j.mcp.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Zhai S.L., Zhou X., Zhang H., Hause B.M., Lin T., Liu R. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol. J. 2017;14:222. doi: 10.1186/s12985-017-0892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu P.L., Zhang Y., Zhao Y., Zheng H.H., Han H.Y., Zhang H.X. Detection and phylogenetic analysis of porcine circovirus type 3 in central China. Transbound Emerg. Dis. 2018;65:1163–1169. doi: 10.1111/tbed.12920. [DOI] [PubMed] [Google Scholar]
- 33.Chen G.H., Tang X.Y., Sun Y., Zhou L., Li D., Bai Y. Development of a SYBR green-based real-time quantitative PCR assay to detect PCV3 in pigs. J. Virol. Methods. 2018;251:129–132. doi: 10.1016/j.jviromet.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Wang J., Zhang Y., Wang J., Liu L., Pang X., Yuan W. Development of a TaqMan-based real-time PCR assay for the specific detection of porcine circovirus 3. J. Virol. Methods. 2017;248:177–180. doi: 10.1016/j.jviromet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Miller L.C., Crawford K.K., Lager K.M., Kellner S.G., Brockmeier S.L. Evaluation of two real-time polymerase chain reaction assays for Porcine epidemic diarrhea virus (PEDV) to assess PEDV transmission in growing pigs. J. Vet. Diagn. Invest. 2016;28:20–29. doi: 10.1177/1040638715621949. [DOI] [PubMed] [Google Scholar]
- 36.Wang L., Zhang Y., Byrum B. Development and evaluation of a duplex real-time RT-PCR for detection and differentiation of virulent and variant strains of porcine epidemic diarrhea viruses from the United States. J. Virol. Methods. 2014;207:154–157. doi: 10.1016/j.jviromet.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.R., Park Y.R., Lim D.R., Park M.J., Park J.Y., Kim S.H. Multiplex real-time polymerase chain reaction for the differential detection of porcine circovirus 2 and 3. J. Virol. Methods. 2017;250:11–16. doi: 10.1016/j.jviromet.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Zhou X., Zhang T., Song D., Huang T., Peng Q., Chen Y. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol. Cell. Probes. 2017;33:36–41. doi: 10.1016/j.mcp.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Hong T., Wu J.Y., Shang Y.J., Yan C., Liu X.T. The development of a rapid SYBR one step real-time RT-PCR for detection of porcine reproductive and respiratory syndrome virus. Virol. J. 2010;7:90. doi: 10.1186/1743-422X-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z.Z., Fang W.H., Habib M. vol. 53. 2010. First results of detection of PRRSV and CSFV RNA by SYBR Green I-based quantitative PCR; pp. 461–467. (Zoonoses & Public Health). [DOI] [PubMed] [Google Scholar]
- 42.Parida M., Shukla J., Sharma S., Ranghia Santhosh S., Ravi V., Mani R. Development and evaluation of reverse transcription loop-mediated isothermal amplification assay for rapid and real-time detection of the swine-origin influenza A H1N1 virus. J. Mol. Diagn. 2011;13:100–107. doi: 10.1016/j.jmoldx.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao X., Li Z., Zeng X., Zhang G., Niu J., Sun B. Sequence analysis of the spike gene of Porcine epidemic diarrhea virus isolated from South China during 2011-2015. J. Vet. Sci. 2017;18:237–243. doi: 10.4142/jvs.2017.18.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phan T.G., Giannitti F., Rossow S., Marthaler D., Knutson T.P., Li L. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016;13:184. doi: 10.1186/s12985-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen H., Liu X., Zhang P., Wang L., Liu Y., Zhang L. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg. Dis. 2018 Feb;65(1):264–266. doi: 10.1111/tbed.12639. [DOI] [PubMed] [Google Scholar]
- 46.Guscetti F., Bernasconi C., Tobler K., Van Reeth K., Pospischil A., Ackermann M. Immunohistochemical detection of porcine epidemic diarrhea virus compared to other methods. Clin. Diagn. Lab. Immunol. 1998;5:412–414. doi: 10.1128/cdli.5.3.412-414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodak L., Valicek L., Smid B., Nevorankova Z. An ELISA optimized for porcine epidemic diarrhoea virus detection in faeces. Vet. Microbiol. 2005;105:9–17. doi: 10.1016/j.vetmic.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishikawa K., Sekiguchi H., Ogino T., Suzuki S. Direct and rapid detection of porcine epidemic diarrhea virus by RT-PCR. J. Virol. Methods. 1997;69:191–195. doi: 10.1016/s0166-0934(97)00157-2. [DOI] [PubMed] [Google Scholar]
- 49.Ren X., Li P. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of porcine epidemic diarrhea virus. Virus Gene. 2011;42:229–235. doi: 10.1007/s11262-011-0570-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing N., Guan X., An B., Cui B., Wang Z., Wang X. Ultrasensitive detection of porcine epidemic diarrhea virus from fecal samples using functionalized nanoparticles. PLoS One. 2016;11:e0167325. doi: 10.1371/journal.pone.0167325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan W., Li Y., Li P., Song Q., Li L., Sun J. Development of a nanoparticle-assisted PCR assay for detection of porcine epidemic diarrhea virus. J. Virol. Methods. 2015;220:18–20. doi: 10.1016/j.jviromet.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ririe K.M., Rasmussen R.P., Wittwer C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 53.Wehrle E., Didier A., Moravek M., Dietrich R., Martlbauer E. Detection of Bacillus cereus with enteropathogenic potential by multiplex real-time PCR based on SYBR Green I. Mol. Cell. Probes. 2010;24:124–130. doi: 10.1016/j.mcp.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Masuda T., Tsuchiaka S., Ashiba T., Yamasato H., Fukunari K., Omatsu T. Development of one-step real-time reverse transcriptase-PCR-based assays for the rapid and simultaneous detection of four viruses causing porcine diarrhea. Jpn. J. Vet. Res. 2016;64:5–14. [PubMed] [Google Scholar]
- 55.Li X., Qiao M., Sun M., Tian K. A duplex real-time PCR assay for the simultaneous detection of porcine circovirus 2 and circovirus 3. Virol. Sin. 2018;33:181–186. doi: 10.1007/s12250-018-0025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chumakov K.M. Reverse transcriptase can inhibit PCR and stimulate primer-dimer formation. PCR Methods Appl. 1994;4:62–64. doi: 10.1101/gr.4.1.62. [DOI] [PubMed] [Google Scholar]
- 57.Chen G.H., Mai K.J., Zhou L., Wu R.T., Tang X.Y., Wu J.L. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound Emerg. Dis. 2017;64:1650–1654. doi: 10.1111/tbed.12702. [DOI] [PubMed] [Google Scholar]
- 58.Fu X., Fang B., Ma J., Liu Y., Bu D., Zhou P. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound Emerg. Dis. 2018;65:e296–e303. doi: 10.1111/tbed.12752. [DOI] [PubMed] [Google Scholar]