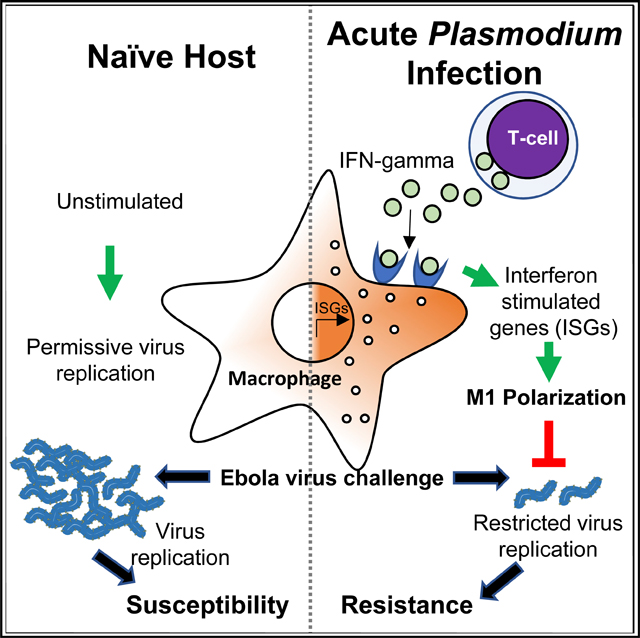
SUMMARY
During the 2013–2016 Ebola virus (EBOV) epidemic, a significant number of patients admitted to Ebola treatment units were co-infected with Plasmodium falciparum, a predominant agent of malaria. However, there is no consensus on how malaria impacts EBOV infection. The effect of acute Plasmodium infection on EBOV challenge was investigated using mouse-adapted EBOV and a biosafety level 2 (BSL-2) model virus. We demonstrate that acute Plasmodium infection protects from lethal viral challenge, dependent upon interferon gamma (IFN-γ) elicited as a result of parasite infection. Plasmodium-infected mice lacking the IFN-γ receptor are not protected. Ex vivo incubation of naive human or mouse macrophages with sera from acutely parasitemic rodents or macaques programs a proinflammatory phenotype dependent on IFN-γ and renders cells resistant to EBOV infection. We conclude that acute Plasmodium infection can safeguard against EBOV by the production of protective IFN-γ. These findings have implications for anti-malaria therapies administered during episodic EBOV outbreaks in Africa.
In Brief
Rogers et al. demonstrate that acute Plasmodium infection protects against lethal Ebola virus challenge. Protection is conferred by Plasmodium-elicited interferon gamma (IFN-γ) that causes M1 polarization of tissue macrophages. These studies provide insight into conflicting clinical data regarding whether malaria protects or sensitizes hosts to Ebola virus.
Graphical Abstract
INTRODUCTION
Ebola virus (EBOV) is a negative-strand RNA virus that triggers severe hemorrhagic fever in primates (Feldmann and Geisbert, 2011). EBOV outbreaks, identified sporadically in Central Africa since 1976, cause severe morbidity and mortality, with case fatalities as high as 90% (Weyer et al., 2015). In late 2013, EBOV was detected for the first time in Western Africa, resulting in the largest outbreak in history with 28,600 confirmed cases of EBOV disease (EVD) and a fatality rate of ~40% (Coltart et al., 2017). Given the magnitude of the West African outbreak, multiple epidemiological studies have identified factors that were positively or negatively associated with EVD outcomes. One study reached the conclusion that EBOV patients co-infected with Plasmodium falciparum, the principal causative agent of malaria in Africa, were less likely to succumb to EVD, with a survival rates 30% higher in patients with high parasite burdens (Rosenke et al., 2016). In contrast, several additional studies report that Plasmodium infection either had no effect on EVD outcomes in pediatric patients or was associated with exacerbated EVD (Smit et al., 2017; Vernet et al., 2017; Waxman et al., 2017). The basis for these disparate conclusions and whether specific host immunity against Plasmodium affects either EBOV infection susceptibility or the course of EVD are not fully understood.
The host immune response to Plasmodium infection is, in part, governed by the complex life cycle of the parasite and involves multiple tissues (Crompton et al., 2014). The parasite is transmitted into the skin by female Anopheline mosquito inoculation of motile sporozoites that results in infection of hepatocytes. These parasites undergo clinically silent differentiation and cell division without triggering robust host cellular immunity. Merozoites are ultimately released from infected hepatocytes and enter a cycle of invasion and asexual replication in red blood cells (RBCs), which elicits the clinical symptoms associated with malarial disease. An initial Plasmodium blood-stage infection often stimulates a pro-inflammatory, febrile illness that is associated with elevated interleukin-1 b (IL-1 b), IL-6, IL-12, interferon gamma (IFN-γ), and tumor necrosis factor (TNF) production (Angulo and Fresno, 2002). In individuals who have either resolved acute febrile illness or have been exposed to repeated Plasmodium infections, the immune response shifts to an immunomodulatory profile characterized by IL-10 and transforming growth factor b (TGF-b) production (Portugal et al., 2014). This temporally regulated and functionally dichotomous immune response to Plasmodium blood-stage infection provides a possible explanation for the resistance of a subset of Plasmodium-infected individuals to EVD and the lack of consensus in the published epidemiological studies of EBOV outbreaks.
Here, we used rodent and non-human-primate models of Plasmodium, mouse-adapted EBOV (Mayinga) (ma-EBOV), and a biosafety level 2 (BSL-2) model virus of EBOV to demonstrate that acute Plasmodium blood-stage infection protects against EBOV infection and disease. We show that protection conferred by Plasmodium is dependent upon IFN-γ production, with loss of IFN-γ signaling abrogating protection.
RESULTS
Acute Plasmodium Infection Protects Mice from Lethal ma-EBOV or Recombinant Vesicular Stomatitis Virus (rVSV)/EBOV Glycoprotein (GP) Challenge
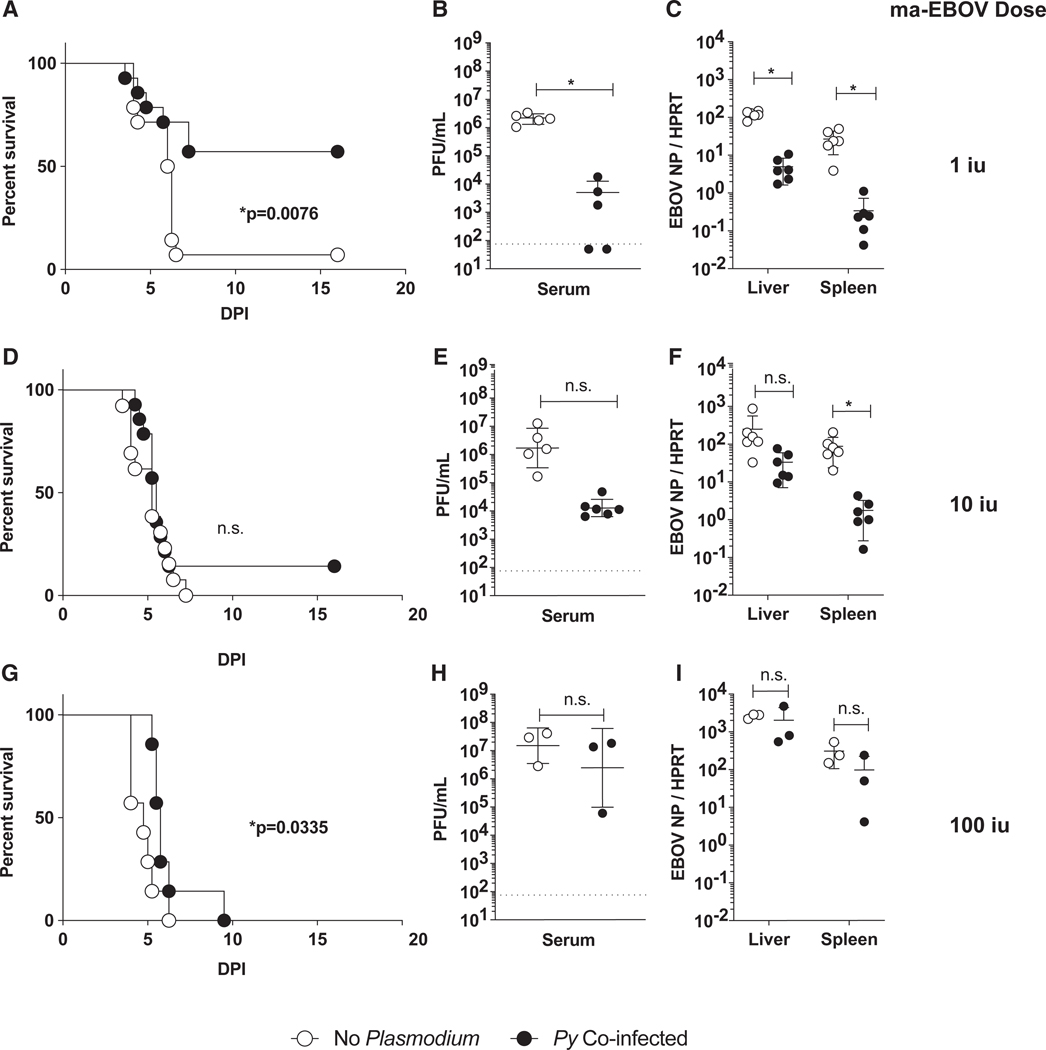
To evaluate the effect of an acute Plasmodium infection on subsequent EBOV infection, BALB/c mice were intravenously (i.v.) administered RBCs (iRBCs) infected with Plasmodium yoelii (17XNL) (Py), a rodent Plasmodium parasite species that models hyperparasitemia and severe malarial anemia (Li et al., 2001). Six days after Py infection, mice were challenged intraperitoneally (i.p.) in a biosafety level 4 (BSL-4) facility with ma-EBOV at a range of infectious doses. At a low, but lethal, dose of 1 infectious unit (iu) of ma-EBOV, Py-infected mice exhibited reduced morbidity (Figures S1A and S1B) and mortality (Figure 1A). Consistent with decreased EVD in Py-infected mice, serum and organs harvested from the co-infected animals at day 3 of infection showed a 3-log decrease in viral titers (Figure 1B) and a 1- to 2-log reduction in viral load in the spleen and liver (Figure 1C). Notably, the amount of ma-EBOV administered to mice was critical, as higher doses of virus overcame the protective effect of Plasmodium in vivo (Figures 1D–1I), although virus load also trended lower in the Py-infected mice administered 10 iu of ma-EBOV.
Figure 1. Acute Plasmodium Infection in Mice Protects against EBOV Challenge.
(A-I) Female BALB/c mice were infected with Plasmodium yoelii (Py) (1 × 106 infected red blood cells [iRBCs]), challenged i.p. 6 days later with 1, 10, or 100 iu mouse-adapted Ebola (ma-EBOV) and assessed for clinical signs and survival daily.
(A, D, and G) Survival curves are shown (n = 14/group in A and D; n = 7/group in G).
(B, E, and H) Day 3 serum titers from individual mice (n = 3–6) are expressed as mean ± SD.
(C, F, and I) Day 3 viral loads in liver and spleen from individual mice (n = 4–6 in B, C, E, and F; n = 3 in H and I) are expressed as mean ± SD.
Experiments in this figure were performed two independent times, and data are pooled (A-F) or once (G-I). For all experiments: n.s., not significant; iu, infectious units; and *p < 0.05.
Also see Figure S1.
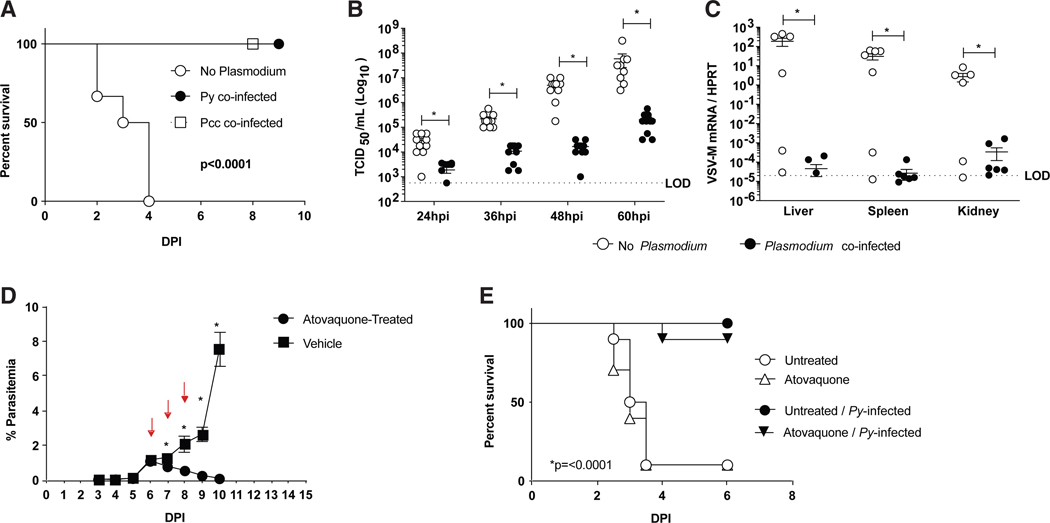
To explore the mechanisms by which Py can protect against ma-EBOV challenge, we used a BSL-2 model of EBOV that challenges IFN-αβ-receptor knockout (Ifnar−/−) mice with a BSL-2 recombinant rVSV engineered to express the EBOV GP (rVSV/ EBOV GP) and green fluorescent protein (GFP) in place of the native GP of VSV. This recombinant virus models EBOV tropism and host cell entry (Takada et al., 1997; Côte et al., 2011; Kondratowicz et al., 2011; Panchal et al., 2014; Wec et al., 2016). Furthermore, infection of Ifnar−/− mice with this virus models the effective control of type I IFN responses by EBOV and the in vivo and ex vivo sensitivity of EBOV to pro-inflammatory and anti-inflammatory cytokines (Takada et al., 1997; Côte et al., 2011; Kondratowicz et al., 2011; Panchal et al., 2014; Rhein et al., 2015; Wec et al., 2016; Rogers et al., 2019). Ifnar−/− mice were infected with either Py or another rodent Plasmodium species, P. chabaudi chabaudi (AS) (Pcc), which is a model of low-grade parasitemia and persistent malaria (Stephens et al., 2012). When challenged 6 days after establishing either Py or Pcc infection, mice were protected from an otherwise lethal dose of rVSV/EBOV GP (Figure 2A). In parallel, we found that Ifnar−/− mice were not protected from rVSV/EBOV GP when given RBCs from naive mice or irradiated Py iRBCs, as previously described (Schmidt et al., 2010) (Figure S2).
Figure 2. Acute Plasmodium Infection of Mice Protects against rVSV/EBOV GP Challenge.
(A) C57BL/6 Ifnar−/− mice were infected with Plasmodium yoelii (Py) or chabaudi (Pcc) (1 × 106 iRBCs) and challenged with rVSV/EBOV GP i.p. 6 days later. Survival curves are shown and analyzed by log-rank (Mantel-Cox) test (n = 12 in no-Plasmodium group; n = 11 in Py-co-infected group; n = 8 in Pcc-co-infected group).
(B and C) C57BU6 Ifnar−/− mice were infected with Py or left untreated and challenged with rVSV/EBOV GP i.p. 6 days later. Each point represents a single sample from an individual mouse.
(B) Serum titers at 12-h intervals assessed as tissue culture infectious dose (TCID50)per mL on Vero cells.
(C) Viral load in organs at 60 hours post infection (hpi), as assessed by qRT-PCR.
(D and E) C57BL/6 Ifnar−/− micewere infected with Py or left untreated. Atovaquonewas administered to both treatment groups on days 6–8 post-Py (red arrows indicate treatments). Parasitemia was monitored daily (n = 10/group).
(D) Mice were infected i.p. with rVSV/EBOV GP on day 9 post-Py when the parasite was eradicated in atovaquone-treated mice.
(E) Mice were monitored for survival (n = 10/group).
For all experiments: *p < 0.05. LOD, limit of detection. Error is expressed as mean ± SEM.
Also see Figures S2 and S3.
Because both Py and Pcc rendered mice resistant to rVSV/EBOV GP pathogenesis, we conducted subsequent studies by using Py, as it more faithfully recapitulates severe acute malarial anemia that would be observed following a primary Plasmodium exposure in malaria-naive humans in that it is cleared by the host and does not recrudesce, unlike Pcc. Experiments showed that co-infected mice had ~15- to 260-fold lower viremia over the first 60 h of virus infection than mice solely infected with rVSV/EBOV GP (Figure 2B). Similarly, virus loads in the livers, spleens, and kidneys at 60 h of infection were 103- to 106-fold lower in the Plasmodium-infected mice than in non-parasitemic mice, although viral burden was variable in some Plasmodium-naive mice (Figure 2C). The low-to-undetectable levels of virus in the peripheral tissues of parasitemic mice at 60 h of infection suggested that virus failed to spread beyond the peritoneal cavity in Py-infected animals. At 21 days after virus challenge, half of co-infected mice produced greater than 100 mg/ml of anti-EBOV GP antibodies, immunoglobulin (Ig) concentrations that we previously showed to be protective against subsequent ma-EBOV challenge (Figure S3) (Lennemann et al., 2017). Less than 1 mg/ml of anti-EBOV GP Ig was detected in some of rVSV/EBOV GP-challenged mice, suggesting either that virus titers were insufficient to elicit a significant humoral response in these animals or Py infection interfered with anti-EBOV GP antibody production. Together, these data show that an active blood-stage Plasmodium infection protects mice from challenge with lethal doses of either ma-EBOV or rVSV/EBOV GP. Importantly, the rVSV/EBOV GP experimental system using an Ifnar−/− host excludes a role for type 1 IFN signaling in the protection conferred by Plasmodium infection.
To determine whether parasite infection was directly facilitating protection, we infected mice with Py and, on days 6–8 post-inoculation (p.i.), administered the anti-malarial drug atovaquone to parasitemic and naive control mice. Parasitemia was quantified daily, and mice were challenged with rVSV/EBOV GP upon successful eradication of Py (Figure 2D). Strikingly, Py-infected mice treated with atovaquone remained protected from rVSV/EBOV GP, suggesting that the protection against lethal virus challenge mediated by acute Py infection was more likely to be due to the host immune response rather than the presence of the parasite per se (Figure 2E). Notably, atovaquone treatment did not impact the disease course in Plasmodium naive mice.
Peritoneal Macrophages Are Crucial for Plasmodium-Elicited Protection from rVSV/EBOV GP
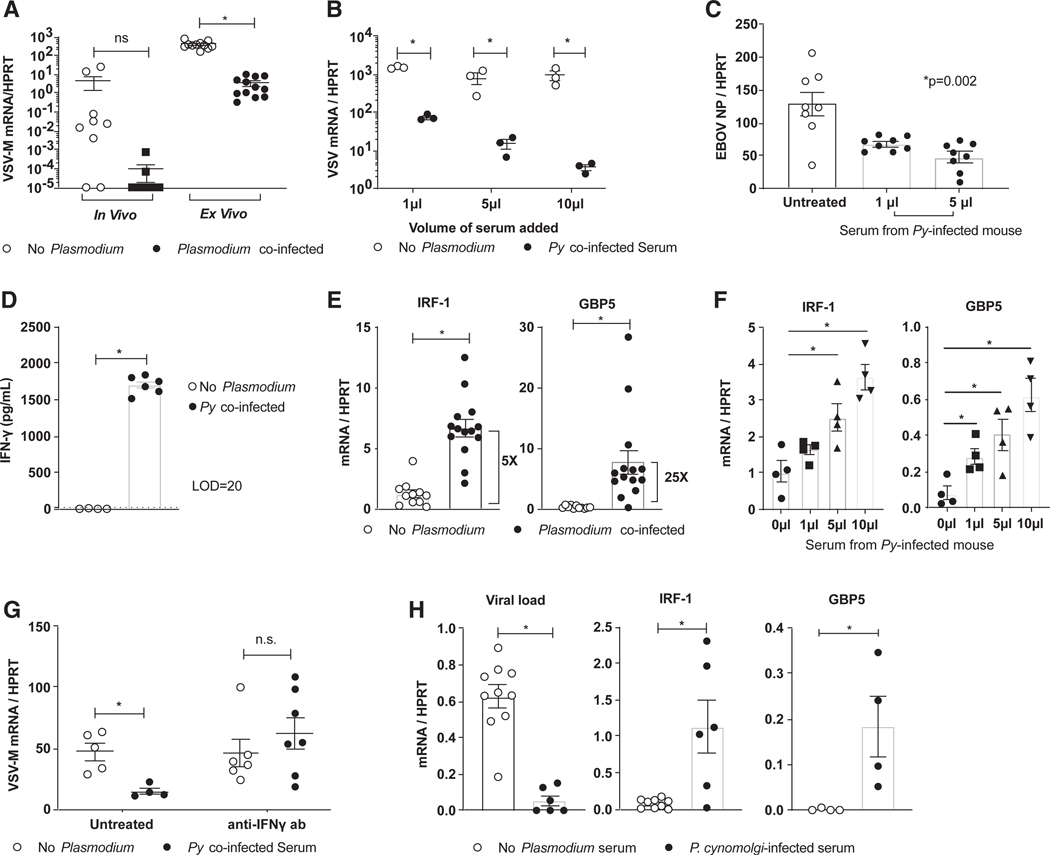
EBOV is primarily a myeloid cell-tropic pathogen (Connolly et al., 1999; Geisbert et al., 2003; Bray and Geisbert, 2005; Rogers and Maury, 2018), and peritoneal macrophages (pmacs) are the primary cell population initially infected during i.p. administration of maEBOV or rVSV/EBOV GP (Mahanty et al., 2003; Rhein et al., 2015; Rogers et al., 2019). To address whether acute experimental malaria impacts virus infection of these critical target cells, Py-infected and naive mice were challenged i.p. with rVSV/EBOV GP. Peritoneal cells were harvested at 24 h and assessed for viral load. Although most naive mice showed high levels of viral RNA in peritoneal cells, virus was largely undetectable in cells recovered from Py-infected mice (Figure 3A, left). Consistent with these in vivo data, ex vivo virus challenge of pmacs that were obtained from Py-infected mice and enriched by adherence showed a ~30-fold decrease in viral load at 24 h of infection compared with pmacs obtained from naive mice (Figure 3A, right). These data support that in vivo Py infection restricts viral infection of a primary cell target of EBOV, pmacs, despite the fact that Py merozoites do not infect macrophages but specifically target RBCs.
Figure 3. Peritoneal Macrophages Are Critical Mediators of Plasmodium-Elicited Protection from rVSV/EBOV GP.
(A-G) C57BL/6 Ifnar−/− mice were inoculated with 1 × 106 Py iRBCs or left untreated, and all experiments were performed 6 days after Py was administered.
(A) For in vivo infections, mice were challenged with a lethal dose of rVSV/EBOV GP, pmacswere harvested at 24 hpi, and viral replication was assessed by qRT-PCR (in vivo). For ex vivo infections, pmacs were harvested from mice, infected with rVSV/EBOV GP (multiplicity ofinfection [MOI] = 1), and assessed by qRT-PCR at 24 hpi (ex vivo).
(B) Pmacs from naive mice were harvested, treated for 24 h with serum from either naive mice or Py-infected mice, and infected with rVSV/EBOV GP (MOI = 1). Viral loads were assessed 24 hpi.
(C) Pmacs were treated with serum from Py-infected mice and infected with wild-type (WT) EBOV under BSL-4 conditions. Infection was quantified by qRT-PCR for EBOV NP gene expression at 24 hpi.
(D) IFN-γ levels in serum on day 6 were assessed by ELISA.
(E and F) M1 polarization markers were assessed in untreated or Py-infected serum exposed pmacs by qRT-PCR (E). A dose-response curve of Py-infected serum added to pmacs (F).
(G) Pmacs exposed to Py-infected serum in the presence or absence of anti-IFN-γ antibody for 24 h and then infected with rVSV/EBOV GP (MOI = 1). Infection was assessed by qRT-PCR 24 hpi.
(H) Human MDMs were incubated with serum from Cynomolgous macaques infected with Plasmodium cynomolgi. Cells were challenged with rVSV/EBOV GP (MOI = 5), and infection was quantified at 24 hpi by qRT-PCR. Uninfected pmacs treated with serum from naive or P. cynomolgi-infected macaques were assessed for M1 polarization markers by qRT-PCR.
For all experiments: *p < 0.05. Error is expressed as mean ± SEM.
Also see Figure S4.
The studies with an anti-malarial agent demonstrating that mice cleared of Py-infected RBCs remained protected against virus challenge raised the possibility that the host inflammatory responses to blood-stage Plasmodium infection may program resistance to ma-EBOV and rVSV/EBOV GP infection. To test this, we incubated pmacs from naive donors with dose titrations of serum from either Py-infected or naive mice for 24 h and then challenged the cells with wild-type EBOV or rVSV/EBOV GP. Serum from Py-infected animals protected pmacs from virus infection in a dose-dependent manner (Figures 3B and 3C).
We previously showed that IFN-γ can protect macrophages from EBOV infection (Rhein et al., 2015). Thus, we postulated that Plasmodium-induced IFN-γ is mechanistically linked to Plasmodium-infection-induced protection against EBOV and rVSV/EBOV GP challenge that was observed both in vivo and ex vivo. In support of this, serum levels of IFN-γ were markedly elevated as early as 4 days after Py infection in Ifnar−/− and wild-type mice, peaking at day 6, whereas no detectable IFN-γ was observed in naive serum (Figure 3D; Figure S4A). Inline with these elevated serum IFN-γ levels, transcriptional changes in pmacs recovered from Py-infected mice exhibited a proinflammatory macrophage M1 phenotype characterized by enhanced expression of the well-established IFN stimulated genes (ISGs) IRF1 and GBP5 (Figure 3E). Similarly, the addition of serum from Py-infected donors stimulated dose-dependent increases in IRF1 and GPB5 expression in pmacs harvested from naive mice (Figure 3F). Importantly, the ex vivo protective effects of serum from Py-infected donors was abolished by neutralizing IFN-γ (Figure 3G).
To extend these studies to a model that more directly approximates human Plasmodium infection, we next evaluated the capacity of serum obtained from rhesus macaques acutely infected with Plasmodium cynomolgi (Collins et al., 1999) to program virus resistance in human monocyte-derived macrophages (MDMs), as many human and non-human primate (NHP) cytokines are known to cross react (Scheerlinck, 1999). Consistent with the results from our rodent studies, viral replication was 20- to 30-fold lower in the MDMs exposed to serum from acutely Plasmodium-infected macaques (Figure 3H, left). Serum exposure also elevated M1 markers associated with IFN-γ signaling (Figure 3H, right). Collectively, these data suggest that IFN-γ produced in response to acute Plasmodium infection protects human and mouse macrophages from EBOV infection.
Plasmodium-Elicited IFN-γ Production Is Necessary for In Vivo Protection from rVSV/EBOV GP
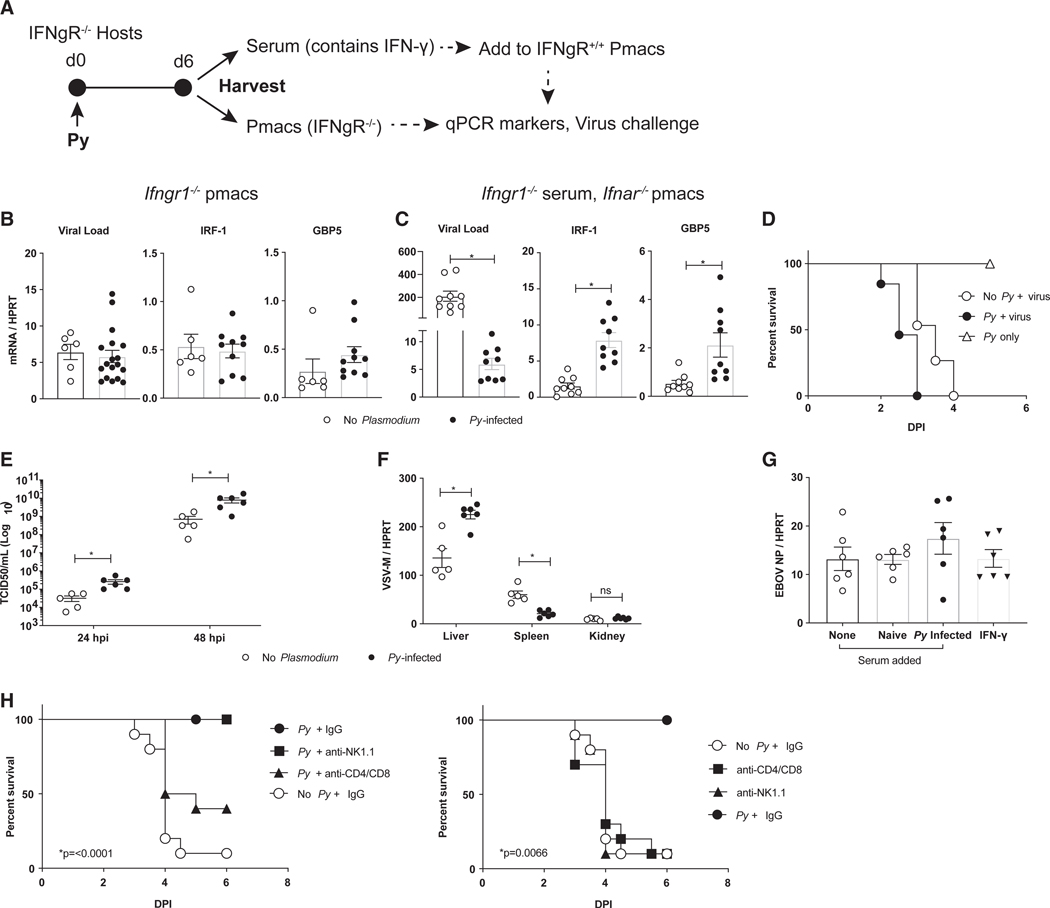
We next sought to determine if IFN-γ signaling was responsible for the in vivo protection mediated by Py infection. To test this, we infected IFN-γ-receptor-null (Ifngr1−/−) mice with Py and isolated serum and pmacs 6 days after establishing blood-stage Plasmodium infection (Figure 4A). The pmacs of Py-infected and naive Ifngr1−/− mice showed no difference in levels of infection upon rVSV/EBOV GP challenge and exhibited no evidence of M1 polarization (Figure 4B). However, Ifngr1−/− mice retained the capacity to secrete IFN-γ, as Ifnar−/− pmacs exposed to the serum of Py-infected Ifngr1−/− animals exhibited reduced viral loads and expressed the M1-polarized ISGs IRF-1 and GBP5 (Figure 4C). To evaluate the role of IFN-γ signaling in vivo, we generated and infected Ifnar−/−Ifngr1−/− mice with Plasmodium. Double-knockout mice were used, as VSV-based recombinant viruses are not virulent in mice with intact IFNAR signaling due to an inability to antagonize type I IFN (Müller et al., 1994). We found that Ifnar−/−Ifngr1−/− mice produced robust levels of IFN-γ when exposed to Py (Figure S4B) but were not protected from rVSV/EBOV GP challenge (Figure 4D). Although parasitemia during early Py and Pcc infection of Ifngr1−/− mice has been shown to be elevated compare to wild-type mice (Su and Stevenson, 2000), the Py parasite burden of Ifnar−/−Ifngr1−/− mice did not alter their survival if these mice were not challenged with virus (Figure 4D). Virus infection of these mice resulted in higher systemic viremia at 24 h and 48 h than virus-challenged naive Ifnar−/−Ifngr1−/− mice (Figure 4E) and higher viral loads in the liver but lower loads in the spleen (Figure 4F), suggesting that, in the absence of type II IFN signaling, acute Py infection enhances overall virus load. Moreover, neither Py-infected mouse serum nor recombinant IFN-γ protected Ifngr1−/− pmacs against challenge with the BSL-4 EBOV (Mayinga) (Figure 4G).
Figure 4. Plasmodium-Dependent Protection against EBOV and rVSV/EBOV GP Requires IFN-γ/IFN-γR Interactions.
(A-C) Pmacs from Ifngr1−/− mice are not protected against rVSV/EBOV GP. Ifngrl−/− mice were inoculated with 1 × 106 Py iRBCs or left untreated, and 6 days later, sera and pmacs were harvested and cells were infected ex vivo.
(A) A schematic of the experiment.
(B) Parasitemic or naive Ifngr1−/− pmacs were infected ex vivo with rVSV/EBOV GP (MOI = 5), and infection was quantified at 24 hpi or left uninfected and assessed for expression of M1 markers.
(C) Serum from Ifngr1−/− micewas added to Ifnar−/− pmacs for 24 h. Cells were either infected ex vivo with rVSV/EBOV GP (MOI = 1) with infection quantified at 24 hpi by qRT-PCR or left uninfected and assessed for expression of M1 markers.
(D-F) Ifnar/Ifngr1−/− mice were inoculated with either 1 × 106 Py iRBCs or left untreated and 6 days later challenged i.p. with a lethal dose of rVSV/EBOV GP.
(D) Survival curves following virus challenge (n = 15 in no Py + virus group; n = 13 in Py + virus group; n = 3 in Py-only group).
(E) Serum virus titers at 24 and 48 hpi.
(F) Viral loads in organs harvested 48 hpi.
(G) Ifngr1−/− pmacs were treated as shown and infected with WT EBOV under BSL-4 conditions, and infection was quantified by qRT-PCR for EBOV NP gene expression at 24 hpi.
(H) C57BL/6Ifnar−/− mice were infected with Py (1 × 106 iRBCs), treated with 200 μg of the indicated antibodies 24 hpi, and challenged with rVSV/EBOV GP i.p. on day 6 post-infection. The x axis represents days following virus challenge. Survival was monitored (n = 10/group). Controls are included as a separate panel (right) to facilitate interpretation.
For all experiments: *p < 0.05. Error is expressed as mean ± SEM.
Finally, we sought to determine the cells responsible for IFN-γ production due to Py infection. As it has been reported that both natural killer (NK) cells and T cells are the source of IFN-γ (De Souza et al., 1997; Villegas-Mendez et al., 2012; King and Lamb, 2015), we performed cell-depletion studies and found thatT cells, but not NK cells, are critical for Py-mediated protection from rVSV/EBOV GP (Figure 4H). Together, these data support that IFN-γ and its interactions with the IFN-γ receptor are critical for Plasmodium-mediated protection against EBOV infection.
Plasmodium-Elicited Protection from rVSV/EBOV GP Wanes over Time
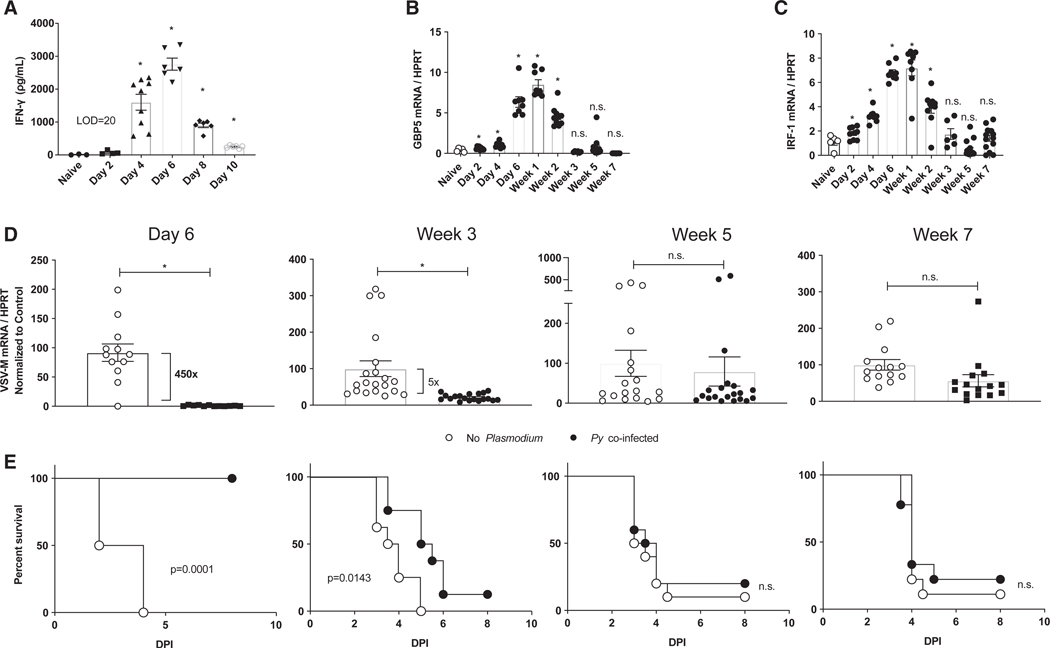
Given that the host response to acute blood-stage Plasmodium infection evolves from pro-inflammatory to immunomodulatory after resolution of infection, we evaluated the durability of Py-infection-induced protection against rVSV/EBOV GP challenge. IFN-γ in the serum of Py-infected Ifnar−/− mice peaked on day 6 p.i. and fell below the limit of detection after day 10 p.i. (Figure 5A). In contrast, M1 markers on pmacs recovered from Py-infected mice remained upregulated after IFN-γ was no longer detectable in peripheral blood (Figures 5B and 5C). To probe the relationship between IFN-γ-induced M1 pmac polarization and protection against virus challenge, we isolated pmacs at weeks 1,3,5, and 7 following Py infection and challenged the cells with rVSV/EBOV GP ex vivo. In parallel experiments designed to evaluate the durability of in vivo protection, we also challenged Py-infected mice at weeks 1,3,5, and 7 following Py infection with rVSV/EBOV GP. Strikingly, both Py-infected mice and pmacs harvested from Py-infected mice were protected from virus challenge for 3 weeks, but protection waned by 5 weeks after Py infection (Figures 5D and 5E). Thus, the in vivo and ex vivo consequences of M1 polarization due to experimental Py infection of the Ifnar−/− host persist for at least 3 weeks. Notably, administration of a single high dose of recombinant IFN-γ only protected mice when administered within 24 h of rVSV/EBOV challenge (Figure S5A), suggesting that either persistent, sub-patent Plasmodium infection and/or sustained, low-grade IFN-γ production imprints more strongly on macrophages than a single bolus of recombinant cytokine. Indeed, the protective effects of recombinant IFN-γ were abrogated when mice were challenged with a 10- to 100-fold higher dose of rVSV/ EBOV GP (Figure S5B). Similarly, IFN-γ did not provide protection to pmacs challenged with a 10-fold higher dose of wild-type EBOV (Figure S5C). These data are consistent with our in vivo ma-EBOV findings (Figure 1) and recent reports from others (Rosenke et al., 2018) showing that experimental malaria fails to modulate the course of EVD when mice are challenged with large doses (eg, 100 lethal dose 50 [LD50]) of ma-EBOV.
Figure 5. Durability of Plasmodium-Mediated Protection to rVSV/EBOV GP.
For all experiments, C57BL/6 Ifnar−/− mice were inoculated with 1 × 106 Py iRBCs or left untreated. All experiments were performed at the indicated time after Py infection.
(A) Serum was harvested and IFN-γ levels were quantified by ELISA.
(B and C) Pmacs were harvested, RNA was isolated, and expression of GBP5 (B) and IRF-1 (C) was quantified by qRT-PCR.
(D) Pmacs were harvested at times noted and infected ex vivo with rVSV/EBOV GP (MOI = 1). Virus infection was quantified 24 hpi by qRT-PCR.
(E) Mice were challenged i.p. with a lethal dose of rVSV/EBOV GP (n = 8–10) at times noted in (D).
For all experiments: *p < 0.05. Error is expressed as mean ± SEM.
Also see Figure S5.
DISCUSSION
Our studies demonstrate that acute infection with Plasmodium transiently protects mice from EBOV challenge. Our findings in mice support the epidemiological study by Rosenke et al. (2016) that observed that during the 2013–2016 EBOV outbreak, overt Plasmodium infection was associated with protection against disease symptoms and fatalities associated with EBOV infection. We elucidated the mechanism by which Plasmodium facilitates protection against EBOV, identifying that Plasmodium infection stimulates IFN-γ production, with serum levels peaking at day 6 following Plasmodium infection. This pro-inflammatory environment polarized tissue macrophages, resulting in inhibition of virus infection. Eliminating IFN-γ/IFNgR interactions abrogated the protection conferred by Plasmodium against rVSV/EBOV GP, strengthening the mechanistic link. We previously reported that IFN-γ administration abrogates virus infection of pmacs and protects mice from an otherwise lethal challenge with ma-EBOV or rVSV/EBOV GP (Rhein et al., 2015), which is consistent with our ex vivo BSL-4 data demonstrating a dependence on IFN-γ production. The data presented here further implicate IFN-γ as a potential EBOV antiviral as a therapeutic strategy at early times of virus infection.
Our findings demonstrate that acute Plasmodium infection generates sufficient IFN-γ to protect against a low, but uniformly lethal, dose of EBOV; however, we found that it is not protective against higher EBOV doses. The inability of Py infection to overcome higher doses of EBOV is consistent with an earlier report that showed that Py infection does not alter morbidity or mortality associated with the administration of 100 LD50 of EBOV to mice (Rosenke et al., 2018). Recombinant IFN-γ administration also protected against low doses, but not high doses, of rVSV/ EBOV GP in Ifnar−/− mice, in a manner similar to protection conferred by acute Py in ma-EBOV studies. These studies provide supporting evidence that our BSL-2 model virus serves as an appropriate model system for dissecting these mechanisms of protection against EVD. Acceptance of BSL-2 models overcomes space and cost restrictions associated with BSL-4 studies.
An important question that bears on the relevance of our studies and others is what is the dose of EBOV to which individuals are likely to be exposed? Furthermore, what dose of virus is required for an individual to become viremic and manifest symptoms? Although answers to these questions are not known, as we show here, the use of a low but predictably lethal dose in an animal model can provide important insight and understanding into the ability of the immune system to successfully overcome these highly virulent infections under some conditions. Additionally, such doses may more accurately reflect exposure within the field.
Some epidemiological studies from the 2013–2016 outbreak found that co-infection with Plasmodium enhanced EVD (Smit et al., 2017; Vernet et al., 2017; Waxman et al., 2017). We propose that these conclusions were drawn from studies that lack adequate stratification of malaria-infected patients based on cytokine profiles. It is well-established that immune responses to acute versus chronic (or repeated) Plasmodium infections differ. Although an acute Plasmodium challenge of a naive individual is known to elicit a strong type 1 immune response, subsequent or chronic infection of an individual shifts the immune response to an anti-inflammatory type 2 response (Angulo and Fresno, 2002). Thus, many parasitemic patients in endemic regions have sub-clinical infections with elevated levels of TGF-b and IL-10 and an absence of pro-inflammatory cytokines (Portugal et al., 2014). As a consequence, the presence or absence of parasites in the blood of patients entering an Ebola treatment unit (ETU) may be an insufficient clinical marker to predict the severity and clinical course of EVD. Instead, serum cytokine profiles may better serve as a predictor of outcome. Hence, our studies suggest that it would be valuable for clinical protocols in ETUs to distinguish Plasmodium-infected patients by their cytokine profile. Although such clinical tests may currently be difficult to perform in resource-poor regions, the current approach of treating all RBC Plasmodium-positive individuals in ETUs may reduce beneficial pro-inflammatory cytokines present at early times in some EBOV-infected patients.
To control for pre-exposure and Plasmodium infection timelines and data, we chose to work with two animal models of acute Plasmodium infection. The results of longitudinal serum draws from Py-infected mice challenged with rVSV/EBOV GP indicate that Py-infected mice become viremic, albeit at much lower levels than Py-naive mice, and that some of these mice develop a robust antibody response to EBOV GP following virus challenge. However, other mice in this treatment group produced few to no detectable anti-EBOV GP antibodies, suggesting that the virus antigens in those mice may have been insufficient to stimulate a humoral response or that Plasmodium co-infection blocked antibody development, as has been suggested by others (Muellenbeck et al., 2013; Scholzen and Sauerwein, 2013).
Evidence of EBOV GP antibody seropositivity in the absence of clinical disease provides potential insights into recent reports that have identified a surprising number of individuals in sub-Saharan Africa who have no known contact with EBOV and, yet, are seropositive for anti-EBOV antibodies. Recent estimates range from 0%−24% of patient populations (Mulangu et al., 2016; Bower and Glynn, 2017; Steffen et al., 2019). Given the pervasive presence of malaria in regions where EBOV is endemic, Plasmodium-elicited subclinical co-infections with EBOV may occur, resulting in a small percentage of seropositive individuals who may be protected against subsequent EBOV infection.
The effects of IFN-γ on macrophages, and the resulting M1 phenotype it induced, lasted several weeks. Although the peak production of IFN-γ occurred at day 6, M1 markers remained elevated for an additional week, with intermediate levels of protection in mice and macrophages observed up to 2 weeks later. This is of interest, as little has been done to investigate the durability of the phenotypes induced by polarizing agents in primary macrophages or in vivo. Furthermore, the absolute levels of IFN-γ achieved in the serum during Plasmodium infection are much lower than the dose we had previously administered directly to the peritoneal cavity to elicit protection (Rhein et al., 2015); yet, they were protective. This could indicate that lower levels of sustained IFN-γ have a long-lasting protective effect, allowing for lower doses administered and, thus, more desirable side effect profiles. Although it is difficult to compare the production of cytokines such as IFN-γ across taxa from mice to humans, numerous groups have shown that human patients produce large amounts of IFN-γ in response to Plasmodium infection, and the role of IFN-γ during clinical malaria is well characterized (Rhodes-Feuilletteet al., 1985; Medina et al., 2011; Perlazaet al., 2011; Nasr et al., 2014; King and Lamb, 2015). For instance, Medina et al. (2011) report a range of serum IFN-γ levels from 30 to 76 ng/ml in humans acutely infected with Plasmodium vivax.
Taken together, these data indicate that acute Plasmodium infection protects against EBOV infection in a mouse model by eliciting the production of IFN-γ. Furthermore, our rVSV/EBOV GP studies demonstrated that IFN-γ signaling through IFNgR is required for this protection. This leads to pro-inflammatory polarization of macrophages that restricts EBOV replication in key cell populations. Future studies to examine additional IFN-γ signaling requirements and downstream IFN-stimulated genes mediating IFN-γ effects would further our understanding of this protection. An earlier study demonstrated that Py infection protects mice against acute chikungunya virus viremia and pathology in an IFN-γ-dependent manner (Teoetal., 2018). This argues that Plasmodium-elicited proinflammatory responses may inhibit a variety of acute viral infections. In combination, these findings suggest that similar studies in virus-infected NHP models are warranted to determine if protection would translate to humans. Furthermore, re-evaluation of the current strategy of administration of anti-malarial therapeutics upon all parasitemic patients at ETUs may be warranted.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
No unique reagents were generated for this study. All materials and protocols are available upon request from the Lead Contact, Wendy Maury (wendy-maury@uiowa.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Wild-type BALB/cJ mice were purchased from The Jackson Laboratory (stock #000651). C57BL/6 IFN-α/β receptor-deficient (Ifnar−/−) were purchased from The Jackson Laboratory (stock #028288). Wild-type C57BL/6 mice were a kind gift from Dr. John Harty, University of Iowa. C57BL/6 Ifnar−/− Ifngr1−/− mice were generated by crossing C57BL/6 Ifnar−/− and C57BL/6 Ifngr1−/− mice (Ifngr1tm1Agt/J Jackson Labs stock #003288). Genotyping was performed using primers and standard PCR conditions from Jackson labs. This study was conducted in strict accordance with the Animal Welfare Act and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The University of Iowa (UI) Institutional Assurance Number is #A3021-01. The Emory University Institutional Assurance Number is A3180-01. The Texas Biomedical Research Institute (TBRI) Institutional Assurance Number is A3082-01. All mouse procedures performed at the UI were approved by the UI Institutional Animal Care and Use Committee (IACUC) which oversees the administration of the IACUC protocols and the study was performed in accordance with the IACUC guidelines (Protocol #8011280). All mouse procedures performed at TBRI were approved by their Institutional Animal Care and Use Committee (IACUC) which oversees the administration of the IACUC protocols and the study was performed in accordance with the IACUC guidelines (Protocol #1645MU). All NHP procedures were performed at the Yerkes National Primate Research Center (YNPRC), an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) international-certified institution. All NHP procedures were reviewed and approved by Emory University’s IACUC (Protocol #Y2003225).
Nonhuman primate Plasmodium cynomolgi infections
NHP sera used in this study came from NHPs that were experimentally infected with P. cynomolgi at the Yerkes National Primate Research Center (YNPRC), an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) international-certified institution. All procedures were reviewed and approved by Emory University’s IACUC, and all NHPs were socially housed in pairs during infections. All housing was in accordance with Animal Welfare Act regulations as well as the Guide for the Care and Use of Laboratory Animals. A detailed description of the experimental design, infections, clinical data, etc., for the samples that were used in this manuscript were previously described in Joyner etal. (2016). Briefly, five malaria-naive, male, rhesus macaques (Macaca mulatta) of Indian origin were infected with 2,000 P. cynomolgi M/B strain sporozoites and parasitemia monitored daily by light microscopy up to 100 days post-inoculation. Plasma was isolated from blood collections prior to inoculation (i.e., Pre-Infection) and during acute, primary infections after performing a Lymphoprep PBMC isolation according to the manufacturer’s suggested protocol. All samples were aliquoted and stored at −80°C until needed for experiments.
rVSV/EBOV GP stocks
Recombinant vesicular stomatitis virus encoding and expressing the glycoprotein from EBOV (Mayinga) was generated as previously described (Ebiharaetal., 2007). Virus was propagated by infecting Vero cells at low MOI (~0.05) and collecting supernatantsat 48hpi. The resulting supernatants were filtered through a 45 micron filter and purified by ultra-centrifugation (28,000 g, 4°C, 2 hr) through a 20% sucrose cushion. The resulting stocks were resuspended in a small volume of PBS and those used for in vivo studies were further purified by treatment with an endotoxin removal kit (Detoxi-Gel Endotoxin Removing Gel, ThermoFisher Scientific 20339) before being aliquoted and stored at −80°C until use. All viral titers were determined by TCID50 assay on Vero cells.
WT EBOV stocks
All experiments with replication-competent EBOV were performed in the Animal Biosafely Level 4 (ABSL4) laboratory at the TBRI (San Antonio, TX). The wild-type EBOV and mouse-adapted EBOV (ma-EBOV), both variant Mayinga (NCBI accession numbers NC_001608 and AF_499101, respectively), were obtained from the virus repository at TBRI. EBOV stocks were generated and characterized as previously described (Bray et al., 1998; Shtanko et al., 2018).
Plasmodium infections and parasite quantification
Plasmodium yoelii (clone 17XNL, obtained from MR4, ATCC) and Plasmodium chabaudi chabaudi AS(Pyr1) (clone MRA-747, obtained from beiresources) infections were initiated by a serial transfer of 106 parasitized red blood cells (pRBCs) i.v. Control RBCs were isolated from naive C57BL/6 mice aged 6–8 weeks, 106 RBCs were transferred. To inactive iRBCs, 5mLof iRBCs were irradiated with 200 Gy by cesium irradiation. Parasitemia was measured using flow cytometry as previously described (Malleret et al., 2011). Briefly, 1 mL of blood was obtained by milking the tail vain and mixed to 100uL of PBS. The cells were spun down and resuspended in 100 mL of staining cocktail containing Hoechst-34580 (1:1000, Thermo Fisher Scientific), Dihydroethidium (1:500, Thermo Fisher Scientific), CD45.2-FITC (1:200, clone:104, Biolegend) and TER-119-APC (1:400, clone: TER-119, Tonbo) and incubated for 30 minutes at 4°C. The cells were washed with PBS and spun down at 200 x g for 5 minutes and run on FACSVerse after resuspending in 100mL of PBS. Percentage RBCs staining positive for Hoechst 34580 and Dihydroethidium indicative of parasitized cells were calculated using FlowJo software (Treestar Inc.).
METHOD DETAILS
BSL-4 mouse experiments
Study #1 consisted of 6 groups of 10 five-week-old female BALB/c mice. On day −6 relative to ma-EBOV exposure, 3 groups of mice were injected with 106 Plasmodium yoelii-infected RBCs as described above. Subsequently, parasite-exposed and naive animals were transferred to the ABSL4 for acclimation. On day −1, blood was collected from a submandibular vein of a subset of mice in each group to confirm parasitemia. On day 0, groups were challenged with either 1, 10, or 100 plaque-forming units (PFU) of ma-EBOV (as accessed on Vero cells) via the intraperitoneal route. Animals were observed at least twice daily for morbidity and mortality for 18 days after virus challenge. Group clinical scores were recorded as the sum of all clinical observations for the group. If a clinical score of > 12 was reached, the animal was considered “terminally ill” and euthanized. On day 3, three animals from each group were euthanized, and blood, liver and spleen were collected to determine viremia and virus loads. The viral titers in the blood was determined after serum isolation, using the neutral red plaque assay, and virus load in tissues by qRT-PCR as described below. The remaining 7 mice in each group served to determine animal survival.
Study #2 consisted for 4 groups of 10 five-week-old female BALB/c mice. Protocols were identical to those described above, except only the two lower doses of ma-EBOV were used. Data from the two different 1 and 10 iu challenge studies were pooled for analysis.
BSL-2 mouse experiments
For in vivo infections with rVSV/EBOV GP, the lowest dose of virus providing consistent lethality was determined in C57BL/6 Ifnar−/− mice. This was found to be 102 iu for males and 5×102 iu for females. Virus was administered in 100 mL sterile PBS by intraperitoneal injection. For atovaquone treatments, drug was administered at 25 mg/kg by ip injection on days 6–8. For experiments with IFN-γ, drug was administered at the indicated dose and time suspended in 100 mL sterile PBS. For depletion studies, mice were given 200 mg of the indicated antibodies 24 hours after administration of Py.
Antibodies: anti-mouse NK1.1 BioXCell clone PK136, anti-mouse CD4 BioXCell clone YTS 191, anti-mouse CD8 alpha BioXCell clone 2.43, IgG controls Rat IgG2b BioXCell clone RG7/11.1.
Titer assay (BSL-2)
To obtain titers, serum samples were filtered through 45 mm filters and serially diluted before being added onto Vero cells. Titers were calculated as 50% tissue culture infectious dose per milliliter of serum (TCID50/mL) according to the Reed-Muench method (Reed and Muench, 1938).
Organ Harvest
Mice were anesthetized using isoflurane and perfused through the left ventricle with 10 mL cold sterile PBS prior to being euthanized by rapid cervical dislocation in accordance to our IACUC protocol. Organs were harvested and snap frozen in liquid nitrogen to preserve virus. Organs were homogenized in 1mL TRizol regent using the “gentleMACS Dissociator” with M tubes (Miltenyi Biotec).
qRT-PCR
RNA was isolated using the TRIzol reagent from Invitrogen. All steps were performed according to the manufacturer’s specifications. RNA was subsequently converted to cDNA with the High Capacity cDNA RevTrans Kit (#4368814) from Applied Biosystems. A total of 1 mg of RNA was used as input for each reaction. Quantitative PCR was performed using POWER SYBR Green Master Mix (#4367659) from Applied Biosystems according to the manufacturer’s instructions and utilizing a 7300 real time PCR machine from Applied Biosystems. 20ng of cDNA were used in each well. Primers are available in Table S1.
ELISAs
Interferon gamma was detected using the “Mouse IFN-γ ELISA MAX” kit from Biolegend (#430805) in accordance with manufacturer’s instructions. Detection of anti-EBOV GP antibodies was performed by coating optical plates overnightwith soluble EBOV GP (50 ml/well at 10 mg/ml). Wells were washed 2x with PBST (0.015% Tween 20), blocked for 1 hour (PBS 2%BSA), washed 3x with PBST, and incubated overnight in 4°C with either a standard curve composed of fractionated mouse immunoglobulin (Immunore-agents #Mu-003-B) or serum samples. Following incubation wells were washed 4x with PBST and incubated with 50 mL rabbit anti-mouse IgG heavy and light chain-HRP (10 mg/ml Pierce #31457) for 1 hr at RT. Wells were then washed 5x with PBST, incubated with 50 mL of HRP substrate (BD OptEIA #555214). The reaction was stopped with 50 mL 2M H2SO4 and absorbance at 450nm was measured on Synergy H1 hybrid reader. Total concentration of anti-EBOV GP antibodies was quantified by comparison to the standard curve.
Serum isolation and ex vivo treatments
Whole blood was obtained by facial vein puncture in accordance with IACUC guidelines (https://animal.research.uiowa.edu/iacuc-guidelines-blood-collection). Serum was isolated by centrifugation (90 s, 8000 x g) in serum separator tubes (BD Microtainer #365967). Serum was passed through 45 mm filters prior to use. For ex vivo experiments, serum was either added directly to macrophages or pretreated for 15 minutes with 1 mL LEAF anti-mouse IFN-γ antibody (1mg/ml) (Clone AN-18, Biolegend #517903). Serum with or without antibody was left on macrophages for 24 hours and removed at the time of infection.
Macrophage isolations
Peritoneal cells were obtained from mice by peritoneal lavage with 10mLof RPMI + 1% pen/strep. For studies utilizing resident peritoneal macrophages, cells were washed once with PBS and resuspended in RPMI containing 10%FBS, 1% pen/strep, 1% non-essential amino acids (NEAA), and 1% sodium pyruvate. After 48 hours, cells were washed with PBS which removed most of the non-adherent cells. This generated a macrophage enriched population of cells.
Human macrophages were matured from monocytes obtained from leukocyte reduction cones containing peripheral blood from healthy donors at the DeGowin Blood Center at University of Iowa Hospitals and Clinics. Peripheral blood mononuclear cells were purified by Ficoll gradient, and monocytes were enriched by adherence to tissue culture flasks coated in 2% gelatin and pre-treated with human plasma. Following isolation, monocytes were plated in RPMI with 10%FBS, 1% pen/strep, 1% non-essential amino acids (NEAA), 1% sodium pyruvate, and 20ng/mL human MCSF. Cells were allowed to mature for 6 days at which point they were washed with PBS to remove non-adherent cells.
QUANTIFICATION AND STATISTICAL ANALYSIS
In vivo experiments: significance was defined as p < 0.05 given alpha = 0.05 using Log-rank (Mantel-Cox) test. N is indicated on the figure legends. Ex vivo experiments: significance was determined by two tailed unpaired Student’s t test (alpha = 0.05). For panels where multiple comparison were made, a line is shown to indicate the comparison made. Individual data points are shown. All statistics were calculated using GraphPad Prism 8 software (GraphPad Software, Inc.).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse NK1.1 | BioXCell | Cat: BE0036 |
| RRID: AB_1107737 | ||
| Clone: PK136 | ||
| Anti-mouse CD4 | BioXCell | Cat: BE0119 |
| RRID: AB_10950382 | ||
| Clone: YTS 191 | ||
| Anti-mouse CD8α | BioXCell | Cat: BE0061 |
| RRID: AB_1125541 | ||
| Clone: 2.43 | ||
| Rat IgG2b | BioXCell | Cat: BE0252 |
| RRID: AB_2687733 | ||
| Clone: RG7/11.1 | ||
| LEAF anti-mouse interferon gamma | Biolegend | Cat: 517903 |
| Clone: AN-18 | ||
| Bacterial and Virus Strains | ||
| rVSV/EBOV GP | Ebihara et al., 2007 | N/A |
| EBOV: Mayinga variant | Texas Biomedical Research Institute | GenBank: NC_001608 |
| Mouse-adapted EBOV: Mayinga variant | Texas Biomedical Research Institute | GenBank: AF_499101 |
| Biological Samples | ||
| Human monocyte derived macrophages | DeGowan Blood Center, Univ. Iowa | https://uihc.org/degowin-blood-center |
| Mouse (C57BL/6J) peritoneal macrophages | The Jackson Laboratory | Resident cells isolated from the peritoneal compartment |
| Mouse (C57BL/6J) Ifnar−/− peritoneal macrophages | The Jackson Laboratory | Resident cells isolated from the peritoneal compartment |
| Mouse (B6.129S7) Ifngr1tm1Agt/J peritoneal macrophages | The Jackson Laboratory | Resident cells isolated from the peritoneal compartment |
| Mouse (C57BL/6J) Ifnar/Ifngr−/− peritoneal macrophages | Bred in house | Resident cells isolated from the peritoneal compartment |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Atovaquone | Sigma-Aldrich | Cat: A7986 |
| Critical Commercial Assays | ||
| ELISA MAX Deluxe Set Mouse Interferon Gamma | Biolegend | Cat: 430805 |
| Detoxi-Gel Endotoxin Removing Gel | ThermoFisher Scientific | Cat: 20339 |
| Soluble trimerized Zaire Ebola Glycoprotein | John Dye, USAMRIID | N/A |
| Experimental Models: Cell Lines | ||
| Vero E6 | ATCC | Cat: CRL-1586 |
| Experimental Models: Organisms/Strains | ||
| Mouse: BALB/cJ | The Jackson Laboratory | JAX: 000651 |
| Mouse: B6.129S7-Ifngr1tm1Agt/J | The Jackson Laboratory | JAX: 003288 |
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| John Harty, Uiowa | ||
| Mouse: C57BL/6J Ifnar−/− | The Jackson Laboratory | JAX: 028288 |
| Mouse: C57BL/6J Ifnar/Ifngr−/− | Bred in house | N/A |
| Oligonucleotides | ||
| See Table S1 | N/A | |
Highlights.
Acute Plasmodium infection protects mice against lethal Ebola virus challenge
Protection is conferred by Plasmodium-elicited IFN-γ polarization of tissue macrophages
Protection is transient, and supraphysiological EBOV doses abrogate animal protection
Some Plasmodium-protected mice elicit a robust antibody response against the virus
ACKNOWLEDGMENTS
This study was supported by AI139902 to W.M. N.S.B. also was supported by the NIH grant numbers AI125446 and AI127481. M.R.G. and C.J.J. were supported by NIH/NIAID contract number HHSN272201200031C and NIH Office of Research Infrastructure Programs/OD P51OD011132. K.J.R. was supported by NIH training grants T32 GM007337 and T32 GM 067795. Analyses were confirmed by the Institute for Clinical and Translational Science Biostatistics Core that was supported by NIH UL1TR002537.
Footnotes
DATA AND CODE AVAILABILITY
All data are available from the Lead Contact upon reasonable request.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.104.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Angulo I,and Fresno M(2002). Cytokines in the pathogenesis of and protection against malaria. Clin. Diagn. Lab. Immunol 9, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower H, and Glynn JR (2017). A systematic review and meta-analysis of seroprevalence surveys of ebolavirus infection. Sci. Data 4, 160133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, and Geisbert TW (2005). Ebola virus: the role of macrophages and dendritic cells inthe pathogenesisofEbola hemorrhagicfever. Int. J. Biochem. Cell Biol 37, 1560–1566. [DOI] [PubMed] [Google Scholar]
- Bray M, Davis K, Geisbert T, Schmaljohn C, and Huggins J. (1998). A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J. Infect. Dis 178, 651–661. [DOI] [PubMed] [Google Scholar]
- Collins WE, Warren M, and Galland GG (1999). Studies on infectionswith the Berok strain of Plasmodium cynomolgi in monkeys and mosquitoes. J. Parasitol 85, 268–272. [PubMed] [Google Scholar]
- Coltart CE, Lindsey B, Ghinai I, Johnson AM, and Heymann DL (2017). The Ebola outbreak, 2013–2016: old lessons for new epidemics. Philos. Trans. R. Soc. Lond. B Biol. Sci 372, 20160297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly BM, Steele KE, Davis KJ, Geisbert TW, Kell WM, Jaax NK, and Jahrling PB (1999). Pathogenesisofexperimental Ebolavirus infection in guinea pigs. J. Infect. Dis 179 (Suppl 1), S203–S217. [DOI] [PubMed] [Google Scholar]
- Côte M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, and Cunningham J. (2011). Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton PD, Moebius J, Portugal S, Waisberg M, Hart G, Garver LS, Miller LH, Barillas-Mury C, and Pierce SK (2014). Malaria immunity in man and mosquito: insights into unsolved mysteries ofadeadly infectious disease. Annu. Rev. Immunol 32, 157–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza JB, Williamson KH, Otani T, and Playfair JH (1997). Early gamma interferon responses in lethal and nonlethal murine blood-stage malaria. Infect. Immun 65, 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara H, Theriault S, Neumann G, Alimonti JB, Geisbert JB, Hensley LE, Groseth A, Jones SM, Geisbert TW, Kawaoka Y, and Feldmann H. (2007). In vitro and in vivo characterization of recombinant Ebola viruses expressing enhanced green fluorescent protein. J. Infect. Dis 196 (Suppl 2), S313–S322. [DOI] [PubMed] [Google Scholar]
- Feldmann H, and Geisbert TW (2011). Ebola haemorrhagic fever. Lancet 377, 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, and Davis KJ (2003). Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol 163, 2347–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner C, Moreno A, Meyer EV, Cabrera-Mora M, Kissinger JC, Barnwell JW, and Galinski MR; MaHPIC Consortium (2016). Plasmodium cynomolgi infections in rhesus macaques display clinical and parasitological features pertinent to modelling vivax malaria pathology and relapse infections. Malar. J 15, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T, and Lamb T. (2015). Interferon-γ: The Jekyll and Hyde of Malaria. PLoS Pathog. 11,e1005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, et al. (2011). T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA 108, 8426–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennemann NJ, Herbert AS, Brouillette R, Rhein B, Bakken RA, Perschbacher KJ, Cooney AL, Miller-Hunt CL, Ten Eyck P, Biggins J, et al. (2017). Vesicular Stomatitis Virus Pseudotyped with Ebola Virus Glycoprotein Serves as a Protective, Noninfectious Vaccine against Ebola Virus Challenge in Mice. J. Virol 91, e00479–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Seixas E, and Langhorne J. (2001). Rodent malarias: the mouse as a model for understanding immune responses and pathology induced by the erythrocytic stages of the parasite. Med. Microbiol. Immunol. (Berl.) 189, 115–126. [DOI] [PubMed] [Google Scholar]
- Mahanty S, Gupta M, Paragas J, Bray M, Ahmed R, and Rollin PE (2003). Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology 312, 415–424. [DOI] [PubMed] [Google Scholar]
- Malleret B, Claser C, Ong AS, Suwanarusk R, Sriprawat K, Howland SW, Russell B, Nosten F, and Renia L. (2011). A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Sci. Rep 1, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina TS, Costa SP, Oliveira MD, Ventura AM, Souza JM, Gomes TF, Vallinoto AC, Povoa MM, Silva JS, and Cunha MG (2011). Increased interleukin-10 and Interferon-γ levels in Plasmodium vivax malaria suggest a reciprocal regulation which is not altered by IL-10 gene promoter polymorphism. Malar. J 10, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellenbeck MF, Ueberheide B, Amulic B, Epp A, Fenyo D, Busse CE, Esen M, Theisen M, Mordmüller B, and Wardemann H. (2013). Atypicaland classicalmemory B cells produce Plasmodium falciparum neutralizing antibodies. J. Exp. Med 210, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S, Borchert M, Paweska J, Tshomba A, Afounde A, Kulidri A, Swanepoel R, Muyembe-Tamfum JJ, and Van der Stuyft P. (2016). High prevalence of IgG antibodies to Ebola virus in the Efe pygmy population in the Watsa region, Democratic Republic of the Congo. BMC Infect. Dis 16,263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, and Aguet M. (1994). Functional role of type I and type II interferons in antiviral defense. Science 264, 1918–1921. [DOI] [PubMed] [Google Scholar]
- Nasr A, Allam G, Hamid O, and Al-Ghamdi A. (2014). IFN-γamma and TNF associated with severe falciparum malaria infection in Saudi pregnant women. Malar. J 13,314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal RG, Mourich DV, Bradfute S, Hauck LL, Warfield KL, Iversen PL, and Bavari S. (2014). Induced IL-10 splice altering approach to antiviral drug discovery. Nucleic Acid Ther. 24, 179–185. [DOI] [PubMed] [Google Scholar]
- Perlaza BL, Sauzet JP, Brahimi K, BenMohamed L, and Druilhe P. (2011). Interferon-γ, a valuable surrogate marker of Plasmodium falciparum pre-erythrocytic stages protective immunity. Malar. J 10, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal S, Moebius J, Skinner J, Doumbo S, Doumtabe D, Kone Y, Dia S, Kanakabandi K, Sturdevant DE, Virtaneva K, et al. (2014). Exposure-dependent control of malaria-induced inflammation in children. PLoS Pathog. 10, e1004079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, and Muench H. (1938). A simple method of estimating fifty percent endpoints. Am. J. Epidemiol 27, 493–497. [Google Scholar]
- Rhein BA, Powers LS, Rogers K, Anantpadma M, Singh BK, Sakurai Y, Bair T, Miller-Hunt C, Sinn P, Davey RA, et al. (2015). Interferon-γ Inhibits Ebola Virus Infection. PLoS Pathog. 11, e1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes-Feuillette A, Bellosguardo M, Druilhe P, Ballet JJ, Chousterman S, Canivet M, and Peries J. (1985). The interferon compartment of the immune response in human malaria: II. Presence of serum-interferon gamma following the acute attack. J. Interferon Res 5, 169–178. [DOI] [PubMed] [Google Scholar]
- Rogers KJ, and Maury W. (2018). The role of mononuclear phagocytes in Ebolavirus infection. J. Leukoc. Biol 104, 717–727. [DOI] [PubMed] [Google Scholar]
- Rogers KJ, Brunton B, Mallinger L, Bohan D, Sevcik KM, Chen J, Ruggio N, and Maury W. (2019). IL-4/IL-13 polarization of macrophages enhances Ebola virus glycoprotein-dependent infection. PLoS Negl. Trop. Dis 13, e0007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K, Adjemian J, Munster VJ, Marzi A, Falzarano D, Onyango CO, Ochieng M, Juma B, Fischer RJ, Prescott JB, et al. (2016).Plasmodium Parasitemia Associated With Increased Survival in Ebola Virus-Infected Patients. Clin. Infect. Dis 63, 1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K, Mercado-Hernandez R, Cronin J, Conteh S, Duffy P, Feldmann H, and de Wit E. (2018). The Effect of Plasmodium on the Outcome of Ebola Virus Infection in a Mouse Model. J. Infect. Dis 218 (suppl_5), S434–S437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerlinck JP (1999). Functional and structural comparison of cytokines in different species. Vet. Immunol. Immunopathol 72, 39–44. [DOI] [PubMed] [Google Scholar]
- Schmidt NW, Butler NS, Badovinac VP, and Harty JT (2010). Extreme CD8T cell requirements for anti-malarial liver-stage immunity following immunization with radiation attenuated sporozoites. PLoS Pathog. 6, e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen A, and Sauerwein RW (2013). How malaria modulates memory: activation and dysregulation of B cells in Plasmodium infection. Trends Parasitol. 29, 252–262. [DOI] [PubMed] [Google Scholar]
- Shtanko O, Reyes AN, Jackson WT, and Davey RA (2018). Autophagy-Associated Proteins Control Ebola Virus Internalization Into Host Cells. J. Infect. Dis 218 (suppl_5), S346–S354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit MA, Michelow IC, Glavis-Bloom J, Wolfman V, and Levine AC (2017). Characteristics and Outcomes of Pediatric Patients With Ebola Virus Disease Admitted to Treatment Units in Liberia and Sierra Leone: A Retrospective Cohort Study. Clin. Infect. Dis 64, 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen I, Lu K, Yamamoto LK, Hoff NA, Mulembakani P, Wemakoy EO, Muyembe-Tamfum JJ, Ndembi N, Brennan CA, Hackett J Jr., et al. (2019). Serologic Prevalence of Ebola Virus in Equatorial Africa. Emerg. Infect. Dis 25, 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R, Culleton RL, and Lamb TJ (2012). The contribution of Plasmodium chabaudi to our understanding of malaria. Trends Parasitol. 28, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, and Stevenson MM (2000). Central role of endogenous gamma interferon in protective immunity against blood-stage Plasmodium chabaudi AS infection. Infect. Immun 68, 4399–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, and Kawaoka Y. (1997). A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94, 14764–14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo TH, Lum FM, Ghaffar K, Chan YH, Amrun SN, Tan JJL, Lee CYP, Chua TK, Carissimo G, Lee WWL, et al. (2018). Plasmodium co-infection protects against chikungunya virus-induced pathologies. Nat. Commun 9, 3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet MA, Reynard S, Fizet A, Schaeffer J, Pannetier D, Guedj J, Rives M, Georges N, Garcia-Bonnet N, Sylla AI, et al. (2017). Clinical, virological, and biological parameters associated with outcomes of Ebolavirus infection in Macenta, Guinea. JCI Insight 2, e88864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas-Mendez A, Greig R, Shaw TN, de Souza JB, Gwyer Findlay E, Stumhofer JS, Hafalla JC, Blount DG, Hunter CA, Riley EM, and Couper KN (2012). IFN-γ-producing CD4+ T cells promote experimental cerebral malaria by modulating CD8+ T cell accumulation within the brain. J. Immunol 189, 968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman M, Aluisio AR, Rege S, and Levine AC (2017). Characteristics and survival of patients with Ebola virus infection, malaria, or both in Sierra Leone: a retrospective cohort study. Lancet Infect. Dis 17, 654–660. [DOI] [PubMed] [Google Scholar]
- Wec AZ, Nyakatura EK, Herbert AS, Howell KA, Holtsberg FW, Bakken RR, Mittler E, Christin JR, Shulenin S, Jangra RK, et al. (2016). A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses. Science 354, 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer J, Grobbelaar A, and Blumberg L. (2015). Ebola virus disease: history, epidemiology and outbreaks. Curr. Infect. Dis. Rep 17, 480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.