Abstract
We present a case of severe pneumonia, associated with a prolonged infection by a species C rhinovirus (HRV) in a 3-week old neonate. HRV RNA was identified in nasal and nasopharyngeal secretions, bronchoalveolar lavage and bronchial specimens, stool and urine, collected from the patient during a one-month period. No other viral or bacterial agents were detected. Sequence analysis of two regions of the viral genome, amplified directly from the clinical specimens revealed a novel HRV-C variant. These observations highlight the occurrence of severe neonatal infections caused by HRVs and the need of rapid viral diagnostics for their detection.
Abbreviations: HRV, human rhinovirus; RT, reverse transcription; PCR, polymerase chain reaction; qPCR, quantitative PCR; 5′NCR, 5′noncoding region
Keywords: Rhinovirus, Respiratory, Infection, Pneumonia, Neonatal
1. Why this case is important
Human rhinoviruses (HRVs) are the most common cause of both upper and lower acute respiratory tract illnesses. Although HRVs are often associated with acute otitis media, sinusitis, bronchiolitis, asthma and pneumonia in children, they are mostly regarded as the cause of common colds with their economic significance at the community level in mind.1
On the basis of their genomic sequences, HRV types have been classified into three species: 74 types belong to HRV A, 25 types to HRV B, and newly discovered, phylogenetically related types are classified in HRV C species.2 HRV C infections may be more severe than those caused by the other HRVs and often require hospitalization.3, 4 However, neonatal HRV infections may be under diagnosed and regarded as an unlikely cause of severe manifestations.
We report here a case of a life-threatening HRV C pneumonia in an otherwise healthy infant.
2. Case report
2.1. Case description
In March 2009, a 3-week-old male infant was admitted to the Department of Pediatrics of Turku University Hospital with dyspnea and toxic appearance. During the past 24 h, he had developed symptoms including nasal discharge, fever, and cough. He had been otherwise healthy except, at the age of two days, he had conjunctivitis caused by Streptococcus agalactiae and was treated with intravenous penicillin G for 5 days with full recovery. On admission to the hospital, the infant was cyanotic and irritable. Tachycardia (182/min) was recorded, the extremities were cold and his temperature was 37.9 °C. He had tachypnea (59/min), severe dyspnea, and arterial oxygen saturation was low (59% by pulse oxymetry). Bilateral crackles were heard on chest auscultation. Chest radiographs revealed non-symmetric atelectasis and diffuse apical alveolar pneumonic infiltrates suggesting pneumonia. Intravenous ampicillin and gentamicin were administered.
Laboratory tests revealed a white blood cell count of 11.3 × 109/l and a serum C-reactive protein level of 108 mg/l. Cerebrospinal fluid was normal. Point-of-care tests for human influenza A and B viruses, respiratory syncytial virus and adenovirus were negative.
The infant was intubated and connected to a respirator due to hypercapnia (CO2 14.9 kPa) and acidosis (pH 7.13). As he was hypotensive (non-invasive blood pressure 33 mmHg) a noradrenalin-infusion was commenced, and due to hyponatremia (133 mmol/l), hydrocortisone substitution was initiated (5 mg/m2/day). In addition, total parenteral nutrition was initiated. Cardiac ultrasound examination exhibited normal heart structure and function.
Ampicillin and gentamicin were discontinued at day 3 due to poor clinical response and intravenous cefuroxime was administered for additional 8 days. Oral azithromycin was added for the treatment of pneumonia. Blood, cerebrospinal fluid and urine samples for bacterial cultures, collected before antibiotic therapy, remained negative.
After 5 days of treatment, the infant became afebrile, the breathing difficulties resolved, and he was extubated. Serum C-reactive protein concentration decreased from 117 to 15 mg/l (days 2–5; respectively) and the patient recovered completely. An analysis of his immunological status revealed no detectable defects in immunoglobulin levels, IgG subclasses, total hemolytic complement activity, or classical, alternative and lectin-dependent complement activation pathways. No virus could be cultured when several of the samples were tested in various cell lines at various temperatures. In addition, Mycoplasma pneumoniae and Chlamydia pneumoniae antibodies remained negative on day 39. During subsequent follow-up, at the age of one year, he had one common cold but no other clinical illnesses.
2.2. Nucleic acid testing and analyses
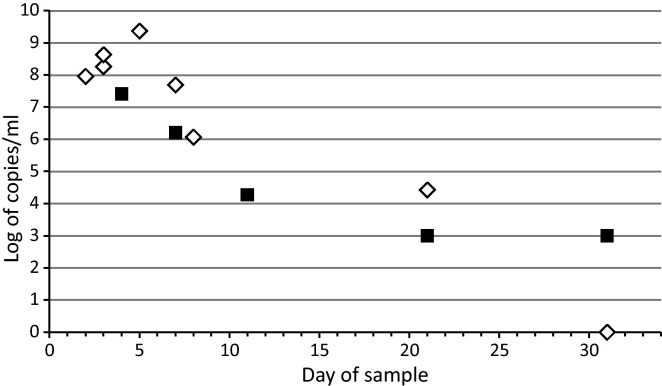
Various specimens were obtained during a 7-week time period from the patient (Table 1 ). Nucleic acids were extracted using the NucliSens EasyMag automated extractor (bioMèrieux, Boxtel, the Netherlands). The extracts were analyzed and quantified with reverse transcription polymerase chain reaction (RT-qPCR) specific for the 5′noncoding region (5′NCR) of rhino- and enteroviruses5 using dilutions of plasmid DNAs with the genome of HRV-85 as the standard. They were also tested for human bocavirus by qPCR and respiratory syncytial virus by RT-qPCR methods as well as for adenovirus, coronaviruses 229E/NL63 and OC43/HKU1, parainfluenza virus types 1–3, influenza A and B viruses, respiratory syncytial group A and B viruses, and HRV A/B with Seeplex RV12 ACE (Seegene, Seoul, South Korea) and ScreenTape system (Lab901, Loanhead, UK). PCR detection was also performed for Bordetella pertussis and M. pneumoniae in bronchoscopic aspiration fluid and for Chlamydia trachomatis in ocular secretions. We detected HRV RNA in nasal secretion and nasopharyngeal aspirate, bronchial secretion and bronchoalveolar lavage, urine, and feces (Table 1, Fig. 1 ). The cereprospinal fluid and blood samples remained negative. None of the tests for other viral or bacterial pathogens was positive.
Table 1.
Human rhinovirus (HRV) detection in a neonate with severe pneumonia.
| Day of samplea | Sample type | Diagnostic result | HRV RNA (log10 of copies/ml) |
|---|---|---|---|
| 0 | Cerobrospinal fluid | HRV− | – |
| 2 | Nasopharyngeal secretion | HRV+ | 8.0 |
| 3 | Nasal secretion | HRV+ | 8.3 |
| 3 | Bronchoalveolar lavage | HRV+ | 8.6 |
| 4 | Feces | HRV+ | 7.4 |
| 4 | Blood | HRV− | – |
| 5 | Bronchial secretion | HRV+ | 9.4 |
| 7 | Feces | HRV+ | 6.2 |
| 7 | Blood | HRV− | – |
| 7 | Nasal secretion | HRV+ | 7.7 |
| 7 | Urine | HRV+ | <3 |
| 9 | Nasopharyngeal secretion | HRV+ | 6.1 |
| 10 | Nasal secretion | HRV+ | No quantitation |
| 11 | Feces | HRV+ | 4.3 |
| 13 | Mucosal secretion | HRV− | – |
| 21b | Feces | HRV− | – |
| 21b | Nasopharyngeal swab | HRV+ | 4.4 |
| 31b | Feces | HRV+ | <3 |
| 51b | Nasal swab | HRV+c | 3.3 |
| 51b | Feces | HRV+c | 3.9 |
At day 0, when the child was admitted to the hospital, his age was 23 days.
Follow-up samples at asymptomatic stage.
Not the same type as in previous samples.
Fig. 1.
Copy number measurements in different respiratory specimens (triangles) and feces (squares) from the patient after a severe infection with human rhinovirus species C. Details of the respiratory specimens are indicated in Table 1.
A 397-bp long amplicon of the 5′NCR region was amplified from the HRV-positive cDNA samples as described earlier.5 An additional 541-bp long amplicon from the VP4/2 gene region was amplified with nested-PCR using primers described by Wisdom et al.6 The PCR products were purified using Nucleo-Spin Extract II (Macherey-Nagel, Düren, Germany) and sequenced in the DNA Sequencing Service Laboratory of the Turku Center for Biotechnology. The sequences were analyzed by BLAST for closest nucleotide identity. The BLAST matches were selected for the phylogenetic analysis with programs of the MEGA software version 4.7
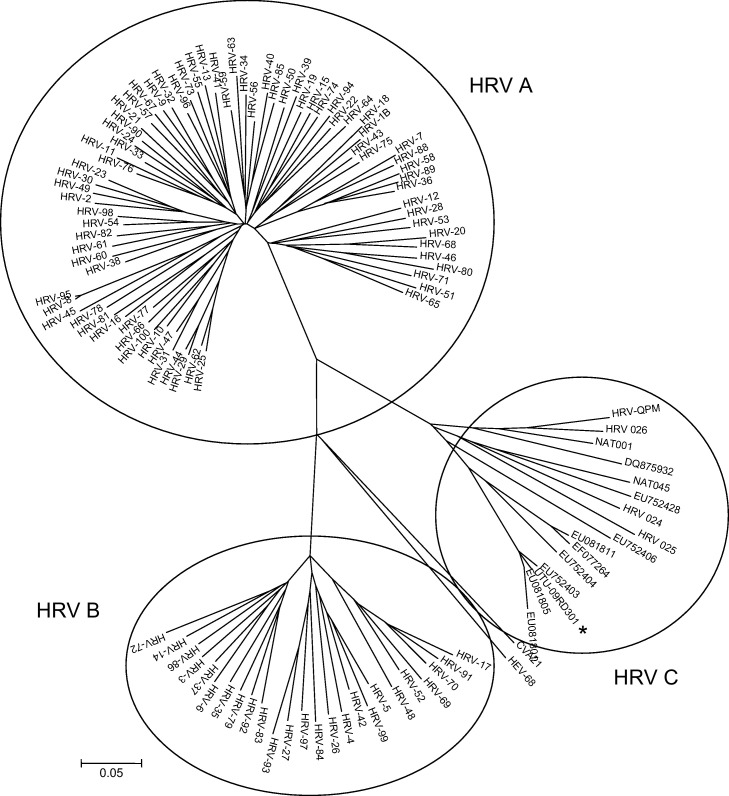
The alignments of the partial sequences of the 5′NCR (GenBank accession number HQ714957) and VP4/2 (HQ714958) showed that all the samples collected during days 2–31 contained a distinct HRV variant, not fully matching with any published sequence. The last two samples from day 51 were of another HRV type, showing closest 5′NCR alignment with HRV-33 (species A). In the VP4/2 region, samples of the first HRV infection had 63.2% (range 61–67%) nucleotide identity with HRV A types, 63.9% (range 62–66%) with HRV B types and 80.5% (range 70–96%) with the previously identified HRV C variants. The patient virus, designated UTU-09RD301, showed a rarely identified genotype, clustering closely with strains EU752403 (95% nucleotide identity) found in the USA,8 and EU081805 (96%) and EU081801 (91%) found in Germany3 (Fig. 2 ). This cluster may represent a HRV C type.2
Fig. 2.
Phylogenetic analysis of the VP4/2 coding region of the patient case (UTU-09RD301, GenBank accession number HQ714958) and selected published HRV sequences. For the HRV C cluster, the GenBank numbers of the partial sequences are shown. The evolutionary history was inferred using the neighbor-joining method. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. Virus sequences with >98% nucleotide identities were considered same. Coxsackievirus A 21 and HRV-68 sequences were used as outgroups.
We further confirmed the identity of the UTU-09RD301 as an HRV C strain by locating the cis-acting replication element (cre) within VP2 of the translated sequence.9 The UTU-09RD301 cre corresponding the amino acid sequence C-G-F-S-D-R-L-K-Q-I-T-I-G-N-S-T showed 100% identity with the HRV C type cre motifs published by others.10
In 2009, specimens from 63 children <3 months of age, who presumably had a community acquired respiratory infection requiring hospitalization, were tested for HRV RNA in our laboratory. The specimens were obtained from 3 tertiary care hospitals with a population base of 1 million people and 13,000 live births in 2009. The rate of HRV detections in <1 month, 1–2 months, and 2–3 months old children was 6/23 (26%), 11/24 (46%), and 7/16 (44%), respectively. Thus, even without clinical data, except for the current case, these numbers emphasize the importance of HRV in neonatal respiratory infections.
2.3. Other similar cases in the literature
In hospitalized children with pneumonia, the detection rates of HRV in nasopharyngeal samples by nucleic acid detection methods have ranged from 20% to 45%.11, 12 A recent study by Louie and colleagues demonstrated that HRV was present in almost half of children with lower respiratory tract infection and admitted to intensive care unit.13 In another recent study, severe HRV infection was detected in 9 preterm and 2 full-term infants with respiratory distress requiring mechanical ventilation.14 A careful review of the literature, covering 4279 episodes of community acquired viral pneumonia, revealed no studies addressing HRV-induced pneumonia in neonates.15
3. Discussion
Our case represents a neonate with obvious life-threatening HRV pneumonia. Our observations provide support for the routine inclusion of HRV detection in etiologic studies of pneumonia in infants and young children.
The investigation revealed that our patient was infected with a virus belonging to the HRV C species. Since HRV C strains have not been cultivable thus far in current cell culture systems, they were only recently identified and have boosted the general interest in HRV.1, 3 In our hands, utilization of the 5′NCR primer sites (“3+” and “4−”) in RT-PCR allow the most sensitive primary detection of rhino- and enteroviruses, including HRV species C-like strains.16, 17, 18 Albeit HRV C viruses can cause merely mild common cold symptoms, like other HRVs, they have been suggested to be more frequently associated with severe complications including pneumonia, bronchiolitis, and exacerbations of asthma.3, 4, 7, 19
The significance of the detection of HRV in nasopharynx has been questioned as HRVs are frequently detected in nasal samples in asymptomatic children. In our patient, however, viral RNA was found in several body sites including nasal and nasopharyngeal secretions, bronchoalveolar lavage and bronchial specimens, feces and urine, indicating direct association between HRV and the severe symptomatic infection, even though no viremia was detected. In our patient, the RNA load in the feces on day 4 was only one log lower than in the bronchoalveolar lavage, unlikely a result of spillover from the respiratory tract. Fecal secretion of HRV suggesting enteric infection has been demonstrated before.19, 20 This could possibly be a result of temporarily low stomach acid.
Interestingly, the child was infected with the same virus for at least a month, but the last two follow-up samples on day 51, containing another type of HRV, were collected when the child was, and remained, asymptomatic. Thus, our case also demonstrates that persistence of a strain, to exclude re-infection with another, must be confirmed with specific measures. In older children and in adults, HRV shedding lasts usually 1–2 weeks.4, 21 Immunological immaturity of a neonate may have contributed to the lengthy shedding. Earlier, persistent HRV shedding has been recorded in immunocompromised lung transplant patients and in hypogammaglobulinemic patients.22, 23
In conclusion, our observations support the view that specific laboratory diagnosis of HRV infections should be used in children with severe pneumonia. On the basis of VP4/2 sequence analysis, the fulminant and long lasting infection in our immunocompetent neonate was caused by a type of HRV C species. The role of HRVs in severe infections in children, either alone or in co-infections with other viruses or bacteria, may be much more important than currently appreciated.
Conflicts of interest
Authors have no conflicts of interest to report.
Acknowledgement
We thank Ms. Tiina Ylinen for technical assistance.
References
- 1.Mackay I.M. Human rhinoviruses: the cold wars resume. J Clin Virol. 2008;42:297–320. doi: 10.1016/j.jcv.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmonds P., McIntyre C., Savolainen-Kopra C., Tapparel C., Mackay I.M., Hovi T. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J Gen Virol. 2010;91:2409–2419. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed] [Google Scholar]
- 3.Renwick N., Schweiger B., Kapoor V., Liu Z., Villari J., Bullmann R. A recently identified rhinovirus genotype is associated with severe respiratory tract infection in children in Germany. J Infect Dis. 2007;196:1754–1760. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo C., Garcia M.L., Pozo F., Reyes N., Perez-Brefia P., Casas I. Role of rhinovirus C in apparently life-threatening events in infants, Spain. Emerg Inf Dis. 2009;15:1506–1508. doi: 10.3201/eid1509.090453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peltola V., Waris M., Österback R., Susi P., Ruuskanen O., Hyypiä T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 6.Wisdom A., Leitch E.C., Gaunt E., Harvala H., Simmonds P. Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4–VP2 typing reveals high incidence and genetic diversity of HRV species C. J Clin Microbiol. 2009;47:3958–3967. doi: 10.1128/JCM.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura K., Dudley J., Nei M., Kumar S. MEGA 4 molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 8.Miller E.K., Edwards K.M., Weinberg G.A., Iwane M.K., Griffin M.R., Hall C.B. A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol. 2009;123:98–104. doi: 10.1016/j.jaci.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordey S., Gerlach D., Junier T., Zdobnov E.M., Kaiser L., Tapparel C. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA. 2008;14:1568–1578. doi: 10.1261/rna.1031408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piralla A., Rovida F., Campanini G., Rognoni V., Marchi A., Locatelli F. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Microbiol. 2009;45:311–317. doi: 10.1016/j.jcv.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Juvén T., Mertsola J., Waris M., Leinonen M., Meurman O., Roivainen M. Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J. 2000;19:292–298. doi: 10.1097/00006454-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Cilla G., Oñate E., Perez-Yarza E.G., Montes M., Vicente D., Perez-Trallero E. Viruses in community-acquired pneumonia in children aged less than 3 years old: high rate of viral coinfection. J Med Virol. 2008;80:1843–1849. doi: 10.1002/jmv.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Louie J.K., Roy-Burman A., Guardia-Labar L., Boston E.J., Kiang D., Padilla T. Rhinovirus associated with severe lower respiratory tract infections in children. Pediatr Infect Dis J. 2009;28:337–339. doi: 10.1097/INF.0b013e31818ffc1b. [DOI] [PubMed] [Google Scholar]
- 14.van Piggelen R.O., van Loon A.M., Krediet T.G., Verboon-Maciolek M.A. Human rhinovirus causes severe infection in preterm infants. Pediatr Infect Dis J. 2010;29:364–365. doi: 10.1097/INF.0b013e3181c6e60f. [DOI] [PubMed] [Google Scholar]
- 15.O. Ruuskanen, E. Lahti, L.C. Jennings, D.R. Murdoch, Viral pneumonia, Lancet, in press. [DOI] [PMC free article] [PubMed]
- 16.Santti J., Hyypiä T., Halonen P. Comparison of PCR primer pairs in the detection of human rhinoviruses in nasopharyngeal aspirates. J Virol Methods. 1997;66:531–539. doi: 10.1016/s0166-0934(97)00049-9. [DOI] [PubMed] [Google Scholar]
- 17.Hyypiä T., Puhakka T., Ruuskanen O., Mäkelä M., Arola A., Arstila P. Molecular diagnosis of human rhinovirus infections: comparison with virus isolation. J Clin Microbiol. 1998;36:2081–2083. doi: 10.1128/jcm.36.7.2081-2083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peltola V., Jartti T., Putto-Laurila A., Mertsola J., Vainionpää R., Waris M. Rhinovirus infections in children: a retrospective and prospective hospital-based study. J Med Virol. 2009;81:1831–1838. doi: 10.1002/jmv.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tapparel C., L’Huillier A.G., Rougemont A.-L., Beghetti M., Barazzone-Argiroffo C., Kaiser L. Pneumonia and pericarditis in a child with HRV-C infection: a case report. J Clin Virol. 2009;45:157–160. doi: 10.1016/j.jcv.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blomqvist S., Savolainen-Kopra C., Paananen A., Hovi T., Roivainen M. Molecular characterization of human rhinovirus field strains isolated during surveillance of enteroviruses. J Gen Virol. 2009;90:1371–1381. doi: 10.1099/vir.0.008508-0. [DOI] [PubMed] [Google Scholar]
- 21.Winther B., Hayden F.G., Hendley J.O. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006;78:644–650. doi: 10.1002/jmv.20588. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser L., Aubert J.D., Pache J.C., Deffernez C., Rochat T., Garbino J. Am J Respir Crit Care Med. 2006;174:1392–1399. doi: 10.1164/rccm.200604-489OC. [DOI] [PubMed] [Google Scholar]
- 23.Kainulainen L., Vuorinen T., Rantakokko-Jalava K., Osterback R., Ruuskanen O. Recurrent and persistent respiratory tract viral infections in patients with primary hypogammaglobulinemia. J Allergy Clin Immunol. 2010;126:120–126. doi: 10.1016/j.jaci.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]