Abstract
We report the discovery of a novel enterovirus C (EV-C118) identified in two Israeli children hospitalised for acute otitis media and community-acquired pneumonia. The highest pair-wise sequence identity scores with the EV-C109 and EV-C117 reference strains were, respectively, 63.5% and 63.6% nucleotide identity, and 82.5% and 79.9% amino acid identity.
Keywords: Acute otitis media, Children, Community-acquired pneumonia, Enterovirus, Picornavirus, Respiratory viruses
1. Why this case is important
New data have recently underlined the clinical importance of enteroviruses (HEV) and new strains have been identified, sometimes associated with severe outbreaks.1, 2 We here report the discovery and molecular and clinical characterisation of a novel HEV type within species HEV-C (designated EV-C118), which was identified in two Israeli children enrolled in the community-acquired pneumonia paediatric research initiative (CAP-PRI) study for the evaluation of the aetiology of community-acquired pneumonia (CAP).
2. Case description
A 21-month-old female child with an unremarkable previous medical history had been suffering from rhinitis and cough for 7 days. The subsequent appearance of high fever (>39 °C) led to a diagnosis of acute otitis media (AOM) by her primary care paediatrician. The patient was treated symptomatically, but the fever remained >39 °C and the cough became more prominent. After 4 days, O2 saturation went down to 91% and the child was admitted to hospital in Beer-Sheva, Israel, in January 2011. Upon admission, bilateral AOM was confirmed, and auscultation revealed reduced breath sounds at the base of both lungs. The patient's body temperature was 39 °C, her respiratory and heart rates were, respectively, 44/min and 132/min, and O2 saturation was 91%. No other abnormality was found upon physical examination. A nasopharyngeal sample was collected using a flexible pernasal flocked swab and immediately placed in a mini-tube containing 1 mL of universal transport medium (UTM-RT Kit Cat. No. 360c, Copan Italia, Brescia, Italy) for research purposes. The sample was stored at 4 °C in the hospital laboratory until it was sent in a refrigerated package to the central laboratory (Pediatric Clinic 1, University of Milan, Italy). Other routine laboratory tests showed haemoglobin 9.9 g/dL, white blood cells (WBCs) 29,960/mm3 (neutrophils 81.3%), platelets 610,000/mm3, glucose 126 mg/dL, and C-reactive protein (CRP) 41.3 mg/L. Blood culture was negative for aerobic and anaerobic bacteria as well as for fungi, as were all of the other viral assays (respiratory virus panel [RVP] fast assay for influenza viruses, parainfluenza viruses, respiratory syncytial virus, adenovirus, human metapneumovirus, coronaviruses 229E, NL63, OC43 and HKU1, enterovirus/rhinovirus, and human bocavirus) and bacterial tests (polymerase chain reaction for detection of Streptococcus pneumoniae and untypeable Haemophilus influenzae) of respiratory secretions. Chest X-ray findings led to a diagnosis of left lobar pneumonia, and amoxicillin 80 mg/(kg day) was started. The child recovered after 48 h of hospitalisation and was discharged home with a recommendation to complete 7 days’ antibiotic treatment. She experienced no recurrence and no further hospitalisation was needed in the following 12 months.
The second case involved a 15-month-old Bedouin male who was admitted to the same hospital in Beer-Sheva, Israel, in the same period because he had been suffering from a high fever (40 °C), cough, vomiting, diarrhoea and apathy with grunting for 4 days. His primary care paediatrician diagnosed AOM and the patient was treated with amoxicillin for one day before admission. On the day of admission, the diarrhoea and vomiting stopped. Upon admission, left AOM was confirmed and auscultation revealed reduced breath sounds and crackles at the base of the left lung. The patient's body temperature was 39 °C, his respiratory and heart rates were, respectively, 64/min and 136/min, and O2 saturation was 95%. Also in this case, a nasopharyngeal sample was collected for research purposes as in the previous case, which was stored and sent to the central laboratory in the same manner. In this case, routine laboratory tests showed haemoglobin 9.9 g/dL, WBCs 21,200/mm3 (neutrophils 78.0%), platelets 466,000/mm3, and CRP 48.5 mg/L. Blood culture was negative, as were all the other viral and bacterial tests of respiratory secretions (the tests for viruses and bacteria were those performed in the first case). Chest X-ray findings led to a diagnosis of left lobar pneumonia with pleural effusion, and so a chest drain was inserted. The pleural fluid culture (available 5 days after admission) was positive for Streptococcus pneumoniae serotype 22F, which was sensitive to all of the tested antibiotics. The child was initially treated with cefuroxime i.v. (100 mg/(kg day)), which was changed to ceftriaxone i.v. (100 mg/(kg day)) plus amoxicillin-clavulanic acid i.v. (80 mg/(kg day)) after 3 days. When the results of the pleural fluid culture became available, the antibiotic treatment was changed to ampicillin i.v. (80 mg/(kg day) i.v.). The child underwent chest drainage for a total of 14 days, and was discharged after 15 days of hospitalisation in good condition with a recommendation to continue oral amoxicillin treatment for a further week. He experienced no respiratory problems in the following 12 months.
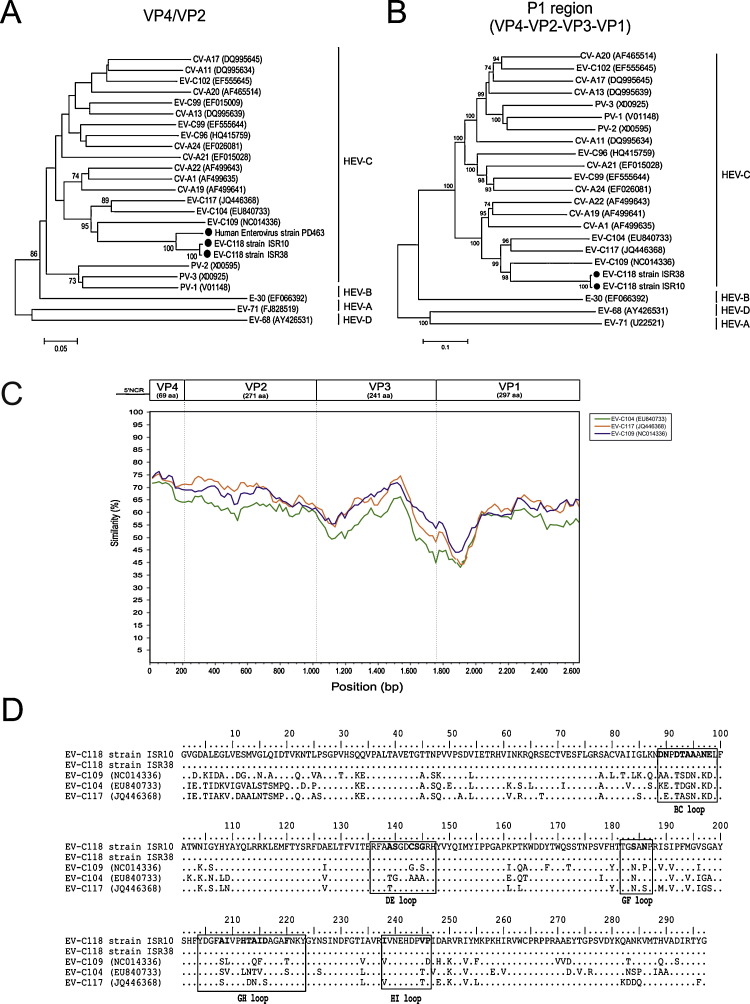
In the central laboratory, the viral nucleic acids extracted from the nasopharyngeal swabs were tested for respiratory viruses using the RVP Fast assay (Luminex Molecular Diagnostics, Inc., Toronto, Canada).3 As both samples were positive for enterovirus/rhinovirus, they were retested in order to identify the rhinovirus and amplify the VP4/VP2 region as previously described.4, 5 Phylogenetic analysis of the VP4/VP2 region showed that both nucleotide sequences belonged to the HEV-C species. A partial VP1 sequence was obtained using the primers described by Nix et al.6 The remaining sequence of the P1 capsid region of both samples was obtained using “in-house” amplification protocols (available upon request). The amplicons of expected sizes were gel purified and sequenced. The complete VP1 sequence of ISR-10 was submitted to the Picornaviridae Study Group (http://www.picornastudygroup.com) and designated a proposed new enterovirus type (EV-C118; accession number JQ768163). The amplified partial VP4/VP2 sequence of the EV-C collected from these two children had 67.8% nucleotide identity and 90.5% amino acid identity with EV-C109 (Fig. 1 A), whereas the amplified VP1 regions had 69.9% nucleotide identity and 77.4% amino acid identity.
Fig. 1.
Phylogenetic relationships of HEV-C and the new EV-C118 based on (A) VP4/VP2 region sequences and (B) complete capside protein coding region sequences (P1). The phylogeny of the nucleotide sequences was reconstructed using maximum likelihood methods and Tamura's 3-parameter evolutionary model with heterogeneous rates among sites, and a gamma distribution for the relative rate. Branch support was assessed by means of bootstrap analyses of 1000 replicates, with a bootstrap value of 70% being used as the cut-off point for cluster analysis. EV-68 and EV-70 were used as the outgroups. (C) Similarity plot of the complete EV-118 capsid-coding sequence using a sliding window of 200 nt moving in 20 nt steps. (D) Deduced amino acid sequence of the complete VP1 gene of EV-C118 (JQ768163), EV-C117 (JQ446368), EV-C104 (EU840733), and EV-C109 (NC014336). The amino acid changes within the loop regions are in bold.
3. Other similar and contrasting cases in the literature
The complete P1 capsid region of these samples was compared with the matching region of all of the complete enterovirus genomes available in the GenBank database as of 30 March 2012. On the basis of the nucleotide sequences, a phylogenetic tree was reconstructed using maximum likelihood methods and Tamura's 3-parameter evolutionary model.7 As shown in Fig. 1B, the closest genotypes were EV-C109 (NC014336) and EV-C117 (JQ446368). The highest pair-wise sequence identity scores with the EV-C109 and EV-C117 reference strains were, respectively, 63.5% and 63.6% nucleotide identity, and 82.5% and 79.9% amino acid identity.
The nucleotide sequence of the EV-C118 P1 region consists of 2634 nucleotides. The base composition of the full P1 EV-C118 ISR10 strain (JQ922256) is 27.5% A, 24.9% C, 23.9% G, and 23.7% U. Sixty-nine of the 878 predicted amino acids are within the VP4 gene, 271 within the VP2 gene, 241 within the VP3 gene, and 297 within the VP1 gene. The complete P1 region of ISR38 was different for 16 nt with no amino acid changes.
The capsid regions of three EV-C prototype strains were analysed by means of Simplot using the P1 peptide sequence of the EV-C118 strain ISR10 as the query sequence. As shown in Fig. 1C, the VP4 gene was the genomic region with the greatest identity with other HEV strains, whereas the lowest identity was within the VP1 coding region (the closest relative is EV-C109 with 30.1% nt and 22.6% aa difference in VP1 (Fig. 1D)).
The amino acid sequence of the complete VP1 gene was determined and compared with those of the most closely related enteroviruses (Fig. 1D). A total of 67 amino acid changes were observed in this region in comparison with EV-C109, a number of which were found in the loop regions (BC, DE, GF, GH, and HI) associated with viral antigenicity.
4. Discussion
We report two interesting cases of children with AOM and CAP associated with an infection due to a new species of HEV which has never been described before. It is related to the previously reported EV-C109 and EV-C117, but differ from them in terms of nucleotide and amino acid composition by, respectively, more than 25% and 12%. It can therefore be considered a novel virus type on the basis of either the more or the less stringent classification requirements for designating new EV species.8
AOM and CAP can be due to viruses, bacteria or both. In our cases, the WBC, neutrophil and CRP values in both children were very high and suggest a bacterial co-infection. Moreover, the pleural fluid culture of one of the children revealed S. pneumoniae, thus confirming the presence of a concomitant bacterial infection. In both children, the manifestation of the disease started with upper respiratory tract signs (associated with gastrointestinal symptoms in one case), and lower respiratory tract involvement occurred later. This suggests a possible initial role of EV-C118 and subsequent superimposed infection due to bacterial pathogens that could have been the main cause of the lower respiratory tract involvement. Further studies are needed to define the etiological and pathogenic role of this new virus but it may be similar to EV-109, which has been associated with severe lower respiratory tract infections requiring hospitalisation.9 Furthermore, other EVs have been found in patients with major respiratory problems.10, 11
Starting from these cases, we can conclude that EV-C118 seems to be associated with AOM and CAP. Given the potentially negative impact of some HEVs, HEV infections should be closely monitored. First of all, it is important to know the epidemiology of the different strains and, in the case of EV-C118, whether it can be found in countries other than Israel. Moreover, the molecular characteristics of EVs should be adequately analysed as this could significantly aid the preparation of appropriate prophylactic and therapeutic measures.
Funding
This review was supported by a grant from the Italian Ministry of Health (Bando Giovani Ricercatori 2007).
Competing interests
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Ethical Committee of Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy and the Soroka University Medical Center, Beer-Sheva, Israel.
References
- 1.Oberste M.S., Maher K., Flemister M.R., Marchetti G., Kilkpatrick D.R., Pallansch M.A. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J Clin Microbiol. 2000;38:1170–1174. doi: 10.1128/jcm.38.3.1170-1174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Clusters of acute respiratory illness associated with human enterovirus 68 – Asia, Europe, and United States, 2008–2010. MMWR Morb Mortal Wkly Rep. 2011;60:1301–1304. [PubMed] [Google Scholar]
- 3.Pabbaraju K., Wong S., Tokaryk K.L., Fonseca K., Drews S.J. Comparison of the Luminex xTAG respiratory viral panel with xTAG respiratory viral panel fast for diagnosis of respiratory virus infections. J Clin Microbiol. 2011;49:1738–1744. doi: 10.1128/JCM.02090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu X., Holloway B., Dare R.K., Kuypers J., Yagi S., Williams J.V. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esposito S., Daleno C., Tagliabue C., Scala A., Tenconi R., Borzani I. Impact of rhinoviruses on pediatric community-acquired pneumonia. Eur J Clin Microbiol Infect Dis. 2011;31:1637–1645. doi: 10.1007/s10096-011-1487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nix W.A., Oberste M.S., Pallansch M.A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown B.A., Maher K., Flemister M.R., Naraghi-Arani P., Uddin M., Oberste M.S. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J Gen Virol. 2009;90:1713–1723. doi: 10.1099/vir.0.008540-0. [DOI] [PubMed] [Google Scholar]
- 9.Yozwiak N.L., Skewes-Cox P., Gordon A., Saborio S., Kuan G., Balmaseda A. Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J Virol. 2010;84:9047–9058. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piralla A., Rovida F., Baldanti F., Gerna G. Enterovirus genotype EV-104 in humans, Italy, 2008–2009. Emerg Infect Dis. 2010;16:1018–1021. doi: 10.3201/eid1606.091533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahamat-Langendoen J., Riezebos-Brilman A., Borger R., van der Heide R., Brandenburg A., Schölvinck E. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J Clin Virol. 2011;52:103–106. doi: 10.1016/j.jcv.2011.06.019. [DOI] [PubMed] [Google Scholar]