Highlights
-
•
We developed one step real-time RT-PCR assays to discriminate two lineages of influenza B viruses.
-
•
The developed assays were evaluated using in vitro transcribed control RNA, clinical specimens, and clinical isolates.
-
•
The assays were shown to have high sensitivity and high specificity.
-
•
The results from the assays were consistent with those from a hemagglutination inhibition (HI) test, which is a standard method to define the lineage of influenza B virus.
-
•
The developed assays will be useful for the diagnosis and surveillance of influenza B viruses.
Keywords: Influenza B virus, Real-time RT-PCR
Abstract
Since the late 1980s, two genetically and antigenically distinct lineages of influenza B virus, namely, B/Victoria/2/87-like (B/Victoria) and B/Yamagata/16/88-like (B/Yamagata), have co-circulated. In this study, one-step real-time reverse transcription-PCR (rRT-PCR) assays were developed to differentiate B/Victoria and B/Yamagata lineages. The assays were evaluated using in vitro transcribed control RNA, isolated viruses, and other respiratory pathogenic viruses, and were shown to have high sensitivity, good linearity (R2 = 0.99), and high specificity. Using the developed rRT-PCR assays, 169 clinical specimens collected between 2010 and 2013 were then tested, resulting in the identification of 20 clinical specimens as positive for influenza B virus. Of these, 14 and 6 samples were identified as positive for the B/Victoria and B/Yamagata lineages, respectively, whereas 149 samples were negative for the influenza B virus. The rRT-PCR assays were also examined using 20 clinical isolates from 20 influenza B virus-positive specimens, revealing that there was no discrepancy between the results from the rRT-PCR assays and the hemagglutination inhibition (HI) test, with the exception that one clinical isolate with different antigenicity could not be discriminated by the HI test. The present results suggest that these highly sensitive and specific assays are useful not only for diagnosing influenza viruses but also for their surveillance.
1. Introduction
Influenza A, B, and C viruses are members of the Orthomyxoviridae family of RNA viruses, circulate in humans, and cause respiratory illness every year (Wright et al., 2007). Influenza A viruses comprise a number of subtypes, whereas influenza B and C viruses are not divided into subtypes (Wright et al., 2007). Since the late 1980s, two genetically and antigenically distinct lineages of influenza B virus, namely, B/Victoria/2/87-like (B/Victoria lineage) and B/Yamagata/16/88-like (B/Yamagata lineage), have co-circulated (Kanegae et al., 1990, Rota et al., 1990). In Japan, the majority of circulating influenza B viruses during the 2010/2011 influenza season was of the B/Victoria lineage. During the 2011/2012 and 2012/2013 seasons, both lineages co-circulated; the B/Victoria lineage was dominant in the 2011/2012 season, whereas the B/Yamagata lineage was dominant in the 2012/2013 season (http://www.who.int/influenza/gisrs_laboratory/flunet/en/). The trivalent vaccines currently licensed for use in Japan include only one lineage of the influenza B virus; therefore, it is important to conduct surveillance to know which lineage is predominant at any one time to select a candidate virus for the generation of vaccines.
Conventionally, the two lineages of the influenza B virus are differentiated antigenically by the hemagglutination inhibition (HI) test with immune sera raised against each lineage. The HI test is a conventional method to define the type, subtype, and lineage of clinical isolates; however, it is a time-consuming and complicated process that includes the isolation of viruses from clinical specimens and the preparation of immune sera. Recently, PCR-based assays for the rapid diagnosis of influenza viruses have become the gold standard and many real-time RT-PCR (rRT-PCR) assays have been reported (Wang and Taubenberger, 2010). Compared with the HI test, rRT-PCR assays have the advantage of being able to be performed rapidly, sensitively, and specifically using purified RNA from clinical specimens without requiring a viral isolation step.
In Japan, several rRT-PCR assays for typing influenza A and B viruses and subtyping influenza A viruses were developed using minor groove binder (MGB) probes; the assay scheme was established and shared with 74 prefectural and municipal public health institutes for the diagnosis and surveillance of influenza viruses (Nakauchi et al., 2011b). However, rRT-PCR assay for universally detecting influenza B viruses, whose target region was the non-structural protein (NS) gene (B/NS rRT-PCR; the sequences of the primers and probe are listed in Table 1 ), was not able to discriminate between the B/Victoria and B/Yamagata lineages.
Table 1.
Primers and probes.
| Names | Sequences (5′–3′) | Orientation | Positiona |
|---|---|---|---|
| Primers and probe for B/NS rRT-PCR | |||
| NIID-Type B TMPrimer-F1 | GGAGCAACCAATGCCAC | + | 43–59 |
| NIID-Type B TMPrimer-R1 | GTKTAGGCGGTCTTGACCAG | − | 138–147 |
| NIID-Type B Probe 1 | (FAM)ATAAACTTTGAAGCAGGAAT(MGB) | + | 61–80 |
| Primers and probe for B/Vic rRT-PCR | |||
| F3vic v2 | CCTGTTACATCTGGGTGCTTTCCTATAATG | + | 310–339 |
| R3vic v2 | GTTGATARCCTGATATGTTCGTATCCTCKG | − | 378–407 |
| FAM-Type B HA Victoria | (FAM)TTAGACAGCTGCCTAACC(MGB) | + | 356–373 |
| Primers and probes for B/Yam rRT-PCR | |||
| F3yam v2 | CCTGTTACATCCGGGTGCTTYCCTATAATG | + | 310–339 |
| R3yam v2 | GTTGATAACCTKATMTTTTCATATCCTCTG | − | 378–407 |
| FAM-Type B HA Yamagata | (FAM)TCAGGCAACTASCCAATC(MGB) | + | 356–373 |
| VIC-Type B HA Yamagatab | (VIC)TCAGGCAACTASCCAATC(MGB) | + | 356–373 |
| Sequencing primers | |||
| BHA1-N | AATATCCACAAAATGAAGGC | + | Noncoding-1–8 |
| BHA1-187F | TGCAAATCTCAAAGGAACAA | + | 168–187 |
| BHA1-400R | GTCCTCCTGGTGCCTTTTCT | − | 426–445 |
| BHA1-703-722F | CCTCAAAAGTTCACCTCATC | + | 673–692 |
| BHA1-802R | GCACCATGTAATCAACAACA | − | 783–802 |
| BHA1-C | AGCAATAGCTCCGAAGAAAC | − | 1107–1088 |
The nucleotide positions of the NS and HA genes are based on cRNA sequences obtained from the GISAID database. The isolate ID number for the NS gene of B/Florida/4/2006 is EPI_ISL_22808, and those for the HA genes of B/Brisbane/60/2008 and B/Massachusetts/02/2012 are EPI_ISL_24365 and EPI_ISL_117042, respectively.
The VIC-labeled probe was used for the duplex one-step rRT-PCR assay.
An rRT-PCR assay for discriminating the B/Victoria and B/Yamagata lineages was first reported by Biere et al. (2010). However, the sequences of the forward primer and the two probes used in the study of Biere et al. had low homology with the recent influenza B viruses circulating in Japan. Another rRT-PCR assay for discriminating these two lineages was reported by Zhang et al. (2012); however, this method did not use MGB probes, resulting in different conditions for this assay compared to the assay used in Japan.
To monitor rapidly and simply the circulating situation of these two lineages of influenza B virus and as a complement to the HI test, rRT-PCR assays for discriminating the B/Victoria and B/Yamagata lineages (B/Vic rRT-PCR and B/Yam rRT-PCR, respectively) were developed by using MGB probes in this study. The highly sensitive and specific rRT-PCR assays for detecting and discriminating these two lineages of influenza B virus will be useful for the diagnosis and surveillance of influenza B viruses.
2. Materials and methods
2.1. Primer and probe design
The nucleotide sequences of the hemagglutinin (HA) gene from influenza B viruses posted over the past 5 years in the Global Initiative on Sharing Avian Influenza Data (GISAID) database were aligned using ClustalW software (Larkin et al., 2007). On the basis of the sequences of the HA gene, 2 probes were designed to discriminate between the B/Victoria and B/Yamagata lineages in which 6 nucleotides (nt) out of the 18 nt probe sequence were different between these lineages. The sequences and positions of the primers and probes are listed in Table 1.
2.2. Viruses
All influenza viruses included in this study were isolated using Madin–Darby Canine Kidney (MDCK) cells. In addition to B/Florida/4/2006 (B/Yamagata), B/Brisbane/60/2008 (B/Victoria), and B/Massachusetts/2/2012 (B/Yamagata), several influenza A viruses were used to validate the specificity of the assays: A/Uruguay/716/2007 (H3N2), A/Perth/16/2009 (H3N2), A/Narita/1/2009 (H1N1)pdm09, and A/California/07/2009 (H1N1)pdm09.
The following the viral respiratory pathogens (the clinical isolates) were used to validate the specificity of the assays: respiratory syncytial viruses A and B; human parainfluenza virus types 1–4; human rhinovirus types A and B; human metapneumovirus types A1 and B2; human coronaviruses OC43, 229E, HKU1, and NL63; human bocavirus; human enterovirus; and human adenoviruses 2 and 4.
2.3. Preparation of RNA transcript controls
To construct an RNA-positive control for the B/NS, B/Vic, and B/Yam rRT-PCR assays, each target gene segment (described in Table 2 ) was amplified by RT-PCR, and the resulting PCR product, containing the T7 promoter, was then transcribed in vitro. The procedure is described in detailed below.
Table 2.
Detection limits of each assay using serial dilutions of in vitro transcribed control viral RNA.
| Template RNA concentration (copies/reaction) | Number of positive replicates/number of tests for each assay (positive %) |
||
|---|---|---|---|
| B/NSa | B/Vicb | B/Yamc | |
| 10 | 6/6 (100) | 6/6 (100) | 6/6 (100) |
| 5 | 6/6 (100) | 6/6 (100) | 6/6 (100) |
| 1 | 0/6 (0) | 1/6 (16.7) | 3/6 (50) |
| 0.1 | 0/6 (0) | 0/6 (0) | 0/6 (0) |
The template RNA for each assay was the NS gene segment of a B/Florida/4/2006 and the HA gene segments of b B/Brisbane/60/2008 and c B/Massachusetts/02/2012.
The primer Uni8 (5′-GCAGAAGC-3′) (Zou, 1997) was used for RT using a SuperScript® III Reverse Transcriptase Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. The NS gene segment of B/Florida/4/2006 was amplified using the following primer pair: TypeB-NS-F(−22–6) (5′-TAGTCACTGGCAAACAGGAAAAATGGCG-3′) and TypeB-NS-R(+1–+29) + T7 (5′-TAATACGACTCACTATAGGGGTAGTAACAAGAGGATTTTTATTTTAAAT-3′). The HA gene segments of the B/Brisbane/60/2008 (B/Victoria lineage) and B/Massachusetts/2/2012 (B/Yamagata lineage) viruses were amplified using the following primer pairs: FLUB Vic HA-F (5′-ATGAAGGCAATAATTGTACTACTCATGGTAGTAACATCC-3′) and FLUB HA-R T7 (5′-TAATACGACTCACTATAGGGTTATAGACAGATGGAGCA-3′), and FLUB Yam HA-F (5′-ATGAAGGCAATAATTGTACTACTAATGGTAGTAACATCC-3′) and FLUB HA-R T7, respectively. PCR was performed using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific). The PCR product was gel purified using a QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) and transcribed using the T7 RiboMAX™ Express Large Scale RNA Production System (Promega, Madison, WI) according to the manufacturers’ instructions. After TURBO® DNase (Thermo Fisher Scientific) digestion to remove the residues of the RT-PCR products, the transcribed RNA was purified twice using the TRIzol Reagent (Thermo Fisher Scientific), and then the dNTPs and NTPs were removed using MicroSpin G-25 Columns (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. The purified transcribed RNA was quantified by spectrophotometric analysis and its integrity was assessed with a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA). Total RNA was prepared as described in Section 2.6.
2.4. Sequencing of the partial HA gene of a clinical isolate
To confirm the lineage of a clinical isolate, the HA1 region of the HA gene of B/Tokyo/F11-015/2011 was sequenced. To amplify the HA1 region, using extracted RNA, RT and PCR were carried out as described in Section 2.3 with the paired primers BHA1-N and BHA1-C (listed in Table 1) for PCR. The PCR products were purified and then sequenced using 6 primers (listed in Table 1). Total RNA was prepared as described in Section 2.6.
2.5. Clinical specimens
Nasal and/or pharyngeal swabs collected from patients with influenza-like illness and suspended in a UTM 360 C Kit (Copan, Brescia, Italy) were obtained from Showa General Hospital, Japan. These specimens were collected between 2010 and 2013. The study protocol was approved by the Ethics Committee at NIID, and the study was performed in compliance with the Declaration of Helsinki. Informed consent was obtained from all patients.
2.6. RNA preparation
Supernatants from cultured MDCK cells were centrifuged at 10,000 × g for 10 min. Viral RNA was prepared from 140 μL of the supernatant using a QIAamp® Viral RNA Kit (QIAGEN) according to the manufacturer's instructions with the slight modification that the viral RNA was eluted in 70 μL AVE buffer (QIAGEN).
Total RNA was prepared from the clinical specimens using a QIAamp® Viral RNA Kit (QIAGEN) (using 140 μL of clinical specimen) or MagMAX™ 96 Viral Isolation Kit (Thermo Fisher Scientific) (using 50 μL of clinical specimen) with KingFisher Flex (Thermo Fisher Scientific) according to the manufacturers’ instructions. Total RNA from the 140-μL clinical specimens was eluted with 60 μL AVE buffer (QIAGEN), whereas total RNA from the 50-μL clinical specimens was eluted with 30 μL elution buffer (Thermo Fisher Scientific).
2.7. One-step rRT-PCR assay
The reaction was performed using a QuantiTect® Probe RT-PCR Kit (QIAGEN) according to the manufacturer's instructions. Briefly, the 25 μL assay contained 12.5 μL of 2× QuantiTect Probe PCR Master Mix, 0.25 μL QuantiTect RT Mix, 0.1 μL RNase Inhibitor (Thermo Fisher Scientific), 1.5 μL of 10 μM forward primer, 1.5 μL of 10 μM reverse primer, 0.5 μL of 5 μM probe, 3.65 μL distilled water, and 5 μL RNA template. Cycling was performed as follows: 30 min at 50 °C to activate RT, followed by an initial denaturation step for 15 min at 95 °C and 45 cycles of amplification (denaturation at 95 °C for 15 s and annealing as well as extension at 56 °C for 75 s) using a LightCycler® 480 (Roche Molecular Biochemicals, Basel, Switzerland). Fluorescent signals were collected during the annealing and extension steps, and the amplification data were analyzed using Light Cycler® 480 SW1.5 software according to the manufacturer's instructions. The rRT-PCR assays for typing influenza A and B viruses and subtyping H1pdm and H3 (Nakauchi et al., 2011b) were also performed for the diagnosis of the clinical specimens.
2.8. One-step duplex rRT-PCR assay
The reaction was performed using a QuantiTect® Virus + ROX Vial Kit (QIAGEN) according to the manufacturer's instructions and as described previously (Nakauchi et al., 2011a). For testing the clinical isolates using cultured medium, the 20 μL assay contained 4 μL of 5× QuantiTect Virus NR Master Mix, 0.2 μL QuantiTect Virus RT Mix, 1.2 μL each of two 10 μM forward primers, 1.2 μL each of two 10 μM reverse primers, 0.4 μL each of two 5 μM probes, 7.6 or 8 μL distilled water, and 2 μL culture medium. For testing the clinical specimens using extracted RNA, 5 μL of RNA template were used. Cycling was performed as follows: 20 min at 50 °C to activate RT, followed by an initial denaturation step for 5 min at 95 °C and 45 cycles of amplification (denaturation at 95 °C for 15 s and annealing as well as extension at 56 °C for 45 s) using a LightCycler® 480 (Roche Molecular Biochemicals). Fluorescent signals were collected during the annealing and extension steps, and the amplification and endpoint data were analyzed using Light Cycler® 480 SW1.5 software according to the manufacturer's instructions.
2.9. HI test
Ferret antisera raised against MDCK-grown B/Brisbane/60/2008 (B/Victoria lineage) and B/Massachusetts/2/2012 (B/Yamagata lineage) viruses were treated with RDEII (Denka Seiken Co., Tokyo, Japan) to remove nonspecific inhibitors. A 1:10 dilution of the treated sera was prepared with phosphate-buffered saline. Two-fold serial dilutions of sera were mixed with 4 HA units of antigen virus per well and preincubated for 60 min in 96-well plates at room temperature; 0.5% turkey red blood cells were added, and the plate was incubated for 45 min at room temperature. HI titers of the sera were determined as the highest dilution that did not display hemagglutinating activity (http://apps.who.int/iris/bitstream/10665/44518/1/9789241548090_eng.pdf:1-153).
3. Results
3.1. Development of the rRT-PCR assays
The specificity of the B/NS rRT-PCR assay for detecting both lineages of influenza B virus, and the B/Vic and B/Yam rRT-PCR assays for discriminating the two lineages of influenza B virus were evaluated using B/Victoria, B/Yamagata, A/H3N2, and A/H1N1pdm influenza viruses and other viral respiratory pathogens. The B/NS rRT-PCR assay reacted specifically with both lineages of influenza B virus (data not shown). The B/Vic rRT-PCR assay reacted specifically with B/Victoria, but not with B/Yamagata, whereas the B/Yam rRT-PCR assay reacted specifically with B/Yamagata, but not with B/Victoria (data not shown). All three assays showed no cross-reactivity against influenza A viruses or other viral respiratory pathogens (data not shown).
The detection limit of each assay was determined by performing serial dilutions of in vitro transcribed control viral RNA for 6 replicates in each assay. The detection limits of the B/NS, B/Vic, and B/Yam rRT-PCR assays were determined to be 8.27, 4.31, and 3.85 copies/reaction, respectively, calculated by a Probit analysis using the results shown in Table 2. A standard curve of each assay was also generated (Fig. 1 ). The standard curve showed a linear relationship between the log of the viral titer and the crossing point (Cp) value for all assays (Fig. 1). The correlation coefficient of the standard curve was 0.99 for all assays, indicating a precise log–linear relationship between the viral titer and Cp value (Fig. 1).
Fig. 1.
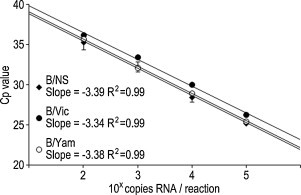
Standard curves of each assay. Ten-fold serial dilutions of synthesized RNA were used for each rRT-PCR assay performed with six replicates. The standard curves were generated using the average Cp values obtained from the assay performed with six replicates. The correlation coefficients and slope of the standard curves are represented in the graph. The standard curves were made based on the B/NS rRT-PCR assay performed using a synthesized NS gene segment of B/Florida/4/2006 (diamonds) and the B/Vic and B/Yam rRT-PCR assays performed using a synthesized HA gene segment of B/Brisbane/60/2008 (filled circles) and B/Massachusetts/02/2012 (open circles), respectively.
3.2. Validation of the rRT-PCR assays using clinical specimens
From 2010 to 2013, clinical specimens (nasal and/or pharyngeal swabs) from patients with an influenza-like illness (n = 169) were obtained, and the B/NS rRT-PCR assay was performed on these samples. As a result, 20 out of 149 samples were identified as positive for influenza B virus. Twenty B/NS-positive samples were then tested using the B/Vic and B/Yam rRT-PCR assays, revealing that 14 and 6 samples were identified as positive for the B/Victoria and B/Yamagata lineages, respectively.
3.3. Validation of the rRT-PCR assays using clinical isolates
Clinical isolates from 20 influenza B virus-positive clinical specimens (listed in Table 3 ) were tested using the B/NS, B/Vic, and B/Yam rRT-PCR assays and also by the conventionally performed HI test. As shown in Table 3, 14 and 6 clinical isolates were identified as positive for the B/Victoria and B/Yamagata lineages, respectively, consistent with the results of the three rRT-PCR assays on the original clinical specimens. Antiserum against B/Massachusetts/2/2012 (B/Yamagata) reacted with all 6 B/Yam-positive clinical isolates. Antiserum against B/Brisbane/60/2008 (B/Victoria) reacted with 13 B/Vic-positive clinical isolates; however, it reacted poorly with one B/Vic-positive clinical isolate, B/Tokyo/F11-015/2011, with 8-fold-reduced HI titers from the homologous titer. To identify the cause of the low reaction of the antiserum against B/Brisbane/60/2008 with B/Tokyo/F11-015/2011, the HA1 region (1–341 amino acids) of the HA gene was sequenced and compared with the HA1 region of B/Brisbane/60/2008. This revealed that B/Tokyo/F11-015/2011 has 8 amino acid mutations (N26D, T52I, K90N, I161V, K180N, K182R, P187S, and A217T).
Table 3.
Comparing the HI test and rRT-PCR assays using clinical isolates.
| Influenza B virus | HI titera of postinfection ferret antisera |
rRT-PCRb |
|||
|---|---|---|---|---|---|
| B/Brisbane/60/2008 | B/Massachusetts/2/2012 | B/NS | B/Vic | B/Yam | |
| Reference virus | |||||
| B/Brisbane/60/2008 | 80 | <10 | + | + | − |
| B/Massachusetts/2/2012 | <10 | 160 | + | − | + |
| Clinical isolate (test virus) | |||||
| B/Tokyo/SH-F11-015/2011 | 10 | <10 | + | + | − |
| B/Tokyo/SH-F11-026/2011 | 40 | <10 | + | + | − |
| B/Tokyo/SH-F11-030/2011 | 80 | <10 | + | + | − |
| B/Tokyo/SH-F11-031/2011 | 80 | <10 | + | + | − |
| B/Tokyo/SH-F11-032/2011 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-068/2012 | 160 | 10 | + | + | − |
| B/Tokyo/SH-F12-082/2012 | 10 | 160 | + | − | + |
| B/Tokyo/SH-F12-099/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-103/2012 | 10 | 160 | + | − | + |
| B/Tokyo/SH-F12-138/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-143/2012 | 10 | 160 | + | − | + |
| B/Tokyo/SH-F12-155/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-157/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-159/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-161/2012 | <10 | 160 | + | − | + |
| B/Tokyo/SH-F12-165/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-170/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F12-173/2012 | 160 | <10 | + | + | − |
| B/Tokyo/SH-F13-066/2013 | 10 | 160 | + | − | + |
| B/Tokyo/SH-F13-068/2013 | 10 | 160 | + | − | + |
The underlined text indicates the homologous titers of the antisera.
“+” and “−” indicate “positive” and “negative” for each rRT-PCR assay.
3.4. Application of the B/VIC and B/Yam RT-PCR assays to the duplex rRT-PCR assay
To facilitate an assay to discriminate between the two lineages, the primers and probes for the B/VIC and B/Yam rRT-PCR assays were mixed and the duplex rRT-PCR assay was constructed. For the duplex rRT-PCR assay, the probe for B/Victoria was labeled with FAM and the probe for B/Yamagata was labeled with VIC (as listed in Table 1). Using 20 clinical specimens and 20 clinical isolates that were identified as positive for influenza B virus, the duplex rRT-PCR assay was evaluated. For the clinical isolates, the assay was performed by using the cultured supernatants from infected MDCK cells directly, without RNA purification, as the template of the assay for screening a large number of clinical isolates. For the clinical specimens, purified RNA was used for the assay. As shown in Fig. 2 , 14 and 6 clinical specimens were determined as B/Victoria and B/Yamagata, respectively. Similarly, 14 and 6 clinical isolates were determined as B/Victoria and B/Yamagata, respectively (Fig. 2).
Fig. 2.
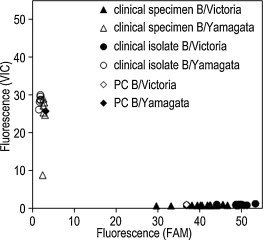
Endpoint fluorescence plot of the duplex rRT-PCR assay using clinical specimens and isolates. The relative fluorescence of B/Victoria (FAM) and B/Yamagata (VIC) is plotted on the x-axis and y-axis, respectively. The clinical specimens discriminated as B/Victoria or B/Yamagata are represented as filled or open triangles, and the clinical isolates discriminated as B/Victoria or B/Yamagata are represented as filled or open circles, respectively. One hundred copies each of the synthesized HA gene segment of B/Brisbane/60/2008 (PC B/Victoria, open diamond) and B/Massachusetts/2/2012 (PC B/Yamagata, filled diamond) were used as positive controls for the assay.
4. Discussion
The B/NS rRT-PCR assay used for detecting influenza B viruses and the B/Vic and B/Yam rRT-PCR assays used for discriminating the two lineages of the influenza B virus, were shown to have good linearity (R 2 = 0.99) and high sensitivity (Table 2 and Fig. 1). No cross-reactivity or nonspecific reactions were observed in any of the assays performed using isolated viruses and clinical specimens (data not shown). In the assay performed using clinical specimens, 20 samples were identified as positive for influenza B virus and 149 samples were negative in the B/NS rRT-PCR assay. Of the 149 B/NS-negative samples, 81 and 3 samples were identified as positive for A/H3 and A/H1pdm by performing the previously developed Type A, H1pdm, and H3 rRT-PCR assays (Nakauchi et al., 2011b), and no influenza A or B viruses were detected in the remaining 65 samples. All 20 B/NS-positive clinical samples were discriminated either as B/Victoria or B/Yamagata by the B/Vic and B/Yam rRT-PCR assays, respectively. These results suggest that these three rRT-PCR assays are highly sensitive and specific for each target gene and should thus be of great use for detecting and discriminating the two lineages of the influenza B virus.
The results of the HI test and the B/Vic and B/Yam rRT-PCR assays were compared using 20 clinical isolates (Table 3) from 20 influenza B virus-positive clinical samples. The results of rRT-PCR assays corresponded to those of the HI test with one exception. B/Tokyo/F11-015/2011 and its original specimen were identified as positive for the B/Victoria lineage by the B/Vic and B/Yam rRT-PCR assays; however, antiserum against B/Brisbane/60/2008 reacted poorly with B/Tokyo/F11-015/2011. It was found that B/Tokyo/F11-015/2011 has eight different amino acids, namely, N26D, T52I, K90N, I161V, K180N, K182R, P187S, and A217T, in the HA1 region when compared with B/Brisbane/60/2008. The amino acid positions at 182 and 217, where B/Tokyo/F11-015/2011 has mutations, were reported as antigenic sites by analyzing escape mutants from monoclonal antibodies (Tung et al., 2004), and the Thr to Ile mutation at position 52 is also known to affect antigenicity (unpublished data from the Influenza Surveillance Group of Japan). Considering these data, B/Tokyo/F11-015/2011 is antigenically different from B/Brisbane/60/2008; thus, the HI test could not identify B/Tokyo/F11-015/2011 as B/Victoria.
The HI test has been performed conventionally for determining type, subtype, and lineage of influenza viruses and for detecting antigenic differences between reference and test viruses. The HI test is a very useful method for monitoring the antigenicity of circulating influenza viruses in humans to select a virus for vaccine production; however, the HI test is not able to detect influenza viruses when the test virus is antigenically different from the reference virus. The combination of the B/NS, B/Vic, and B/Yam rRT-PCR assays developed in this study was able to detect and discriminate both lineages of the influenza B virus, including a virus that was undetected by the HI test, with high sensitivity and specificity (Table 3); however, the rRT-PCR assays cannot be used to compare antigenic differences. Therefore, it is important to use the rRT-PCR assays in cooperation with the HI test for conducting surveillance of influenza B viruses.
To facilitate the assay scheme of the B/Vic and B/Yam rRT-PCR assays, a duplex rRT-PCR assay for discriminating the two lineages of the influenza B virus was also constructed. No discrepancy was found between the results from the duplex rRT-PCR assay and the B/Vic and B/Yam rRT-PCR assays for 20 clinical specimens and 20 clinical isolates (Table 3 and Fig. 2). It is noteworthy that the duplex assay could be performed using clinical isolates without an RNA purification step, since a sufficient amount of viral RNA may be released from the virion under the RT conditions (50 °C, 20 min). When considering the handling of a large number of clinical isolates, this assay can be performed rapidly and simply with a lower risk of cross-contamination because an RNA purification step is unnecessary.
In conclusion, the highly sensitive and specific B/NS, B/Vic, and B/Yam rRT-PCR assays developed in this study will be very useful not only for laboratory diagnostic tests but also for speeding up surveillance by discriminating the lineages of influenza B viruses in clinical specimens. Considering that the trivalent vaccines currently licensed for use in Japan include only one lineage of the influenza B virus, the assays are also powerful tools for the rapid recognition of the predominant lineage of influenza B virus at any one time and helpful to decide which candidate virus should be included in vaccines.
Acknowledgment
We appreciate Drs. Noriko Kishida, Hong Xu, and Takato Odagiri, Influenza Virus Research Center, National Institute of Infectious Diseases for providing ferret antisera and for helpful advice to interpret the results of the HI test.
References
- Biere B., Bauer B., Schweiger B. Differentiation of influenza B virus lineages Yamagata and Victoria by real-time PCR. J. Clin. Microbiol. 2010;48:1425–1427. doi: 10.1128/JCM.02116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae Y., Sugita S., Endo A., Ishida M., Senya S., Osako K., Nerome K., Oya A. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J. Virol. 1990;64:2860–2865. doi: 10.1128/jvi.64.6.2860-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Nakauchi M., Ujike M., Obuchi M., Takashita E., Takayama I., Ejima M., Oba K., Konomi N., Odagiri T., Tashiro M., Kageyama T. Rapid discrimination of oseltamivir-resistant 275Y and -susceptible 275H substitutions in the neuraminidase gene of pandemic influenza A/H1N1 2009 virus by duplex one-step RT-PCR assay. J. Med. Virol. 2011;83:1121–1127. doi: 10.1002/jmv.22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakauchi M., Yasui Y., Miyoshi T., Minagawa H., Tanaka T., Tashiro M., Kageyama T. One-step real-time reverse transcription-PCR assays for detecting and subtyping pandemic influenza A/H1N1 2009, seasonal influenza A/H1N1, and seasonal influenza A/H3N2 viruses. J. Virol. Methods. 2011;171:156–162. doi: 10.1016/j.jviromet.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Wallis T.R., Harmon M.W., Rota J.S., Kendal A.P., Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- Tung C.S., Goodman J.L., Lu H., Macken C.A. Homology model of the structure of influenza B virus HA1. J. Gen. Virol. 2004;85:3249–3259. doi: 10.1099/vir.0.80021-0. [DOI] [PubMed] [Google Scholar]
- Wang R., Taubenberger J.K. Methods for molecular surveillance of influenza. Expert Rev. Anti-Infect. Ther. 2010;8:517–527. doi: 10.1586/eri.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.F., Neumann G., Kawaoka Y. Orthomyxoviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virology. fifth ed. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1691–1740. [Google Scholar]
- Zhang N., Fang S., Wang T., Li J., Cheng X., Zhao C., Wang X., Lv X., Wu C., Zhang R., Cheng J., Xue H., Lu Z. Applicability of a sensitive duplex real-time PCR assay for identifying B/Yamagata and B/Victoria lineages of influenza virus from clinical specimens. Appl. Microbiol. Biotechnol. 2012;93:797–805. doi: 10.1007/s00253-011-3710-8. [DOI] [PubMed] [Google Scholar]
- Zou S. A practical approach to genetic screening for influenza virus variants. J. Clin. Microbiol. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


