Abstract
It is likely that phylogroup 2 lyssaviruses circulate within bat reservoirs. We adapted a pseudotype (pt) neutralisation assay (PNA) to a multiplex format enabling serosurveillance for Lagos bat virus (LBV), Mokola virus (MOKV) and West Caucasian bat virus (WCBV) in a potential reservoir, the African straw-coloured fruit bat, Eidolon helvum. Highly correlated titres were observed between single and multiplex PNAs using ptLBV and ptMOKV (r = 0.97, p < 0.0001), validating its use for bat serosurveillance. Of the bat serum samples screened 56% neutralised ptLBV, 27% ptMOKV and 1% ptWCBV. Mean VNAb titres were 1:266, 1:35 and 1:7 against ptLBV, ptMOKV and ptWCBV respectively. The high seroprevalence estimates suggest that the infection rate of LBV in E. helvum remains high enough to persist in this species. This supports the hypothesis that LBV is endemic in Ghanaian E. helvum and we speculate that LBV may have co-evolved with African megachiroptera.
Keywords: Lyssavirus, Rabies virus, Lagos bat virus, Pseudotype, Bat, Africa, Ghana, Antibody, Neutralisation, Serology
Introduction
Substantial evidence exists to suggest lyssaviruses evolved in Chiropteran (bat) hosts. Infection by each Lyssavirus (Family Rhabdoviridae) causes rabies encephalomyelitis and cases are indistinguishable by clinical presentation, regardless of which species infects the individual. All Lyssavirus species, other than Mokola virus (MOKV), are distributed in New or Old World bats (Fooks, 2004), with rabies virus (RABV) routinely isolated from New World bats. Lyssavirus species isolated from Old World bats include Lagos bat virus (LBV), Duvenhage virus (DUVV), European bat lyssavirus type 1 (EBLV 1) and 2 (EBLV 2), Australian bat virus (ABLV), Irkut virus (IRKV), Khujand virus (KHUV) and West Caucasian bat virus (WCBV). MOKV has been isolated from terrestrial mammals only, although surveillance has been limited (Nel et al., 2000). Recently, a putative new species, Shimoni bat virus (SHIBV), was isolated from a Kenyan bat in 2009 (Kuzmin et al., 2010).
There are approximately 1100 species of bats, comprising over 20% of extant mammalian species, which live on every continent other than Antarctica (Teeling et al., 2005). It has become clear that designated “rabies free” nations, such as the UK and Australia, have endemic lyssaviruses circulating within bat populations (Banyard et al., 2010, Warrilow, 2005). In both nations humans have died from lyssavirus infection transmitted by bats (Fooks et al., 2003, Fraser et al., 1996). The geographic distribution of bats together with the apparent ubiquitous infection of bat populations with lyssaviruses means that, with the exception of Antarctica and some oceanic islands, new lyssaviruses may spill-over from these reservoir populations into new potential reservoirs, such as dogs, or to dead-end hosts, such as humans, anywhere on the globe. The true threat to the human and animal populations posed by the spill-over of these viruses is unknown due to the lack of knowledge at both the epidemiological and molecular level (Fooks, 2004). While reported human infections by lyssaviruses other than RABV are rare (Johnson et al., 2010), they are fatal and the real number of cases is unknown due to limited surveillance and misdiagnosis (Mallewa et al., 2007, Nel and Rupprecht, 2007).
Each Lyssavirus species belongs to one of two phylogroups based on their cross neutralisation profile, pathogenicity and genetic relatedness (Badrane et al., 2001, Kuzmin et al., 2009). LBV and MOKV belong to phylogroup 2, with the others comprising phylogroup 1, apart from SHIBV and WCBV, which are awaiting official classification but have tentatively been placed in phylogroup 2 and a putative phylogroup 3, respectively. The World Health Organisation estimates 55,000 human deaths each year from rabies (World-Health-Organisation, 2008), primarily due to RABV circulating globally in domestic dogs and wild carnivores (such as foxes, skunks and racoons). Potential epidemics of viruses against which current biologicals offer no protection (LBV, MOKV, WCBV and possibly SHIBV (Hanlon et al., 2005, Kuzmin et al., 2010)) can only be determined if the infection dynamics of these viruses are understood within reservoir hosts. The absence of a known MOKV reservoir, for example, is a substantial gap in the understanding of Lyssavirus ecology and evolution. While there is a suggestion that shrews (family Soricidae) are reservoirs, the number of isolations from shrews is currently limited to five (Sabeta et al., 2007), with three isolated in a single study (Shope et al., 1970).
Four recognised Lyssavirus species (RABV, MOKV, DUVV, and LBV) and SHIBV have been isolated from African mammals (King et al., 1993, King et al., 1994, Kuzmin et al., 2010, Kuzmin et al., 2008a, Sabeta et al., 2003, Sabeta et al., 2007, van Thiel et al., 2008). LBV reportedly contains at least two clades that are divergent enough to suggest they may represent different species (Delmas et al., 2008, Markotter et al., 2008). RABV and MOKV have never been isolated from bats in Africa, while LBV, DUVV and SHIBV have, with both LBV and DUVV occasionally isolated from other mammals (Kuzmin et al., 2010, Kuzmin et al., 2008a, Sabeta et al., 2007, van Thiel et al., 2008). Africa therefore possesses the greatest known Lyssavirus diversity, both genetically and serologically. Given that such diversity exists in Africa, it has been hypothesised that early evolution and divergence of lyssaviruses occurred in African bats (Nel and Rupprecht, 2007). Recently, surveillance programs and greater access to serosurveillance techniques have resulted in the discovery of a high seroprevalence against LBV in West (3–37%) and East (29–67%) African fruit bats (Dzikwi et al., 2010, Hayman et al., 2008a, Kuzmin et al., 2008a). WCBV has not been isolated from bats in Africa, the only isolation of WCBV was from Miniopterus bats in the West Caucasus (Botvinkin et al., 2003), however a high seroprevalence of anti-WCBV antibodies was detected in African Miniopterus when the appropriate study was undertaken (Kuzmin et al., 2008b). These reports and others (Cleaveland, 1998, Knobel et al., 2005) highlight the limited understanding of lyssavirus epidemiology.
Recently, Streicker et al. (2010) suggested that the phylogenic distance between bat species is an important factor in cross-species transmission and host shifts of rabies viruses in North America. Coupled with the probable co-evolution of lyssaviruses and bats it is possible that the species specificity shown for those lyssaviruses isolated in Eurasia, WCBV and European bat lyssaviruses 1 and 2, in epidemiological studies to-date could be caused by an intrinsic innate restriction factor(s) similar to those reported for retroviruses (Neil and Bieniasz, 2009). However, given LBV has been isolated from five different species of frugivorous and insectivorous bats, other behavioural, ecological or cell entry restrictions could also be important factors in determining lyssaviruses susceptibility.
Given that bats can be extraordinarily small, with many common species weighing less than 5 g, handling animals and, in particular, bleeding live animals to obtain adequate volumes of sera is particularly difficult. Indeed, several seroprevalence studies report pooling sera as a necessary method to enable serological tests to be undertaken, such as a study of EBLV-2 in Scotland (Brookes et al., 2005). Current studies of bat lyssavirus ecology and evolution in Africa are hampered by resource-limited laboratories and the requirement for large volumes of sera for routine virus neutralising antibody (VNAb) tests. In this study we adapted a lyssavirus pseudotype neutralisation assay (PNA; Wright et al., 2009, Wright et al., 2008) to a multiplex format allowing in-depth serosurveillance to be undertaken. Spill-over of phylogroup two lyssaviruses has the potential to cause fatal zoonotic transmission events to dead-end hosts, such as humans, or emergence in terrestrial carnivores. Due to the limited data available it is unknown whether these viruses are endemic within bat populations or whether there is any species specificity. In order to address this we undertook investigations to determine if the previously reported high seroprevalence of anti-LBV antibodies in Ghanaian fruit bats (Eidolon helvum [E. helvum]) was due to a recent epidemic within the colony or due to endemic infection. In addition, we tested the hypothesis that lyssavirus circulation is species specific so undertook seroprevalence studies of LBV, MOKV (phylogroup 2) and WCBV (putative phylogroup 3) in a potential reservoir, E. helvum, using a novel multiplexed PNA assay.
Results
Characteristics of bats sampled for study
Details of the bats sampled are provided in Table 1 . Briefly, bats were caught in three roosting colony sites in urban (Accra and Kumasi) or rural (Tanoboase) Ghana (Fig. 1A), 184 serum samples were collected by manually restraining the animals (Fig. 1B). Classification, using a dichotomous key as previously described (Hayman et al., 2008a), revealed 183 were from E. helvum and a single sample was from a sexually mature male Hypsignathus monstrosus caught in Tanoboase. Eighty-one percent of the E. helvum sampled for this study were males and the majority of bats, 62%, were sexually mature males or females.
Table 1.
Characteristics of Eidolon helvum caught for this study.
| Sex | Age | Location |
||
|---|---|---|---|---|
| Accra | Kumasi | Tanoboasea | ||
| Female | SI | 5 | 4 | 17 |
| SM | 1 | 2 | 6 | |
| Male | SI | 23 | 4 | 17 |
| SM | 64 | 24 | 16 | |
| Total | 93 | 34 | 56 | |
In addition, a H. monstrosus bat was caught at this location. SI—sexually immature. SM—sexually mature.
Fig. 1.
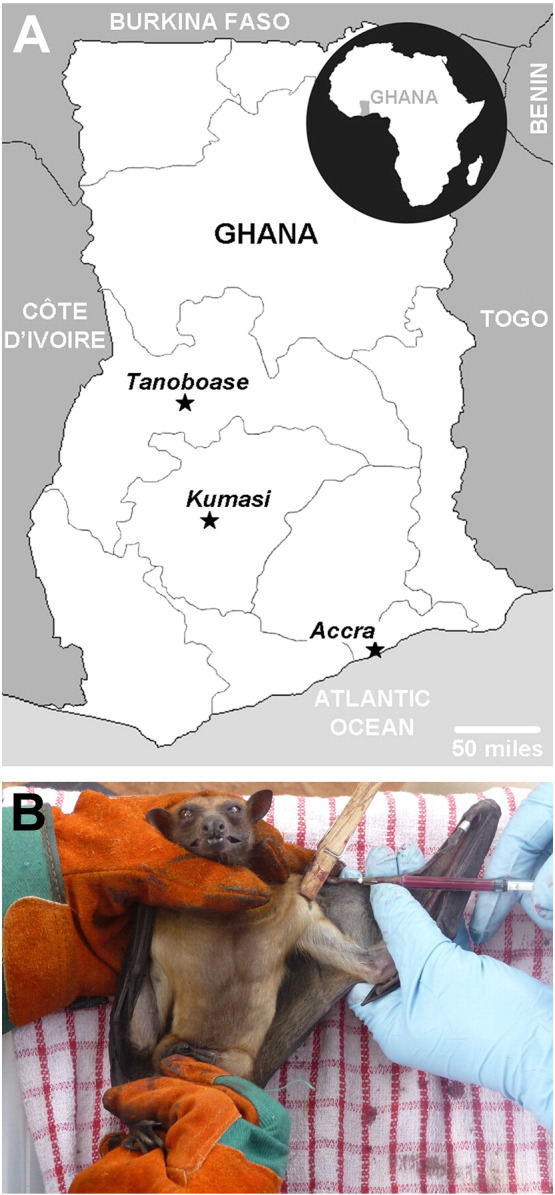
A map of Ghana showing the three Eidolon helvum colonies from which bats were sampled, indicated by black stars (A). Bats were caught in mist nets with blood samples taken from the propatagial vein (B).
Strongly correlated titres between mFAVN and standard and multiplex pseudotype neutralisation assays for LBV and MOKV serology
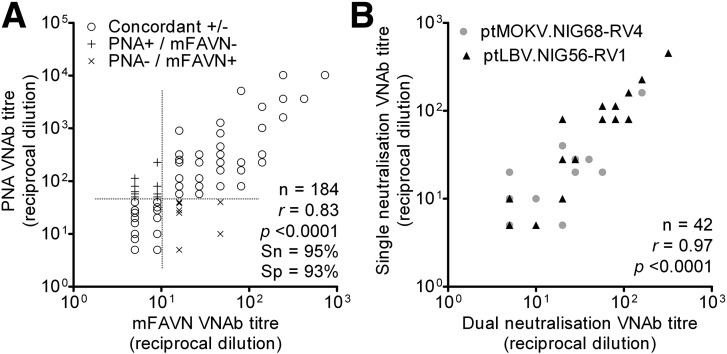
We undertook a definitive study comparing titres achieved in LBV PNA and modified fluorescent antibody virus neutralisation assays (mFAVN) for 184 bat serum samples. Analysis of the VNAb titres obtained for each serum sample using the PNA or mFAVN reveals a correlation of 0.83 (p < 0.0001; Pearson's product-moment correlation; Fig. 2A). As no serum sample neutralised ptRABV challenge virus standard 11 (CVS-11) above a dilution of 1:20 (data not shown), any sample that scored > 1:40 was classified as VNAb positive. Using these parameters, both the sensitivity and specificity of the PNA were high, 95% and 93% respectively, with nine serum samples classified as negative by the PNA but positive by the mFAVN and 12 positive in the PNA but negative by mFAVN (Fig. 2A). It should be noted that discordant results for both assays occur only at low VNAb titres.
Fig. 2.
Comparison of serum VNAb titres achieved using the LBV PNA or mFAVN assays. A high correlation (r), sensitivity (Sn) and specificity (Sp) was observed between the PNA and mFAVN (A). Samples that were classified as positive or negative in both assays (Concordant +/−) are shown by circles. Discordant samples classified as positive in the PNA and negative in the mFAVN or vice versa are highlighted by + and x, respectively. The dotted line corresponds to the cut-off dilution in each assay. (B) VNAb titres strongly correlate between single and dual PNAs. ptLBV.NIG56-RV1 (black triangles) and ptMOKV.NIG68-RV4 (grey circles) VNAb titres were determined in separate wells (single) or in the same well (multiplex). Titres reported are IC100 endpoint reciprocal dilutions r and p values were calculated using Pearson's product-moment correlation.
To obtain as much information as possible from the samples a multiplex platform was incorporated into the PNA. This made it possible to measure VNAb titres against two different lyssaviruses in the same well using an equal volume of sera as the standard PNA. A pilot experiment was undertaken with 21 bat serum samples using ptLBV carrying the renilla luciferase gene and ptMOKV encoding the firefly luciferase gene. Standard and dual PNAs were undertaken for each sample. Analysis of VNAb titres obtained with each assay reveals a strong correlation between the results of the standard and dual PNAs (0.97; p < 0.0001; Pearson's product-moment correlation; Fig. 2B). A strong correlation was also observed when similar experiments were undertaken but this time using ptEBLV-1 (RV9) with the renilla luciferase reporter, ptEBLV-2 (RV1787) with the firefly luciferase reporter and sera that covered a range of titres from dogs and rabbits, previously immunised with commercial RABV vaccines or EBLV-1 and -2 isolates respectively (0.86; p < 0.0001; Pearson's product-moment correlation; Supplementary Fig. S1).
Figure S1.
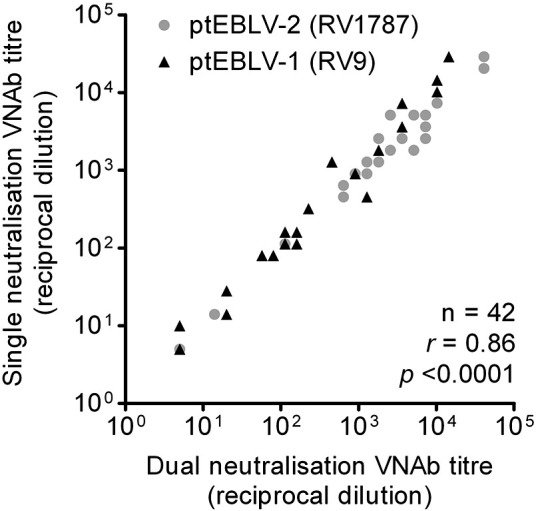
Strong correlation of VNAb titres between single and dual PNA using ptEBLV-1 (RV9; black triangles) and ptEBLV-2 (RV1787; grey circles) pseudotypes carrying the renilla and firefly reporter gene respectively. VNAbs were detected in separate assays/wells (single) or in the same assay/well (dual) and are reported as IC100 endpoint reciprocal dilutions.
As there is cross-neutralisation between lyssavirus species LBV and MOKV we had anticipated greater variation between low VNAb titres obtained using the single or dual assay due to saturation of the antibodies available with the antigen. While the correlation of ptLBV and ptMOKV VNAb titres achieved using the single or dual PNAs for samples with a VNAb titre in the single PNA of less than the mean (1:48) was lower than for those above the mean a significantly strong correlation is maintained (r = 0.69 [p < 0.0001] and 0.92 [p < 0.0001] respectively). With the ptEBLV-1 and ptEBLV-2 single and dual PNA data (Supplementary Fig. S1) samples with a VNAb titre, in the single PNA, of less than the mean (1:4046) had a stronger correlation than for those with a titre greater than the mean (r = 0.86 [p < 0.0001] and 0.77 [p = 0.006], respectively).
Potent neutralising activity of Ghanaian bat sera against phylogroup 2 lyssavirus
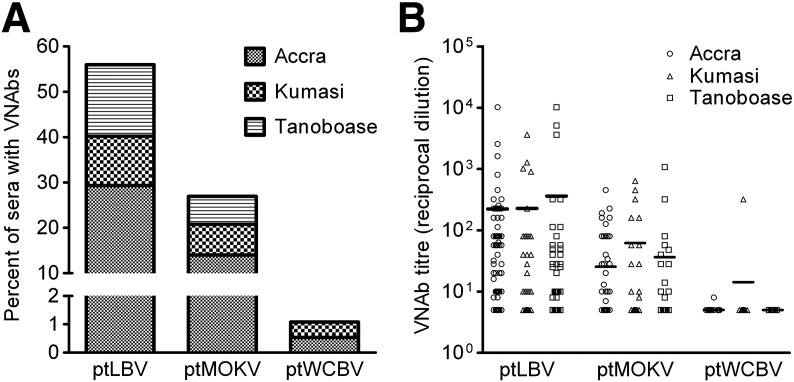
Of the 184 serum samples screened, 56% (n = 103) neutralised ptLBV, 27% (n = 48) neutralised ptMOKV and 1% (n = 2) neutralised ptWCBV (Fig. 3A). In Accra 58%, 27% and 1% of the bat sera neutralised ptLBV, ptMOKV and ptWCBV respectively. In Kumasi and Tanoboase, ptLBV, ptMOKV and ptWCBV were neutralised by 59%, 36% and 3% and 51%, 21% and 0% serum samples respectively. Eleven percent of serum samples neutralised both ptLBV and ptMOKV, 1% of samples neutralised ptLBV and ptWCBV or ptMOKV and ptWCBV, while only 2 samples neutralised all 3 pseudoviruses. The mean VNAb titre across all serum samples tested was 1:266 for ptLBV, 1:35 for ptMOKV and 1:7 for ptWCBV (Fig. 3B). There was no statistical difference in VNAb percentage of neutralising sera or titres among the three sampling sites (p < 0.05; Kruskal–Wallis test).
Fig. 3.
Lyssavirus neutralisation by Eidolon helvum serum samples. Percentage of serum samples that had neutralising activity (A) and VNAb titres (B) against pseudotyped LBV, MOKV or WCBV. Results for Accra, Kumasi and Tanoboase are denoted by small hatching/circles, large hatching/triangles and horizontal lines/squares in A and B, respectively. In (B) horizontal bars represent mean VNAb titres, given as IC100 endpoint reciprocal dilutions. n = 184 for ptLBV and ptWCBV and n = 178 for ptMOKV.
Four (2%) samples neutralised ptMOKV but not ptLBV or ptWCBV and 11 (6%) samples had a higher VNAb titre against ptMOKV than ptLBV. The single H. monstrosus was seropositive against both ptLBV (1:10240) and ptMOKV (1:1076). The two samples that neutralised ptWCBV were collected in Accra and Kumasi and had geometric mean IC100 endpoint reciprocal dilution titres of 1:320 and 1:8. Both also neutralised ptLBV (1:1280 and 1:226 respectively) and ptMOKV (1:640 and 1:10 respectively) but neither sample neutralised ptCVS-11.
Discussion
Where active surveillance of bats is undertaken, detection of VNAb and/or viral RNA suggests that the lyssavirus species detected circulate within the colonies. Understanding virus ecology in bats is particularly difficult given the behaviour and ecology of the hosts (Freuling et al., 2009, Vos et al., 2007). Active infections and antibodies to RABV variants have been reported in numerous bat species in North and South America (Belotto et al., 2005) but not in bats on other continents. In the Old World there are many more species of lyssaviruses, the majority of which have been isolated from bats (Botvinkin et al., 2003, Kuzmin et al., 2005, Kuzmin et al., 2010). Our understanding of how viruses persist in different bat populations is very limited, particularly outside the Americas. The ecology of EBLV-2, for example, is poorly understood but it appears to be endemic in designated “rabies free” nations such as the UK (Banyard et al., 2010, Banyard et al., 2009). In Africa there exists greater lyssavirus diversity, and there is intriguing evidence that LBV, WCBV and SHIBV might be circulating within different bat species in the same roost (Kuzmin et al., 2010, Kuzmin et al., 2008b).
The advantages of using pseudotyped viruses as alternatives to handling live virus have been utilised by many laboratories for over two decades. They have primarily been used as gene delivery or gene therapy vectors (Naldini et al., 1996, Zufferey et al., 1997) but more recently as antigen substitutes in PNAs evaluating antiviral drug potency (Aljofan et al., 2009, Parry et al., 2009, Su et al., 2008) and serum VNAb titres (Temperton et al., 2005, Temperton et al., 2007, Wright et al., 2008), as vaccine immunogens (Szecsi et al., 2006, Szecsi et al., 2009, Yang et al., 2007) or target antigens in antibody binding assays (Wang et al., 2010). Also, PNAs have been shown to aid the identification of more potent and broader cross-neutralising MAbs than has been possible using other neutralisation assays (Corti et al., 2010). The strong correlation (0.83) and the high sensitivity/specificity (95/93%) between LBV PNA and mFAVN VNAb titres observed in this study further validates PNA as a useful alternative to existing serological assays. Similar PNAs are used widely in influenza research to answer important epidemiological and biological questions with equally accurate results as live virus assays (Alberini et al., 2009, Garcia et al., 2009).
We have further added to the flexibility of the lyssavirus PNA scaffold by including a multiplex platform that could prove valuable for serosurveillance in bats. By incorporating different luciferase reporter genes in the viral genome and different envelope glycoproteins on the viral membrane of separate pseudotype preparations it is now possible to detect VNAb titres to two viruses in one serum dilution. This holds great promise for bat serology studies where the scales of investigations are usually limited by the volume of serum collected. With this multiplex PNA it is possible to use ≤ 10 μl of sera to obtain VNAb titres to two viruses, twice as much information than with the standard PNA. We postulate that it is possible to increase this capacity further by including pseudotyped viruses encoding fluorescent reporter genes, such as GFP or RFP, as these signals could be read prior to cell lysis for the luciferase measurements. It would then theoretically be possible to undertake serosurveillance for many highly pathogenic viruses that have been successfully pseudotyped, for example, filoviruses, coronaviruses, henipaviruses and other lyssaviruses, in low containment laboratories using one serum dilution series and a tiny volume of serum compared with that required for conventional assays.
In this study, we observed a similar seroprevalence of LBV and MOKV antibodies in E. helvum as had been reported by other African serologic studies (Hayman et al., 2008a, Kuzmin et al., 2008a) and a low seroprevalence of WCBV in Ghanaian E. helvum. The high percentage of LBV serum positive bats (56%), approximately 2 years after Hayman et al. (2008a) first detected anti-LBV antibodies in E. helvum in Ghana, provide support for our hypothesis that LBV is endemic within this bat population. It is currently unknown whether these seropositive animals have ever been infectious and recovered from a neurological infection or whether the seropositivity is evidence of peripheral infection that has been prevented from reaching the nervous system by the host's immune response. In addition, the difficulties of aging adult wild animals and the problems of undertaking capture-mark-recapture studies in such large populations of flying wild animals currently limit our knowledge regarding age-specific seroprevalence and antibody titre decay rates in this virus–host system. Until these data are available further conclusions regarding virus infection dynamics within this population will be limited, whether for lyssaviruses or other viral infections against which antibodies have been detected in E. helvum (Hayman et al., 2008b, Hayman et al., 2010). However, our results suggest that the risk to humans and domestic animals may be present wherever this species occurs in close proximity to them, especially given that they are a food source across much of Africa (Mickleburgh et al., 2009). A small proportion (≤ 11%) of the bat serum neutralised more than one of the lyssaviruses which is notable. It is probable that some of the ptMOKV neutralisation observed is due to cross-reaction with LBV VNAb as this has been reported previously from this species in Kenya and Nigeria (Dzikwi et al., 2010, Kuzmin et al., 2008a). However, we detected four serum samples which neutralised ptMOKV but not ptLBV and 11 with a higher VNAb titre against ptMOKV than ptLBV. There are different possible explanations for these data. The variable response individuals show heterologous antigens might explain the variation shown with these animals (Kuzmin et al., 2008a, Smith et al., 2004). The dynamic nature of these biological tests might also affect the VNAb titre, with test-to-test variation being possible. Also, at the genomic level the domains containing the major immuno-dominant epitopes might appear different but when folded the antigenic sites might adopt similar conformations. Alternatively, these data might show true evidence of previous exposure to MOKV, although we believe that the variable immune response to heterologous antigen is the most likely reason for these results, particularly given the degree of cross-neutralisation in this and previous studies (Dzikwi et al., 2010, Kuzmin et al., 2008a). It is also interesting to note one strong (1:320) and one weak (1:8) VNAb titre were recorded against ptWCBV for different serum samples. Support for cross-neutralisation as the cause of such observations is provided by the individual with the 1:320 VNAb titre having a very high titre against ptLBV (1:1280), and ptMOKV (1:640), perhaps demonstrating that a strong immune response to LBV can lead to neutralisation of other lyssaviruses. That this represents a seroprevalence of 1% in our sampled group of bats, far below what has been recorded for WCBV in other studies, suggests that even though the same serum samples did not neutralise ptCVS-11, these results do not constitute convincing evidence for the presence of WCBV circulating in Ghanaian bats. Therefore, analysis of our results with those of previous studies suggests possible species specificity for certain bat lyssaviruses. While LBV RNA and antibodies have been detected in a different bat species (Dzikwi et al., 2010, Hayman et al., 2008a, Kuzmin et al., 2008a), MOKV infection has never been reported in bats and WCBV has only been isolated in Miniopterus species (Botvinkin et al., 2003, Kuzmin et al., 2008b). Evidence of LBV and WCBV infection has been reported within the same cave colony but in different bat species (Kuzmin et al., 2008a, Kuzmin et al., 2008b). This may suggest a restricted host range for certain lyssaviruses, possibly due to cellular, behavioural, or ecological restrictions that may have evolved during the co-evolution of the host and virus. However, with more intensive, far reaching virological studies, MOKV and WCBV might be isolated from bats or more than one species respectively. Therefore, seroprevalence studies are needed to elucidate the true distribution of LBV, MOKV and WCBV and we propose the multiplexed PNA we describe will substantially improve surveillance capabilities. Given our studies also provide evidence of endemic circulation of LBV within bat populations in urban centres, further studies are necessary in order to understand both virus dynamics at the population level and species specificity at the cellular level.
Conclusion
The results of this study increase our understanding of the natural history of lyssaviruses, providing the first evidence that Lagos bat virus might be endemic in urban dwelling E. helvum bat populations. Using a novel multiplexed PNA we also detect neutralisation of MOKV and WCBV by LBV antibody positive sera. From these observations, coupled with at least a dozen documented cases of non-RABV lyssavirus spill-over events into humans with a 100% fatality rate (Belikov et al., 2009, Johnson et al., 2010) and that many other cases have probably been unrecorded or misdiagnosed as RABV infections, it is clear that the extent of the public health threat posed by zoonotic transmission of highly pathogenic lyssaviruses from bats is still not entirely understood. In particular, understanding lyssavirus dynamics and whether bat lyssaviruses are restricted in their host species range is necessary in order to understand the potential risk of cross-species transmission and virus spill over. Therefore, further studies leading to a greater understanding of lyssavirus molecular biology and epidemiology are required to address this knowledge gap, which will ultimately aid the design of novel antivirals against these emerging viruses.
Methods
Virus isolates, plasmids and pseudotype production
The LBV Nigeria 1956 (LBV.NIG56-RV1) isolate (Boulger and Porterfield, 1958, Shope et al., 1970) was used in the mFAVN assays. To produce infectious pseudotype particles for use in PNAs the glycoprotein (G-) genes from LBV.NIG56-RV1 (GenBank accession number: HM623779) and a Nigerian MOKV isolated in 1968 ((Kemp et al., 1972, Shope et al., 1970); MOKV.NIG68-RV4; GenBank accession number: HM623780) were amplified using specific primers (Supplementary table S1) and cloned into a pUC-based expression plasmid (pI.18). WCBV G-gene ((Kuzmin et al., 2008c); EF614258) was chemically synthesised (Genescript, USA) and then sub-cloned into pI.18. These were then expressed concurrently with the HIV type 1 gag-pol plasmid (pCMV-∆8.91) and one of the following reporter plasmids: pCSGW (GFP), pCSFLW (firefly luciferase) or pCSRLW (renilla luciferase). Detailed information regarding plasmids, pseudovirus production and titration has been given previously (Wright et al., 2009, Wright et al., 2008) apart from pCSRLW, which comprises the pCSGW backbone with the renilla luciferase gene subcloned in place of the GFP gene (Alberini et al., 2009, Demaison et al., 2002). GFP pseudotype titres were comparable for ptLBV.NIG56-RV1 (2.9 × 105 infectious units (IFU)/ml) and ptWCBV (7.2 × 105 IFU/ml) but a log10 lower for ptMOKV.NIG68-RV4 (3.8 × 104 IFU/ml).
Cell lines
A highly transfectable derivative of the 293 cell line, human embroyonic kidney 293T clone-17 cells (HEK 293T/17; ATCC CRL-11268) were used for pseudotype production and maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with Glutamax and 15% foetal calf serum (FCS) at 10% CO2. Baby hamster kidney-21 clone-13 cells (BHK-21; ATCC CCL-10), used for pseudotype and mFAVN neutralisation assays, were cultured in DMEM with 10% FCS, 1% penicillin/streptomycin and 5% CO2.
Study area and serum samples
In March 2009, 184 serum samples were collected from three large E. helvum colonies within Ghana: Accra (urban; n = 93), Kumasi (urban; n = 34) and Tanoboase (rural; n = 57; Fig. 1A). Ethical approval for the capture and study of the bats was given by the Zoological Society of London Ethics Committee (WLE/0467). Bats were caught using 6- to 18-m mist nets when returning to their roosting sites. Blood samples were taken from the propatagial vein while bats were manually restrained (Fig. 1B). Sera were prepared by centrifugation of blood at 3,000 rpm for 15 min, heat-treated at 56 °C for 30 min before and storage at −70 °C immediately. Six serum samples contained insufficient volumes for testing for MOKV VNAb.
Separate rabbit anti-sera raised against the LBV.NIG56-RV1 isolate and against a Duvenhage virus isolate recovered in a bat from Zimbabwe (RV131;(Johnson et al., 2002)) were used as positive control sera in the PNAs.
Neutralisation assays
Standard, single pseudotype neutralisation assays were conducted as previously described (Wright et al., 2009, Wright et al., 2008). To allow the detection of VNAb to two pseudoviruses in one well of a tissue culture plate using ≤ 10ul of serum, a multiplex or dual assay was developed. This assay was undertaken using the same protocol as the single assay apart from: (1) Two lyssavirus pseudotypes, 100× TCID50 of both, were added to each well of the assay plate, one contained a firefly luciferase reporter gene and the other a renilla luciferase reporter gene, and (2) following the normal 48 h incubation the plates were assayed using the Dual-Glo® reagent (Promega, UK) instead of the Bright-Glo® reagent (Promega, UK), as detailed in the manufacturer's instructions. The Dual-Glo® reagent can differentiate between the firefly and renilla luciferase signal so a VNAb titre for each lyssavirus pseudotype could be recorded for each serum sample concurrently. All neutralisation data reported in this study was undertaken using LBV, MOKV, WCBV or CVS-11 PNA but to allow comparison of VNAb titres samples were also tested in a LBV mFAVNs, which were undertaken as described previously (Hayman et al., 2008a). As with the widely used FAVN assay, titres reported correspond to 100% neutralisation of pseudotype or virus input and are reported as IC100 endpoint reciprocal dilutions, Statistical analyses were undertaken using GraphPad Prism (GraphPad Software, San Diego, CA, USA).
It should be noted that the method for scoring sera as positive or negative by mFAVN or PNA was the same as that used in a previous study in which no sera cross-neutralised with CVS-11 (Hayman et al., 2008a). In the absence of known uninfected E. helvum colonies the sensitivity and specificity of this assay cannot be known, however in Hayman et al. (2008a) a 1:9 dilution was a three-fold dilution above the 1:3 dilution at which there was no virus neutralisation using standard FAVN methods against rabies (CVS-11; Cliquet et al., 1998), and therefore deemed an acceptable cut-off. Similarly we observed no neutralisation of ptCVS-11 above a dilution of 1:20 using the PNA and serum samples collected for the current study (data not shown). Therefore, we considered any mean neutralising titre above the next dilution in the series, 1:40 for the PNA and > 1:9 for the mFAVN, seropositive.
The following are the supplementary materials related to this article.
Primers used to amplify G-protein sequencesa.
Conflict of interest statement
The authors have no commercial or other association that might pose a conflict of interest.
Acknowledgments
We thank Kate Baker for assistance in collecting field samples and supplying the photograph used in this manuscript. This research was supported by the UK Medical Research Council (grant number G0801176), the UK Department for Environment, Food and Rural Affairs (grant number SE0423/SE0424), the Wellcome Trust (DTSH), the Alborada Trust, the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security and the U.S. Fogarty International Center, National Institutes of Health.
Contributor Information
Edward Wright, Email: edward.wright@ucl.ac.uk.
David T.S. Hayman, Email: dtsh2@cam.ac.uk.
Aisling Vaughan, Email: aisling.vaughan@ndm.ox.ac.uk.
Nigel J. Temperton, Email: N.Temperton@kent.ac.uk.
James L.N. Wood, Email: jlnw2@cam.ac.uk.
Andrew A. Cunningham, Email: A.Cunningham@ioz.ac.uk.
Richard Suu-Ire, Email: suurire@hotmail.com.
Robin A. Weiss, Email: r.weiss@ucl.ac.uk.
Anthony R. Fooks, Email: t.fooks@vla.defra.gsi.gov.uk.
References
- Alberini I., Del Tordello E., Fasolo A., Temperton N.J., Galli G., Gentile C., Montomoli E., Hilbert A.K., Banzhoff A., Del Giudice G., Donnelly J.J., Rappuoli R., Capecchi B. Pseudoparticle neutralization is a reliable assay to measure immunity and cross-reactivity to H5N1 influenza viruses. Vaccine. 2009;27(43):5998–6003. doi: 10.1016/j.vaccine.2009.07.079. [DOI] [PubMed] [Google Scholar]
- Aljofan M., Sganga M.L., Lo M.K., Rootes C.L., Porotto M., Meyer A.G., Saubern S., Moscona A., Mungall B.A. Antiviral activity of gliotoxin, gentian violet and brilliant green against Nipah and Hendra virus in vitro. Virol. J. 2009;6:187. doi: 10.1186/1743-422X-6-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrane H., Bahloul C., Perrin P., Tordo N. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 2001;75(7):3268–3276. doi: 10.1128/JVI.75.7.3268-3276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyard A.C., Johnson N., Voller K., Hicks D., Nunez A., Hartley M., Fooks A.R. Repeated detection of European bat lyssavirus type 2 in dead bats found at a single roost site in the UK. Arch. Virol. 2009;154(11):1847–1850. doi: 10.1007/s00705-009-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyard A.C., Hartley M., Fooks A.R. Reassessing the risk from rabies: a continuing threat to the UK? Virus Res. 2010;152(1–2):79–84. doi: 10.1016/j.virusres.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S.I., Leonova G.N., Kondratov I.G., Romanova E.V., Pavlenko E.V. Isolation and genetic characterisation of a new lyssavirus strain in the Primorskiy kray. East Siberian J. Infect. Pathol. 2009;16(3):68–69. [Google Scholar]
- Belotto A., Leanes L.F., Schneider M.C., Tamayo H., Correa E. Overview of rabies in the Americas. Virus Res. 2005;111(1):5–12. doi: 10.1016/j.virusres.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Botvinkin A.D., Poleschuk E.M., Kuzmin I.V., Borisova T.I., Gazaryan S.V., Yager P., Rupprecht C.E. Novel lyssaviruses isolated from bats in Russia. Emerg. Infect. Dis. 2003;9(12):1623–1625. doi: 10.3201/eid0912.030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulger L.R., Porterfield J.S. Isolation of a virus from Nigerian fruit bats. Trans. R. Soc. Trop. Med. Hyg. 1958;52(5):421–424. doi: 10.1016/0035-9203(58)90127-5. [DOI] [PubMed] [Google Scholar]
- Brookes S.M., Aegerter J.N., Smith G.C., Healy D.M., Jolliffe T.A., Swift S.M., Mackie I.J., Pritchard J.S., Racey P.A., Moore N.P., Fooks A.R. European bat lyssavirus in Scottish bats. Emerg. Infect. Dis. 2005;11(4):572–578. doi: 10.3201/eid1104.040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S. Royal Society of Tropical Medicine and Hygiene meeting at Manson House, London, 20 March 1997. Epidemiology and control of rabies. The growing problem of rabies in Africa. Trans. R. Soc. Trop. Med. Hyg. 1998;92(2):131–134. doi: 10.1016/s0035-9203(98)90718-0. [DOI] [PubMed] [Google Scholar]
- Cliquet F., Aubert M., Sagne L. Development of a fluorescent antibody virus neutralisation test (FAVN test) for the quantitation of rabies-neutralising antibody. J. Immunol. Methods. 1998;212(1):79–87. doi: 10.1016/s0022-1759(97)00212-3. [DOI] [PubMed] [Google Scholar]
- Corti D., Suguitan A.L., Jr., Pinna D., Silacci C., Fernandez-Rodriguez B.M., Vanzetta F., Santos C., Luke C.J., Torres-Velez F.J., Temperton N.J., Weiss R.A., Sallusto F., Subbarao K., Lanzavecchia A. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 2010 doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas O., Holmes E.C., Talbi C., Larrous F., Dacheux L., Bouchier C., Bourhy H. Genomic diversity and evolution of the lyssaviruses. PLoS ONE. 2008;3(4):e2057. doi: 10.1371/journal.pone.0002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaison C., Parsley K., Brouns G., Scherr M., Battmer K., Kinnon C., Grez M., Thrasher A.J. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 2002;13(7):803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- Dzikwi A.A., Kuzmin I.I., Umoh J.U., Kwaga J.K., Ahmad A.A., Rupprecht C.E. Evidence of Lagos bat virus circulation among Nigerian fruit bats. J. Wildl. Dis. 2010;46(1):267–271. doi: 10.7589/0090-3558-46.1.267. [DOI] [PubMed] [Google Scholar]
- Fooks A. The challenge of new and emerging lyssaviruses. Expert Rev. Vaccin. 2004;3(4):333–336. doi: 10.1586/14760584.3.4.333. [DOI] [PubMed] [Google Scholar]
- Fooks A.R., McElhinney L.M., Pounder D.J., Finnegan C.J., Mansfield K., Johnson N., Brookes S.M., Parsons G., White K., McIntyre P.G., Nathwani D. Case report: isolation of a European bat lyssavirus type 2a from a fatal human case of rabies encephalitis. J. Med. Virol. 2003;71(2):281–289. doi: 10.1002/jmv.10481. [DOI] [PubMed] [Google Scholar]
- Fraser G.C., Hooper P.T., Lunt R.A., Gould A.R., Gleeson L.J., Hyatt A.D., Russell G.M., Kattenbelt J.A. Encephalitis caused by a Lyssavirus in fruit bats in Australia. Emerg. Infect. Dis. 1996;2(4):327–331. doi: 10.3201/eid0204.960408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling C., Vos A., Johnson N., Fooks A.R., Muller T. Bat rabies—a Gordian knot? Berl. Münch. Tierärztl. Wochenschr. 2009;122(11–12):425–433. [PubMed] [Google Scholar]
- Garcia J.M., Lagarde N., Ma E.S., de Jong M.D., Peiris J.S. Optimization and evaluation of an influenza A (H5) pseudotyped lentiviral particle-based serological assay. J. Clin. Virol. 2009;47(1):29–33. doi: 10.1016/j.jcv.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Hanlon C.A., Kuzmin I.V., Blanton J.D., Weldon W.C., Manangan J.S., Rupprecht C.E. Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Res. 2005;111(1):44–54. doi: 10.1016/j.virusres.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Hayman D.T., Fooks A.R., Horton D., Suu-Ire R., Breed A.C., Cunningham A.A., Wood J.L. Antibodies against Lagos bat virus in megachiroptera from West Africa. Emerg. Infect. Dis. 2008;14(6):926–928. doi: 10.3201/eid1406.071421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T., Suu-Ire R., Breed A.C., McEachern J.A., Wang L., Wood J.L., Cunningham A.A. Evidence of henipavirus infection in West African fruit bats. PLoS One. 2008;3(7):e2739. doi: 10.1371/journal.pone.0002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T., Emmerich P., Yu M., Wang L.F., Suu-Ire R., Fooks A.R., Cunningham A.A., Wood J.L. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS One. 2010;5(8):e11978. doi: 10.1371/journal.pone.0011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N., McElhinney L.M., Smith J., Lowings P., Fooks A.R. Phylogenetic comparison of the genus Lyssavirus using distal coding sequences of the glycoprotein and nucleoprotein genes. Arch. Virol. 2002;147(11):2111–2123. doi: 10.1007/s00705-002-0877-4. [DOI] [PubMed] [Google Scholar]
- Johnson N., Vos A., Freuling C., Tordo N., Fooks A.R., Muller T. Human rabies due to lyssavirus infection of bat origin. Vet. Microbiol. 2010;142(3–4):151–159. doi: 10.1016/j.vetmic.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Kemp G.E., Causey O.R., Moore D.L., Odelola A., Fabiyi A. Mokola virus. Further studies on IbAn 27377, a new rabies-related etiologic agent of zoonosis in nigeria. Am. J. Trop. Med. Hyg. 1972;21(3):356–359. [PubMed] [Google Scholar]
- King A.A., Meredith C.D., Thomson G.R. Canid and viverrid rabies viruses in South Africa. Onderstepoort J. Vet. Res. 1993;60(4):295–299. [PubMed] [Google Scholar]
- King A.A., Meredith C.D., Thomson G.R. The biology of southern African lyssavirus variants. Curr. Top. Microbiol. Immunol. 1994;187:267–295. doi: 10.1007/978-3-642-78490-3_15. [DOI] [PubMed] [Google Scholar]
- Knobel D.L., Cleaveland S., Coleman P.G., Fevre E.M., Meltzer M.I., Miranda M.E., Shaw A., Zinsstag J., Meslin F.X. Re-evaluating the burden of rabies in Africa and Asia. Bull. World Health Organ. 2005;83(5):360–368. [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I.V., Hughes G.J., Botvinkin A.D., Orciari L.A., Rupprecht C.E. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Res. 2005;111(1):28–43. doi: 10.1016/j.virusres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Kuzmin I.V., Niezgoda M., Franka R., Agwanda B., Markotter W., Beagley J.C., Urazova O.Y., Breiman R.F., Rupprecht C.E. Lagos bat virus in Kenya. J. Clin. Microbiol. 2008;46(4):1451–1461. doi: 10.1128/JCM.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I.V., Niezgoda M., Franka R., Agwanda B., Markotter W., Beagley J.C., Urazova O.Y., Breiman R.F., Rupprecht C.E. Possible emergence of West Caucasian bat virus in Africa. Emerg. Infect. Dis. 2008;14(12):1887–1889. doi: 10.3201/eid1412.080750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I.V., Wu X., Tordo N., Rupprecht C.E. Complete genomes of Aravan, Khujand, Irkut and West Caucasian bat viruses, with special attention to the polymerase gene and non-coding regions. Virus Res. 2008;136(1–2):81–90. doi: 10.1016/j.virusres.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Kuzmin I.V., Novella I.S., Dietzgen R.G., Padhi A., Rupprecht C.E. The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infect. Genet. Evol. 2009;9(4):541–553. doi: 10.1016/j.meegid.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Kuzmin I.V., Mayer A.E., Niezgoda M., Markotter W., Agwanda B., Breiman R.F., Rupprecht C.E. Shimoni bat virus, a new representative of the Lyssavirus genus. Virus Res. 2010;149(2):197–210. doi: 10.1016/j.virusres.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Mallewa M., Fooks A.R., Banda D., Chikungwa P., Mankhambo L., Molyneux E., Molyneux M.E., Solomon T. Rabies encephalitis in malaria-endemic area, Malawi, Africa. Emerg. Infect. Dis. 2007;13(1):136–139. doi: 10.3201/eid1301.060810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markotter W., Kuzmin I., Rupprecht C.E., Nel L.H. Phylogeny of Lagos bat virus: challenges for lyssavirus taxonomy. Virus Res. 2008;135(1):10–21. doi: 10.1016/j.virusres.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Mickleburgh S., Waylen K., Racey P. Bats as bushmeat: a global review. Oryx. 2009;43(2):217–234. [Google Scholar]
- Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Neil S., Bieniasz P. Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 2009;29(9):569–580. doi: 10.1089/jir.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel L.H., Rupprecht C.E. Emergence of lyssaviruses in the Old World: the case of Africa. Curr. Top. Microbiol. Immunol. 2007;315:161–193. doi: 10.1007/978-3-540-70962-6_8. [DOI] [PubMed] [Google Scholar]
- Nel L., Jacobs J., Jaftha J., von Teichman B., Bingham J., Olivier M. New cases of Mokola virus infection in South Africa: a genotypic comparison of Southern African virus isolates. Virus Genes. 2000;20(2):103–106. doi: 10.1023/a:1008120511752. [DOI] [PubMed] [Google Scholar]
- Parry C.M., Kohli A., Boinett C.J., Towers G.J., McCormick A.L., Pillay D. Gag determinants of fitness and drug susceptibility in protease inhibitor-resistant human immunodeficiency virus type 1. J. Virol. 2009;83(18):9094–9101. doi: 10.1128/JVI.02356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeta C.T., Bingham J., Nel L.H. Molecular epidemiology of canid rabies in Zimbabwe and South Africa. Virus Res. 2003;91(2):203–211. doi: 10.1016/s0168-1702(02)00272-1. [DOI] [PubMed] [Google Scholar]
- Sabeta C.T., Markotter W., Mohale D.K., Shumba W., Wandeler A.I., Nel L.H. Mokola virus in domestic mammals, South Africa. Emerg. Infect. Dis. 2007;13(9):1371–1373. doi: 10.3201/eid1309.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope R.E., Murphy F.A., Harrison A.K., Causey O.R., Kemp G.E., Simpson D.I., Moore D.L. Two African viruses serologically and morphologically related to rabies virus. J. Virol. 1970;6(5):690–692. doi: 10.1128/jvi.6.5.690-692.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.J., Lapedes A.S., de Jong J.C., Bestebroer T.M., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Streicker D.G., Turmelle A.S., Vonhof M.J., Kuzmin I.V., McCracken G.F., Rupprecht C.E. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329(5992):676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- Su C.Y., Wang S.Y., Shie J.J., Jeng K.S., Temperton N.J., Fang J.M., Wong C.H., Cheng Y.S. In vitro evaluation of neuraminidase inhibitors using the neuraminidase-dependent release assay of hemagglutinin-pseudotyped viruses. Antivir. Res. 2008;79(3):199–205. doi: 10.1016/j.antiviral.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Szecsi J., Boson B., Johnsson P., Dupeyrot-Lacas P., Matrosovich M., Klenk H.D., Klatzmann D., Volchkov V., Cosset F.L. Induction of neutralising antibodies by virus-like particles harbouring surface proteins from highly pathogenic H5N1 and H7N1 influenza viruses. Virol. J. 2006;3:70. doi: 10.1186/1743-422X-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szecsi J., Gabriel G., Edfeldt G., Michelet M., Klenk H.D., Cosset F.L. DNA vaccination with a single-plasmid construct coding for viruslike particles protects mice against infection with a highly pathogenic avian influenza A virus. J. Infect. Dis. 2009;200(2):181–190. doi: 10.1086/599840. [DOI] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O'Brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307(5709):580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Temperton N.J., Chan P.K., Simmons G., Zambon M.C., Tedder R.S., Takeuchi Y., Weiss R.A. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg. Infect. Dis. 2005;11(3):411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperton N.J., Hoschler K., Major D., Nicolson C., Manvell R., Hien V.M., Ha D.Q., Jong M.D., Zambon M., Takeuchi Y., Weiss R.A. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza Respir. Viruses. 2007;1(3):105–112. doi: 10.1111/j.1750-2659.2007.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Thiel P.P., van den Hoek J.A., Eftimov F., Tepaske R., Zaaijer H.J., Spanjaard L., de Boer H.E., van Doornum G.J., Schutten M., Osterhaus A., Kager P.A. Fatal case of human rabies (Duvenhage virus) from a bat in Kenya: The Netherlands, December 2007. Euro Surveill. 2008;13(2) [PubMed] [Google Scholar]
- Vos A., Kaipf I., Denzinger A., Fooks A.R., Johnson N., Muller T. European Bat Lyssaviruses—an ecological enigma. Acta Chiropterol. 2007;9(1):283–296. [Google Scholar]
- Wang W., Xie H., Ye Z., Vassell R., Weiss C.D. Characterization of lentiviral pseudotypes with influenza H5N1 hemagglutinin and their performance in neutralization assays. J. Virol. Methods. 2010;165(2):305–310. doi: 10.1016/j.jviromet.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Warrilow D. Australian bat lyssavirus: a recently discovered new rhabdovirus. Curr. Top. Microbiol. Immunol. 2005;292:25–44. doi: 10.1007/3-540-27485-5_2. [DOI] [PubMed] [Google Scholar]
- World-Health-Organisation (2008). Rabies. Fact Sheet No. 99.
- Wright E., Temperton N.J., Marston D.A., McElhinney L.M., Fooks A.R., Weiss R.A. Investigating antibody neutralization of lyssaviruses using lentiviral pseudotypes: a cross-species comparison. J. Gen. Virol. 2008;89(Pt 9):2204–2213. doi: 10.1099/vir.0.2008/000349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E., McNabb S., Goddard T., Horton D.L., Lembo T., Nel L.H., Weiss R.A., Cleaveland S., Fooks A.R. A robust lentiviral pseudotype neutralisation assay for in-field serosurveillance of rabies and lyssaviruses in Africa. Vaccine. 2009;27(51):7178–7186. doi: 10.1016/j.vaccine.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Wei C.J., Kong W.P., Wu L., Xu L., Smith D.F., Nabel G.J. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science. 2007;317(5839):825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15(9):871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used to amplify G-protein sequencesa.