Abstract
Post-transcriptional control makes a major contribution to the overall regulation of gene expression pathway. Within the cytoplasm this is mediated by a combination of regulatory RNA motifs within the 5′ and 3′ untranslated regions of mRNAs and their interacting protein/RNA partners. One of the most common regulatory RNA elements in mammalian transcripts (present in approximately 40% of all mRNAs) are upstream open reading frames (uORFs). However, despite the prevalence of these RNA elements how they function is not well understood. In general, they act to repress translation of the physiological ORF under control conditions, and under certain pathophysiological stresses this repression can be alleviated. It is known that re-initiation following the translation of an uORF is utilised in some situations however there are numerous alternative mechanisms that control the synthesis of a protein whose mRNA contains uORFs. Moreover, the trans-acting factors that are also involved in this process are not well defined. In this review we summarise our current understanding of this area and highlight some common features of these RNA motifs that have been discovered to date.
Abbreviations: 4EBPs, 4E-binding proteins; AdoMetDCS, adenosyl-methionine decarboxylase; Eif, eukaryotic initiation factor; EMCV, encephalomyocarditis virus; IRES, internal ribosome entry segment; NMD, nonsense mediated decay; RRL, rabbit reticulocyte lysate; SNP, single nucleotide polymorphism; THPO, thrombopoietin; uORF, upstream open reading frame; UTR, untranslated region
Keywords: Protein synthesis, uORF, Upstream open reading frame, Translational control
1. Introduction
Gene expression can be regulated at multiple levels including: transcription, mRNA processing and localisation, protein translation and protein stability. It is now well accepted that control of translation makes a major contribution to the overall regulation of gene expression although in comparison to transcription it has been less extensively studied. Regulation at the level of translation allows a rapid and usually reversible response to various internal or external signals in the life-cycle of a cell.
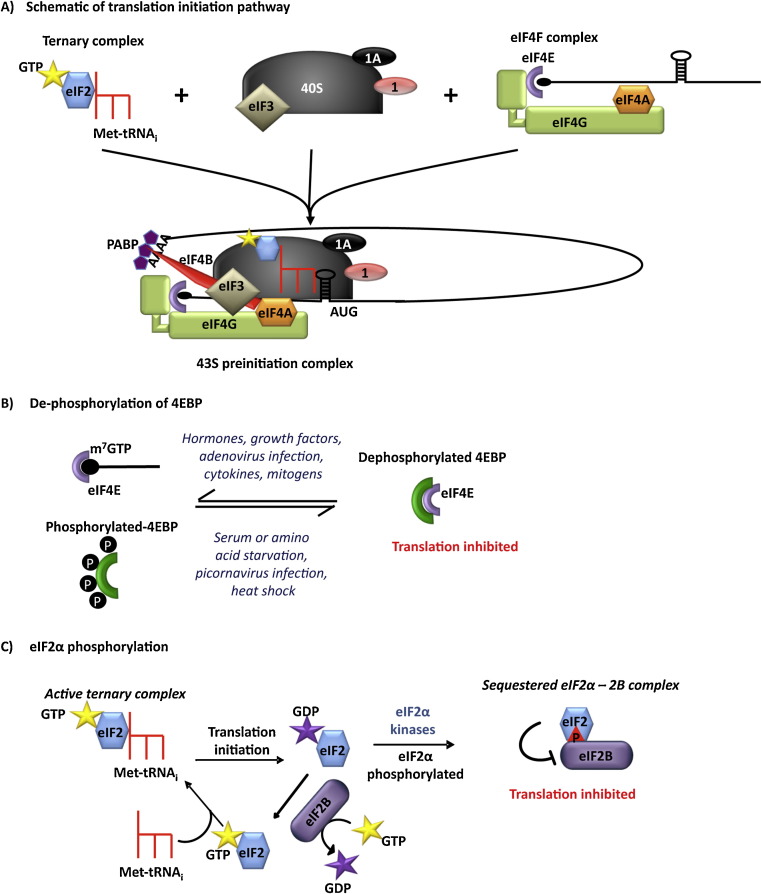
Translation can be considered as a three-step process that is comprised of initiation, elongation and termination. Although all three stages can be regulated, translation is primarily regulated at the initiation stage and this is assumed to be the rate-limiting step of the process (Sonenberg and Hinnebusch, 2009). Translation initiation is an intricate process that culminates in an elongation-competent 80S ribosome forming at the initiation codon. Essentially two complexes are required for this process which function to bind to the mRNA, via the m7GTP cap at the 5′ end of the message, recruit the ribosome and bring the initiator Met-tRNAi to the start codon. The eIF4F complex is required for both cap-binding and ribosome recruitment to the message. This complex is comprised of eIF4E (the cap binding protein), eIF4G (a scaffold protein) and eIF4A (a DEAD-box helicase) (Fig. 1A). eIF4G also contains binding sites for eIF3 and poly A binding protein (PABP). The other complex, the eIF2 ternary complex (eIF2-TC), contains eIF2, GTP and the Met-tRNAi. Once formed this complex is recruited to the 40S ribosomal subunit along with eIF1, eIF1A, eIF3, and possibly eIF5, forming the 43S pre-initiation complex. The 43S complex then interacts with the eIF4F complex via eIF3 binding to eIF4G to form the scanning competent 43S complex (Fig. 1A). This complex is additionally stabilised by the interaction of eIF4G with PABP and of eIF4B with eIF4A and PABP (Sonenberg and Hinnebusch, 2009, Bushell et al., 2001). The scanning model then predicts that the 43S complex progressively scans along the message until a suitable initiation codon is recognised. Although the codon AUG is used primarily as the “start codon” there are many examples of non-AUG start codons that are used including GUG and CUG (Touriol et al., 2003). Once the start codon is recognised and a 48S preinitiation complex has been formed, joining of the 60S ribosomal subunit occurs to form the 80S ribosome (reviewed by Jackson et al., 2010).
Fig. 1.
Translation initiation and its regulation. (A) Schematic of translation initiation pathway. Two complexes are required for initiation of translation; the eIF4F complex and the eIF2-TC. eIF2-TC interacts with the 40S ribosomal subunit to form the 43S preinitiation complex which then interacts (via eIF3 binding to eIF4G) with eIF4F complex to form the initiation competent 43S preinitiation complex. Translation initiation can be controlled by regulating the levels of eIF2-TC or eIF4F complex. (B) eIF4F complex is regulated by controlling the availability of the cap-binding protein eIF4E. Dephosphorylation of the eIF4E binding partners, 4EBPs, allows them to bind to eIF4E and so reduce the amount of this protein that is available for eIF4F complex formation. (C) eIF2-TC is controlled by phosphorylation of the alpha subunit of eIF2 by its kinases which results in sequestration of eIF2 and its GEF eIF2B in an inactive complex. This limits GTP:GDP recycling on eIF2 and therefore reduces the amount of eIF2-TC available.
Overall translation rates can be regulated by controlling the levels and activities of the components of the eIF4F and eIF2-TC complexes. Thus the availability of eIF4E is dependent on the phosphorylation status of its interacting partners, 4E-binding proteins (4EBPs). When phosphorylated they release eIF4E and this increases the net amount of protein that is able to interact with eIF4G to form the eIF4F complex (Fig. 1B). Signalling through the phosphoinositide 3-kinase pathway (PI3K) regulates 4EBP phosphorylation and when activated by growth factors, PI3K phosphorylates Akt, which in turn phosphorylates the mammalian target of rapamycin (mTOR) leading to 4EBP phosphorylation and upregulation of translation. Since there is an increase in both the levels and phosphorylation status of translation initiation factors in cancers, drugs that target these pathways such as rapamycin (which targets mTOR) are being investigated as potential therapeutic agents (Blagden and Willis, 2011).
eIF2-TC formation is regulated by altering the phosphorylation status of eIF2. This protein is comprised of 3 subunits and phosphorylation occurs on the alpha subunit at position Ser51 (Dever and Sicheri, 2007). The phosphorylation of eIF2 inhibits translation since phosphorylated eIF2 binds with high affinity to its guanine nucleotide exchange factor eIF2B that is present in limiting amounts. This prevents eIF2 recycling, lowering the amount of eIF2-TC formation, and inhibits translation (Fig. 1C). There are four kinases that phosphorylate eIF2 that are activated by different external stimuli (Spriggs et al., 2010). For example, GCN2 (General control non-de repressible 2) is activated following nutrient starvation and UVB exposure (Powley et al., 2009, Spriggs et al., 2010), PERK (PKR-like ER kinase) is activated when unfolded proteins accumulate in the cell e.g. in prion disease (Moreno et al., 2012), PKR (RNA-dependent protein kinase) is activated during viral infection, but also during the disruption of systemic metabolic homeostasis (Hotamisligil, 2006) and HRI (Haem-regulated inhibitor) promotes survival of erythroid precursors when iron levels are low (Chen, 2007).
The main protein coding sequence of the mRNA is often flanked by upstream and downstream regions, which are termed untranslated regions (UTRs). These regions may contain regulatory elements such as hairpins, protein binding sites, internal ribosome entry sequences (IRES) elements or upstream open reading frames (uORF) in the 5′UTR or miRNA binding sites, localisation elements and poly-A tail signals in the 3′UTRs (Pichon et al., 2012). Herein, we will focus our attention on discussing the occurrences and behaviours of mammalian uORFs.
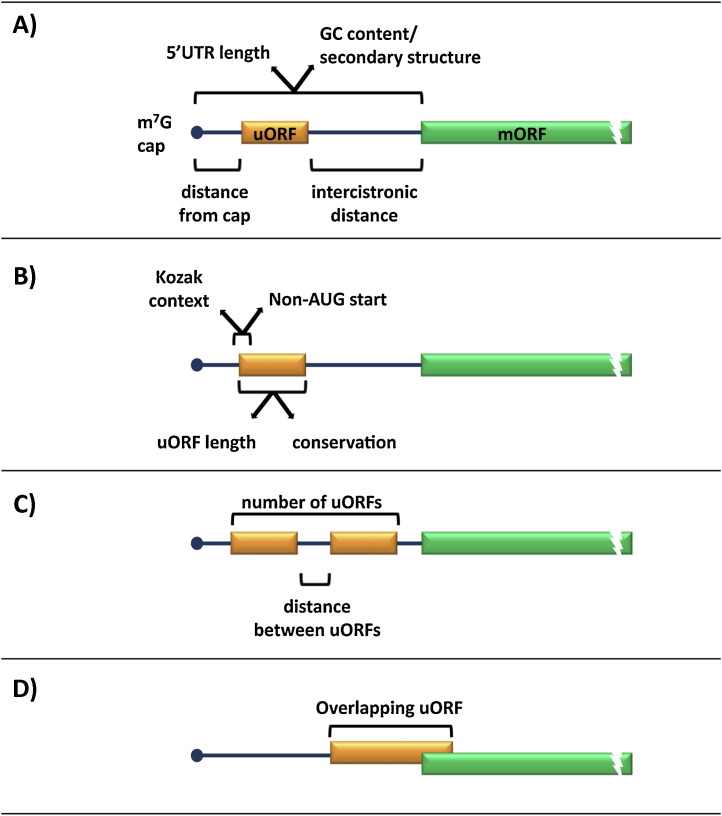
Several bioinformatic and genetic studies revealed that 40–50% of human and rodent mRNAs contain at least one uORF (Calvo et al., 2009, Iacono et al., 2005, Matsui et al., 2007). Transcripts with one uORF generally have a 5′UTR of at least 75 nucleotides. The probability that a mRNA will contain more than one uORF increases with the increasing length of the 5′UTR, although the occurrence of upstream AUGs (uAUGs) does not show any correlation with 5′UTR length (Fig. 2 ) (Iacono et al., 2005). Both uORFs and uAUGs occur less frequently than expected by chance implying that they are under negative selection pressure (Calvo et al., 2009, Iacono et al., 2005, Resch et al., 2009). However, the presence of multiple uORFs seems to be enriched in certain subgroups of mRNAs, including genes coding for growth factors, transcription factors and proto-oncogenes (Davuluri et al., 2000, Kozak, 1987a).
Fig. 2.
Many properties may contribute to an uORF's role in the translational control of a mORF. These include the length of the 5′UTR, the secondary structure and GC content. Consideration of where the uORF is situated, including the distance from the cap and the intercistronic distance between the termination of the uORF and the mORF (A). The sequence of the uORF might be important, whether it is an AUG or non-AUG initiator codon, the strength of the surrounding Kozak context and the uORF length and conservation. Conservation of uORFs may indicate a role for the peptide coding sequence of the uORF (B). Longer 5′UTRs tend to have multiple uORFs, so consideration of the distance between these uORFs is important (C). Lastly, some uORFs do not terminate within the 5′UTR, rather they overlap with the mORF. These uORFs if recognised by ribosomes will cause repression of the mORF (D).
Both uAUG or uORFs (when uAUG is followed by an in-frame stop codon) are major regulatory elements in the 5′UTR regions. According to the scanning model the 43S preinitiation complex enters the mRNA at the 5′ cap of the mRNA and scans sequentially along the 5′UTR until it encounters the first AUG codon (Kozak, 1989b). There are some circumstances, which allow the ribosome to skip the first AUG, e.g. if the first AUG is in a sequence context which is not recognised efficiently by the scanning ribosome causing leaky scanning (discussed below) (Kozak, 1986), or if the first AUG is located too close to the 5′ cap (Kozak, 1991). However, uAUGs/uORFs located in the 5′UTR that are recognised by the 40S ribosomal subunit will by default downregulate the translation from the main open reading frame (mORF). The recent large scale genetic and proteomics study has shown that uORFs reduce protein expression from the downstream mORF by 30–80% (Calvo et al., 2009).
For an uORF to be able to regulate downstream protein expression, it has to be recognised by the 40S ribosome subunit. The most favourable nucleotide context surrounding a start codon has been shown to be GCCA/GCCAUGG (termed Kozak consensus) of which A at −3 and G at +4 (the A at the AUG codon being +1) are the most important nucleotides (Kozak, 1986, Kozak, 1989b). The Kozak context has been shown to be especially important around the non-AUG start sites (Kozak, 1989a, Peabody, 1989). The A at the −3 position was demonstrated to interact with the eIF2α subunit and the G at +4 with helix 44 in the 18S rRNA in experiments using UV-crosslinking and purified factors. Both these interactions are proposed to promote AUG recognition at the start site (Pisarev et al., 2006). However, in addition to AUG context, the secondary structure of the transcript, especially following the start site, has been shown to play an important role in start site selection (Kozak, 1986, Kozak, 1991). Interestingly, it has been shown that about 90% of the mORF AUG start sites are either in a strong (both −3A/+4G) or adequate (either −3 or +4) nucleotide context; there was no likewise ‘bias’ for the start sites of uORFs (Iacono et al., 2005).
The uORFs identified in mRNAs to date vary in a wide number of ways. Thus, they display differences in (i) length, (ii) number, (iii) distance from the cap, (iv) whether the uORF is completely within the 5′UTR or overlapping with the mORF and (v) distance between the uORF stop codon and the start site in the mORF (Fig. 2). How all these different parameters contribute to the down regulation of translation from the mORF is a complex process that is not well understood. In this review, we summarise the current literature in order to generate some common rules from the studies carried out thus far.
2. Reinitiation
The ability of eukaryotic ribosomes to reinitiate after termination was found unexpectedly during studies that gave rise to the scanning ribosome model. An experiment which inserted an uAUG into the 5′UTR of the (pre)proinsulin mRNA reduced the amount of protein expressed from this mRNA. However, inserting an in-frame stop codon after the uAUG increased the yield of (pre)proinsulin compared to the mRNA only having the uAUG (Kozak, 1984). Following studies showed that the reinitiation efficiency at the downstream AUG increased with an increasing intercistronic distance (the distance between the uORF-stop and the ‘reinitiation’ start codon downstream) (Kozak, 1987b). This suggested that the ribosome needed to reacquire some initiation factors during the scanning before becoming competent to allow reinitiation at an AUG codon downstream. One of the obvious factors lacking from the ribosome was eIF2-TC, which is lost from the ribosome during the first initiation event. Subsequent studies both in yeast and mammalian systems have shown that the available eIF2-TC concentration correlates with the distance the 40S ribosomal subunit needs to scan before it is able to reinitiate (Hinnebusch, 2005, Vattem and Wek, 2004; discussed in more detail below).
Reinitiation efficiency is very difficult to determine accurately as it is hard to assess how much is really true reinitiation or a combination of leaky scanning and reinitiation. However, efficient reinitiation has been detected after very short uORFs (up to 50% for GCN4) (Hinnebusch, 2005). The reinitiation efficiency decreases with the increasing length of the uORF (Luukkonen et al., 1995), or if the translation of the uORF is slowed down by strong secondary structure which causes the translating ribosome to pause (Kozak, 1987b, Kozak, 2001). Together these findings suggest that the time taken to translate an uORF is more critical than the length of the uORF per se. This implies that some critical initiation factors needed for reinitiation are lost during translation of the uORF rather than during the ribosome ‘subunit joining’ stage of initiation. A series of experiments were carried out to address whether reinitiation was dependent on the eIFs that initiated at the first uORF start codon. Using an in vitro translation assay in rabbit reticulocyte lysate (RRL) efficient reinitiation occurred at 20–35% when an uORF-containing construct was driven by either scanning dependent mechanism or by the encephalomyocarditis virus IRES (EMCV), which both required the eIF4F complex for initiation (except that EMCV does not need eIF4E) (Pöyry et al., 2004). However, no reintiation was observed if the classical swine fever virus IRES (which does not require any of the eIF4 family members) or the cricket paralysis virus IRES (which is not dependent on any eIFs) were used in the translation assay. When an uORF construct that had 19xCAA-repeats in the 5′UTR, which can initiate translation without the eIF4F complex (Pestova and Kolupaeva, 2002) were used efficient reinitiation only occurred in RRL that contained all eIFs or in an eIF4G-depleted lysate supplemented with recombinant eIF4G p50 fragment (eIF4G central domain which has both eIF3 and eIF4A binding sites). These data led to a model that suggests that reinitiation will only occur if the eIF4F complex participated in the primary initiation event and that an interaction between 40S/eIF3/eIF4F is maintained during the initial translation. Only the 40S ribosome subunits that have kept this interaction at the uORF termination codon are able to resume scanning and reinitiate at a downstream initiation codon (Pöyry et al., 2004).
2.1. eIF2 concentration and reinitiation
As discussed above, phosphorylation of the eIF2α subunit is an important mechanism to regulate protein synthesis in different stress conditions within the cell. However although eIF2α phosphorylation in general reduces protein synthesis in the cell, it simultaneously induces protein synthesis of a subset of mRNAs e.g. ATF4 and ATF5.
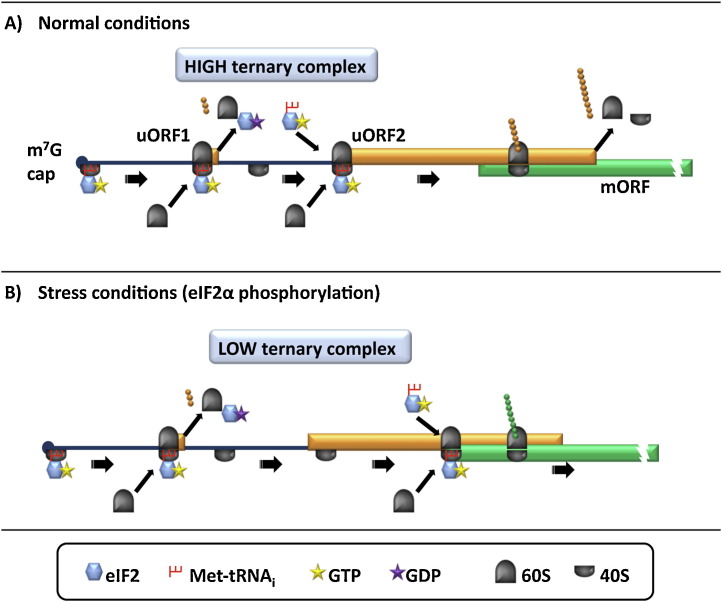
The best understood mechanism of mammalian translational control by uORFs is reinitation on the human ATF4 mRNA. This transcript encodes a basic zipper (bZIP) transcription factor and has 2 uORFs in its 5′UTR of which the uORF1 is only 3 amino acids long and the uORF2 is 59 amino acids long and overlaps with the ATF4 mORF (Fig. 3 ) (Vattem and Wek, 2004). Under normal conditions (eIF2-TC abundant) uORF1 is efficiently translated. Any 40S ribosomal subunits that resume scanning after the uORF1 stop codon will reacquire eIF2-TC before reaching the uORF2 AUG codon and thus reinitiate at uORF2 (Fig. 3A). Because the uORF2 is much longer and more importantly, overlaps with the mORF, the majority of the ribosomes will be recycled at the uORF2 termination codon, leading to repression of the ATF4 mORF. During stress conditions (i.e. ER stress) eIF2α subunit phosphorylation is increased which reduces the eIF2-TC concentration. Now, the 40S ribosomal subunits that resume scanning will bypass the uORF2-AUG codon due to the longer time it takes to reacquire eIF2-TC, thus leading to reinitation at the ATF4 mORF (Fig. 3B) (Vattem and Wek, 2004). Interestingly, in the ATF4 and ATF5 transcripts both the uORFs and the nucleotide distance between them are conserved among different vertebrate species that suggests a common regulatory mechanism in these mRNAs (Vattem and Wek, 2004, Zhou et al., 2008).
Fig. 3.
Reinitation mechanism of ATF4 mRNA translation. (A) Under normal conditions (abundant eIF2-TC), ribosomes that translate uORF1 (3 codons) may reinitate, that is the 40S remains attached to the transcript and resufmes scanning downstream. eIF2B acts as a GEF to eIF2-GDP, exchanging the GDP for GTP. Due to the abundant eIF2-TC availability the 40S ribosome reacquires a ternary complex before uORF2 allowing reinitation at uORF2 AUG codon. Hence, preventing translation of the ATF4 mORF. (B) Under stress conditions, which elevate eIF2α phosphorylation uORF1 in translated as described above. However, phosphorylated eIF2α on Ser 51 binds more strongly to eIF2B inhibiting its GEF function and reducing the available pool of eIF2-TC. This leads to the 40S ribosomes that resume scanning to progress further along the transcript before reacquiring eIF2-TC, thus bypassing the start codon of uORF2 to reinitiate at the ATF4 mORF.
It has been shown that there is coordinated translational upregulation of a subset of mRNAs that contain uORFs following exposure of cells to UVB light. In this case, as part of the stress response, the eIF2α subunit is phosphorylated and the reduced eIF2-TC levels lead to the selective synthesis of proteins that are required as part of the DNA damage response including the critical DNA repair enzymes ERCC1, ERCC5 and DDB1 (Powley et al., 2009). However, this appears to be specific to the DNA repair enzymes since ATF4 was not found to be upregulated (Powley et al., 2009). Further work is needed to understand the precise mechanism of this selective translational control.
In general, most mammalian uORFs are permissive for reinitiation except for some rare cases where, for example, the nascent peptide chain causes ribosome stalling at the termination codon (discussed below). In contrast, most of the uORFs in yeast appear to be nonpermissive for reinitiation. The only native uORFs in yeast that have been shown to be permissive for reinitiation are the first uORF in the GCN4 mRNA and the only uORF in YAP1 mRNA (Hinnebusch, 2005, Munzarova et al., 2011, Vilela et al., 1998). In addition, nucleotide sequences both upstream and immediately downstream of the uORF1 in the GCN4 mRNA have been shown to interact with the yeast eIF3 and that this interaction is important for reinitiation after uORF1 translation (Munzarova et al., 2011, Szamecz et al., 2008). Interestingly, mammalian eIF3 and eIF4F interact directly with each other, and this interaction has been suggested to be important for reinitiation in mammalian systems (Pöyry et al., 2004), but in yeast only an indirect interaction between eIF3 and eIF4F (by eIF1 and eIF5) has been shown (Hinnebusch, 2006).
3. Mechanisms other than reinitiation
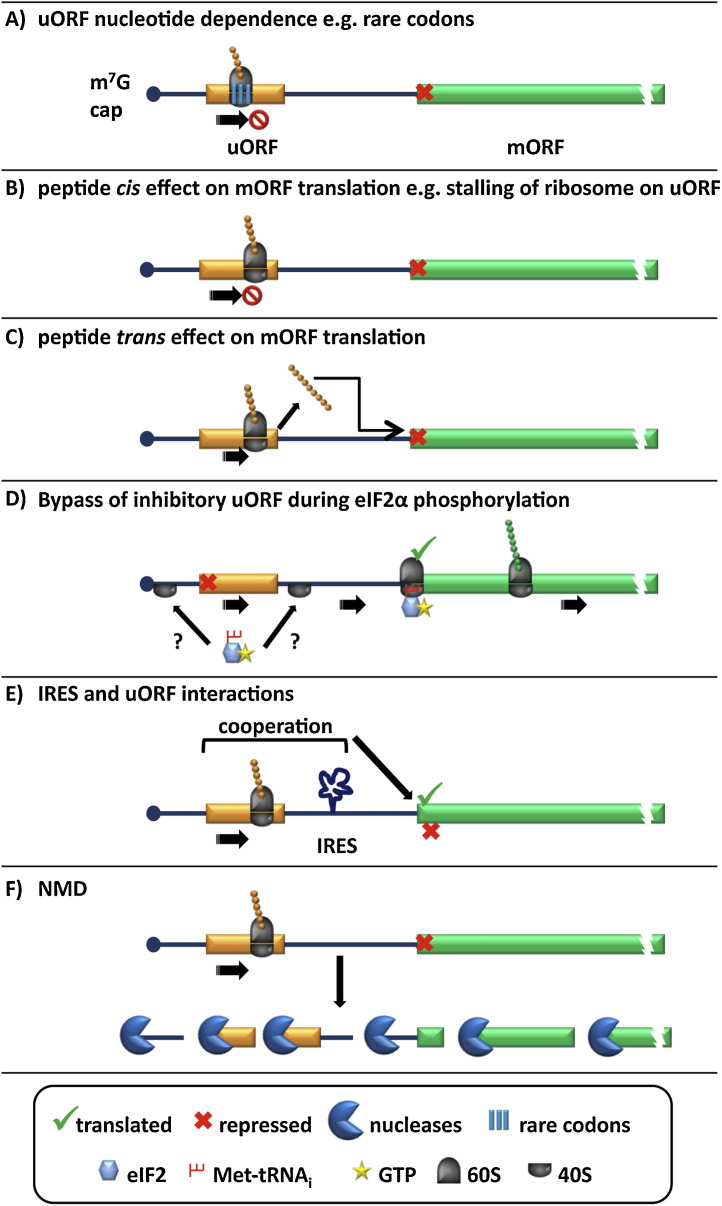
The premise of reinitiation does not account for the observed behaviour of all uORFs. There are also a number of, as yet, not fully understood mechanisms by which an uORF may function (Fig. 4 ). Herein, we discuss the possibilities that can occur concurrently or independently to reinitiation including; uORF nucleotide dependence, uORF peptide expression and functionality, uORF bypass, non-sense mediated mRNA decay (NMD), and interplay with an IRES.
Fig. 4.
The roles of uORFs in translational control. In addition to reinitiation (Fig. 2) uORFs have been characterised to perform a number of different roles. (A) The nucleotide sequence can have a predominant role on uORF translatability, for instance by encoding for rare codons that cause the ribosome to stall (methionine synthase) or the potential involvement of such sequences in secondary structure or RNA motifs (UCP2). The peptide product of uORFs can have cis regulatory functions, for instance causing the stalling of ribosomes (AdoMetDC and CHOP) (B), or by the trans repression of the mORF (AS, β2 adrenergic receptor and vasopressin V1B receptor) (C). (D) Bypass of an inhibitory uORF has been observed under stress conditions and is dependent on eIF2α phosphorylation. An interesting possibility for how this may occur is the loading of the 40S without an eIF2-TC which it acquires as it scans (C/EBP α and β, CHOP, GADD34, protein kinase C isoform η). (E) Interactions that involve both uORFs and IRES elements within 5′UTRs can cause expression of the mORF (Cat-1) or repression of particular splice variants (VEGF-A). (F) Approximately 35–50% of uORF containing transcripts undergo selective degradation by NMD (e.g. ATF4, CHOP and IFDR1).
3.1. Upstream open reading frame nucleotide dependence
For certain uORFs the nucleotide sequence is the most important determinant of uORF behaviour. Methionine synthase contains 2 inhibitory uORFs. Interestingly, the second uORF contains 6 rare codons for either arginine or proline within its 30 codon sequence. This would be predicted to potentially slow down or even stall the ribosome during uORF translation. Indeed, synonymous replacement of the adjacent rare arginine codons in the N-terminus of the peptide with a more abundant codon (either arginine or alanine) alleviated repression (Fig. 4A) (Col et al., 2007). A nucleotide sequence that allowed for RNA secondary structure to form and/or binding motifs within the 5′UTR sequence could also affect uORF behaviour and mRNA translation. For example, the pausing of the ribosome caused by the presence of highly structured RNA could allow increased time to recognise an AUG in a mechanism that could be similar to the way in which pseudoknots are used to achieve frame-shifting in coronaviruses (Roberts et al., 2009). Mutational analysis of the 36 codon uORF inhibitory peptide of UCP2 demonstrated that the uORF C-terminal nucleotide sequence was important for its inhibitory behaviour, although the mechanism is not yet understood (Hurtaud et al., 2006, Pecqueur et al., 2001).
3.2. Upstream open reading frame peptide expression and function
The detection of endogenous uORF peptide expression has been limited. Global screens of small peptide expression within human cells by mass spectrometry only identified 7 uORF peptides (Oyama et al., 2004, Oyama et al., 2007). A more recent peptidomics approach detected 15 uORF peptides (Slavoff et al., 2013). These low numbers suggest that either the majority of uORF products are expressed at levels currently undetectable and/or that they are selectively degraded within cells (Oyama et al., 2007, Slavoff et al., 2013). When uORF peptide expression does occur it may be spatially and/or developmentally regulated. For instance MYCNOT expression, the uORF product of the MYCN Δ1b transcript, was detectable within foetal but not adult brains (Besancon et al., 2009). Further confusion arises as to the function of such uORF peptides, with many containing unique domains and motifs (Besancon et al., 2009, Oyama et al., 2007). Differences in subcellular localisation; the 2 uORF peptides of the MKKS gene both localise to the mitochondria, which is in contrast to the cytoplasmic location of the MKKS protein (Akimoto et al., 2012). Or their association with other cellular factors, the uORF2 peptide of the GR transcript variant 1A interacts with a number of proteins in the cytoplasm but not membrane-bound GR (Diba et al., 2001). Collectively, these observations hint at the prospect that uORF peptides refractory to rapid proteolysis may also fulfil roles additional to translational control.
Functional characterisation has shown that uORF peptide products can produce cis- or trans-regulation on mORF translation (Fig. 4B and C). S-adenosyl-methionine decarboxylase (AdoMetDC) uORF-mediated repression is regulated by polyamine concentration, which determines the duration of ribosomal pausing at the C-terminus of the uORF sequence, thus obstructing passage of ribosomes to the mORF (Law et al., 2001, Nishimura et al., 1999, Raney et al., 2000). AdoMetDC encodes an inhibitory 6 codon uORF (Hill and Morris, 1992). Detailed molecular studies both in cell and cell-free systems support the hypothesis that the AdoMetDC uORF protein sequence is responsible for this inhibition, with the codon identity at the fourth and fifth positions as well as the length of this peptide being essential for its repressive properties (Hill and Morris, 1993, Mize et al., 1998, Raney et al., 2000). Despite the ability of the AdoMetDC uORF to inhibit a reporter construct in yeast, the peptide shows no sequence similarity to yeast uORFs that likewise promote ribosomal pausing/stalling suggesting these peptides associate with different molecular targets within the ribosome or associated factors (Law et al., 2001, Mize et al., 1998).
Several examples of an uORF peptide acting in a trans-manner have been demonstrated. Argininosuccinate synthase (AS) transcripts differ in their 5′UTR length due to the usage of different transcriptional start sites. The major transcripts (present at >90% of all AS transcripts within a cell) contain a short UTR with no uAUG, whereas the longer UTRs of the minor products contain an inhibitory uORF (Pendleton et al., 2002). Transfection of the AS uORF construct into bovine aortic endothelial cells repressed endogenous AS protein expression, with mutational analysis demonstrating that both the uORF peptide sequence and length being essential determinants of repression (Pendleton et al., 2005). Other examples of trans-repression have thus far been demonstrated by the addition of high concentrations of uORF peptides within cell-free systems. These include the mammalian β2 adrenergic receptor uORF peptide sequence, which encodes a number of conserved arginine residues. However, this effect was recreated with an unrelated peptide of equivalent high basic charge, although arginine alone proved insufficient (Parola and Kobilka, 1994). Another example involves the last uORF peptide of Vasopressin V1b receptor (whose endogenous expression has been detected within cells) that was able to re-establish translational inhibition when added exogenously to a cell free system expressing a transcript in which this uORF AUG had been mutated (Rabadan-Diehl et al., 2007). These interesting examples of uORF peptide functionality highlight the important of experimental design and appropriate controls when dissecting out an uORF's behaviour, including the need for consideration of any coexpressed transcript variants.
3.3. Upstream open reading frame bypass
uORF bypass has been used to describe the derepression of a mORF downstream of one or more inhibitory uORFs. It has been suggested that this arises from an increase in leaky scanning of uORF start codons. However, as discussed below the requirement for the uORFs during derepression shows this explanation is likely to be too simplistic (Lee et al., 2009, Raveh-Amit et al., 2009). Current known examples of this phenomenon occur during elevated levels of eIF2α phosphorylation (Fig. 4D) (Calkhoven et al., 2000, Lee et al., 2009, Palam et al., 2011, Raveh-Amit et al., 2009). Whether this mechanism is due to the reduction of eIF2-TC availability or a change in another translation factor is currently unknown.
The conserved CCAAT/enhancer binding protein (C/EBP) α and β isoforms contain a number of potential initiator codons, including a short uORF positioned upstream of the mORF. Unlike the other examples this uORF promotes reinitiation, but its surrounding sequence acts to reduce the likelihood of initiation from the full length mORF in favour of initiation at an internal AUG that encodes a truncated C/EBP product (Calkhoven et al., 1994, Calkhoven et al., 2000, Lincoln et al., 1998). Indeed, in the absence of the uORF no truncated product is produced (Calkhoven et al., 1994, Calkhoven et al., 2000). Translation of the full length or truncated products is regulated by changes in the availability of initiation factors. In addition to eIF2α phosphorylation, a reduction of free eIF4E by rapamycin causes bypass of the uORF AUG in favour of translation from the full length mORF (Calkhoven et al., 2000).
CHOP mRNA contains a conserved repressive uORF, with 2 in-frame AUGs creating a 31 or 34 codon product in rodents and humans (Jousse et al., 2001). Mutagenesis studies have shown that the C-terminal peptide region is essential for the inhibitory properties of this uORF. The absence of a trans-effect led to the supposition that the peptide sequence acts in a cis-manner to impede ribosomal movement either during elongation or termination, although future experiments are needed to confirm such an occurrence (Jousse et al., 2001, Palam et al., 2011). However, following ER stress when eIF2α is phosphorylated, CHOP is translationally upregulated, both in the presence or absence of the uORF. Therefore up-regulation of CHOP protein requires eIF2α phosphorylation, but is not necessarily dependent on the uORFs. It was suggested that under these conditions the uORF start codons (2 in-frame) become less well recognised by the ribosome complex (Palam et al., 2011). Further support for this theory was shown by mutation of the uORF to a strong Kozak consensus (which would be expected to decrease leaky scanning) reduced CHOP translational upregulation (Palam et al., 2011).
Other examples of potential bypass require the presence of an uAUG for the derepression. GADD34 mRNA undergoes both transcriptional and translational upregulation after ER stress. The 5′UTR of GADD34 mRNA contains 2 conserved uORFs that are inhibitory. Derepression after thapsigargin treatment (which induces ER stress PERK activation and eIF2α phosphorylation) was reliant on the presence of the second uORF start codon, whereas the first uORF was dispensable (Lee et al., 2009). Similarly, protein kinase C isoform η mRNA contains 2 conserved uORFs that are inhibitory to mORF translation, especially uORF2 that has a strong Kozak consensus sequence (A−3/G+4). Derepression after amino acid starvation required both GCN2 kinase and the presence of either uORF. Interestingly, mutation of the uORF2 stop codon, so that it now terminated downstream of the mORF start codon had no effect (Raveh-Amit et al., 2009). This implies that it is the start codons of the uORFs that are required and recognised during uORF bypass events, rather than uORF translation.
3.4. Internal ribosome entry segments
IRES elements provide a structured RNA context that can recruit ribosomes to initiate translation internally and thus independently from cap-dependent translational events (see review, Pichon et al., 2012). A couple of examples have been demonstrated in mammalian transcripts where translational control involves coordination between an uORF and an IRES element (Fig. 4E). Cat-1 mRNA undergoes IRES-mediated translational upregulation following amino acid or glucose deprivation. Interestingly, translation of a 48 codon uORF is required for maximal IRES activity (Fernandez et al., 2001, Fernandez et al., 2002a, Fernandez et al., 2002b, Yaman et al., 2003). Detailed mutagenesis studies led to a model which predicts that the Cat-1 IRES is in an inactive conformation which is restructured into the active IRES during translation of the uORF. Although, it is not yet fully understood how the uORF is initially translated (as the sequence of the 5′ end of the UTR is inhibitory to ribosomal movement) or how the IRES is maintained in its active context during stress stimulation (Yaman et al., 2003). The VEGF-A gene undergoes a number of transcriptional and translational regulatory steps that give rise to 9 protein isoforms. The presence of 2 IRES elements within the 5′UTR allows translation initiation from either the mORF AUG or from an uCUG codon that produces a N-terminally extended protein that is subsequently cleaved. VEGF-A isoforms arise from differential splicing, with the 121 amino acid isoform, unlike the other commonly expressed 165 and 189 amino acid isoforms, only being translated from the uCUG codon. Interestingly, cap-independent translation of an uORF upstream (situated between the uCUG and main AUG codons) acts to repress IRES-mediated expression of the 121 isoform from the AUG codon. This demonstrates the occurrence of widespread cis regulatory actions between the uORF, the IRES element and downstream RNA sequences (Bastide et al., 2008).
3.5. Nonsense mediated decay
Nonsense mediated decay (NMD) allows for the elimination of transcripts that encode peptides with a premature termination codon. The present model predicts that during the initial round of translation messages with a stop codon more than 50 nucleotides upstream of an exon–exon junction will not have the exon junction complexes displaced and this directs such messages to be recognised and selectively removed by NMD (Fig. 4F) (Rebbapragada and Lykke-Andersen, 2009). Many transcripts with uORFs would fit this criterion and thus be candidates for this pathway. However, examination of HeLa transcriptome changes following siRNA treatment of UPF1, PNRC2 or CTIF, all involved in NMD, identified 35–50% of upregulated transcripts contained an uORF (Kim et al., 2012, Mendell et al., 2004). However, 40–50% of human and mouse transcripts contain at least one uORF (Calvo et al., 2009, Iacono et al., 2005, Matsui et al., 2007), suggesting that there is no predisposition for NMD by uORF containing transcripts.
ATF4 and CHOP transcripts (ATF4 Reinitiation, CHOP uORF Bypass) are targets of NMD, illustrating the involvement of multifaceted pathways in uORF translational control (Gardner, 2008, Mendell et al., 2004). Interestingly, during hypoxia eIF2α phosphorylation caused a relocalisation of NMD components to stress granules and inhibition of NMD (Gardner, 2008). Similarly, it has been demonstrated that transcript turnover of another NMD target, the uORF containing IFDR1, was reduced following eIF2α phosphorylation (Zhao et al., 2010). Such a mechanism would be expected to augment translation of these proteins during stress conditions.
The very complex UTR of thrombopoietin (THPO) transcript contains multiple uORFs. The seventh uORF, which overlaps with the mORF, fits the criterion for a NMD target. However, this transcript avoids NMD and only by expanding the seventh uORF from 27 to 40 codons was it possible to reduce transcript levels, which were only in part reversible by NMD inhibition (Stockklausner et al., 2006). This observation fits with the hypothesis that proposes that the length of short ORFs may be a key discrimination factor in NMD. Studies using mutant beta globin transcripts demonstrated that shorter ORFs were more insensitive to NMD, and this occurred independently of any reinitation events (Inacio et al., 2004, Silva et al., 2006).
4. Non-AUG upstream open reading frames
Ribosomes have been shown to start translation at non-AUG start codons, although this occurs with much lower efficiency than at AUG codons both within mammalian cells and in vitro translation assays using RRL (Kozak, 1989a, Peabody, 1987, Peabody, 1989). Near-cognate non-AUG start codons (which differ from AUG by one nucleotide) within a strong Kozak context were tested in RRL (Peabody, 1989). All variations caused a certain degree of leaky scanning, with the ACG and CUG being the best recognised initiation codons and AGG and AAG the least recognised. In another study, GUG was found to be the most efficient non-AUG start codon. Furthermore, it was observed that the nucleotide context surrounding the GUG had a stronger influence than for an AUG codon implying that the nearby nucleotides compensate for the weaker codon–anticodon interaction (Kozak, 1989a).
A recent new technology termed ribosome profiling, involves deep sequencing of ribosome-protected mRNA fragments, allowing the detection of ribosome occupancy in any given mRNA genome wide. Thus, in principal one can detect which part of the mRNA is translated (Ingolia et al., 2009, Ingolia et al., 2011). By the use of two drugs; harringtonine, which acts by stalling 80S ribosomes at translation initiation start sites, and cycloheximide that arrests elongating ribosomes, the start sites and reading frames of a transcript can be identified. Ribosome profiling in mouse embryonic stem cells revealed high-level ribosome occupancy in uORFs that had non-AUG start codons (Ingolia et al., 2011). In fact the majority of the all uORFs detected had near-cognate start sites of which; CUG was used the most (30–35%), GUG and AUG (20%), UUG (15%) and ACG (10%). Many mRNAs had multiple uORFs with both AUG and non-AUG start sites. All these uORFs were accompanied by elongating ribosome footprints, which were detected without harringtonine treatment indicating that the uORFs were actively translated (Ingolia et al., 2011). When ATF4 mRNA was examined, ribosome footprints were found at both start codons and within both uORFs, however no ribosome footprints were observed at the mORF start codon indicating translational suppression. In contrast, in mRNAs containing non-AUG uORFs, ribosome footprints were seen at both the start codons and within the reading frames of non-AUG uORFs as well as at the mORFs suggesting that these non-AUG uORFs might have a weaker regulatory function than uORFs with AUG start codons.
A recent proteomics study identified short translated peptides by mass spectrometry and found that some uORF peptides were initiated from a non-AUG start codon. Due to the limitations of the technology, these peptides were on average 75 amino acids. Interestingly, many of these non-AUG uORFs overlapped the mORF. To validate these findings, the uORF and the adjoining mORF were tagged and expression of both the non-AUG uORF and mORF was confirmed using immunofluorescence. Mutation of the non-AUG to an AUG codon, completely repressed translation of the mORF indicating that the mORF was being translated by leaky scanning (Slavoff et al., 2013). How such non-AUG codons are recognised is not fully understood. However, it has been shown that eIF1 plays an important role in start codon selection and that in the absence of eIF1 the scanning ribosome is unable to discriminate between an AUG and non-AUG codon and is also indiscriminate to the surrounding Kozak context (Pestova and Kolupaeva, 2002). More recently, overexpression of eIF1 in cells abolished initiation from non-AUG codons. Furthermore, it was proposed that eIF1 concentration is involved in an autoregulatory feedback loop controlling its own translation, as the mORF AUG codon of the eIF1 transcript is positioned within a poor Kozak context (Ivanov et al., 2010).
5. Upstream open reading frames and disease
As the preceding examples have shown, uORFs can play an integral part in the translational control of gene expression. It is therefore not surprising that an increasing number of diseases are being linked to changes in uORF functionality. Here we will discuss examples of the consequences of alternative 5′UTRs, single nucleotide polymorphisms (SNPs) and uORF mutations in disease and treatment.
Many genes are encoded by alternative transcript isoforms that can arise from the usage of different promoters, transcription start sites and/or differential splicing. Strictly speaking, changes in isoform abundance in cancer are due to aberrations in transcription. However, as isoforms can differ in both their untranslated regions and coding sequences, they may produce a change to translational potential. MDM2, an oncogene that binds and inactivates the tumour suppressor p53, is encoded by 2 transcripts a short and long version, transcribed from different promoters. Translation from the long transcript is dampened by the presence of 2 inhibitory uORFs, which are absent from the shorter transcript. In human soft tissue tumours overexpression of MDM2 protein can occur from enrichment of the more highly translated short transcript (Brown et al., 1999). Altered promoter usage has also been identified in breast cancers that display reduced expression of the DNA repair gene BRCA1. Increased transcription of BRCA1 transcripts with a longer inhibitory 5′UTR are observed in these cells. The lower translation of the mORF in these transcripts arises from both an increase in secondary structure and the presence of 3 uORFs in the 5′UTR (Sobczak and Krzyzosiak, 2002). In head and neck cancer a change in splicing caused the retention of a longer inhibitory 5′UTR of the tumour suppressor hyaluronidase to be the predominant transcript produced in these cancers. This transcript contains numerous uAUGs that reduced expression of the mORF (Frost et al., 2000).
As uORFs often act as insulator elements on the downstream translation of a gene, the possibility of a SNP creating or removing an uORF would be expected to have serious consequences on mORF translation. Over 509 human genes have been identified where SNPs occur to create/delete an uORF. Further testing of 5 of these genes by reporter assay confirmed that the loss of the uORFs in all cases lead to increased translational efficiency (Calvo et al., 2009). The examination of SNPs within uORF sequences also needs future consideration. A SNP that creates a missense change within the second uORF of the serotonin receptor gene HTR3A has been significantly linked with bipolar affective disorder. Reporter assays showed this SNP to double luciferase activity (Niesler et al., 2001).
Familial uORF mutations have been reported for a number of diseases. In hereditary melanoma certain families show a G-T mutation, which creates an uAUG within the Cdk4/Cdk8 kinase inhibitor CDKN2A mRNA. This uORF, which is efficiently recognised by ribosomes, reduces CDKN2A mRNA expression and consequently alleviates its restriction on G1 progression in the cell cycle (Liu et al., 1999). The human hairless gene contains 4 uORFs, but it is the second (and longest) uORF in which a range of mutations has been identified in families with Marie Unna hereditary hypotrichosis. The mutations identified were either missense or nonsense or removed either the uORF initiation or termination codon, as uORF2 is in-frame with the mORF this would produce an N-terminally extended protein. All tested mutations were shown to enhance translation from a reporter gene. Furthermore, the observation that missense mutations augment translation suggests that the peptide sequence of the uORF2 is important for translational repression (Wen et al., 2009). Enhanced translation of the THPO gene is observed in hereditary thrombocythemia, the 5′UTR of which contains 7 uORFs. One familial mutation involves a nucleotide change that creates a new splice donor site, which eliminates all uORFs from the spliced message (Wiestner et al., 1998). Other mutations affect the seventh uORF, which overlaps with the mORF. The introduction of a nonsense mutation upstream of the mORF, allowed reinitiation by ribosomes after uORF7 translation (Ghilardi et al., 1999). Alternatively, a nucleotide deletion in uORF7, which places it in-frame with the coding sequence, produced a N-terminally extended protein (Ghilardi and Skoda, 1999, Kondo et al., 1998).
Collectively these examples highlight the growing emphasis on identifying mutations or SNPs within the 5′UTR that may influence disease progression or aetiology. The numerous ways in which an uORF may regulate translation will require careful study into how aberrant uORF regulation affects translation.
6. Conclusion
Given the prevalence of uORFs in mammalian transcripts it is perhaps surprising that still relatively little is known about how these elements are regulated. The current data strongly suggest that their regulation is likely to be complex and that several independent mechanisms exist to allow their repressive effects under normal conditions and selective translation of subsets of defined mRNAs under situations of pathophysiological cell stress. Further studies are needed to address how mutations/polymorphisms within uORFs/AUGs may contribute to an individual's disease response.
Acknowledgements
We would like to thank Thomas Sbarrato for useful discussions. This work was supported by funding from CRUK (JS), BBSRC (Professorial fellowship AEW), MRC (TP).
Contributor Information
Joanna Somers, Email: jks30@le.ac.uk.
Tuija Pöyry, Email: tp117@le.ac.uk.
Anne E. Willis, Email: aew5@le.ac.uk.
References
- Akimoto C., Sakashita E., Kasashima K., Kuroiwa K., Tominaga K., Hamamoto T. Translational repression of the McKusick-Kaufman syndrome transcript by unique upstream open reading frames encoding mitochondrial proteins with alternative polyadenylation sites. Biochimica et Biophysica Acta. 2012 doi: 10.1016/j.bbagen.2012.12.010. epub pii: S0304-4165(12)00356-X. [DOI] [PubMed] [Google Scholar]
- Bastide A., Karaa Z., Bornes S., Hieblot C., Lacazette E., Prats H. An upstream open reading frame within an IRES controls expression of a specific VEGF-A isoform. Nucleic Acids Research. 2008;36:2434–2445. doi: 10.1093/nar/gkn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besancon R., Valsesia-Wittmann S., Locher C., Delloye-Bourgeois C., Furhman L., Tutrone G. Upstream ORF affects MYCN translation depending on exon 1b alternative splicing. BMC Cancer. 2009;9:445. doi: 10.1186/1471-2407-9-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden S.P., Willis A.E. The biological and therapeutic relevance of mRNA translation in cancer. Nature Reviews Clinical Oncology. 2011;8:280–291. doi: 10.1038/nrclinonc.2011.16. [DOI] [PubMed] [Google Scholar]
- Brown C.Y., Mize G.J., Pineda M., George D.L., Morris D.R. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. doi: 10.1038/sj.onc.1202949. [DOI] [PubMed] [Google Scholar]
- Bushell M., Wood W., Carpenter G., Pain V.M., Morley S.J., Clemens M.J. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. Journal of Biological Chemistry. 2001;276:23922–23928. doi: 10.1074/jbc.M100384200. [DOI] [PubMed] [Google Scholar]
- Calkhoven C.F., Bouwman P.R., Snippe L., Ab G. Translation start site multiplicity of the CCAAT/enhancer binding protein alpha mRNA is dictated by a small 5′ open reading frame. Nucleic Acids Research. 1994;22:5540–5547. doi: 10.1093/nar/22.25.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven C.F., Muller C., Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes and Development. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Pagliarini D.J., Mootha V.K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.J. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col B., Oltean S., Banerjee R. Translational regulation of human methionine synthase by upstream open reading frames. Biochimica et Biophysica Acta. 2007;1769:532–540. doi: 10.1016/j.bbaexp.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Suzuki Y., Sugano S., Zhang M.Q. CART classification of human 5′ UTR sequences. Genome Research. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E., Sicheri F. The eIF2α kinases. In: Mathews M., Sonenberg N., Hershey J.W., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; New York: 2007. pp. 319–344. [Google Scholar]
- Diba F., Watson C.S., Gametchu B. 5′UTR sequences of the glucocorticoid receptor 1A transcript encode a peptide associated with translational regulation of the glucocorticoid receptor. Journal of Cellular Biochemistry. 2001;81:149–161. [PubMed] [Google Scholar]
- Fernandez J., Bode B., Koromilas A., Diehl J.A., Krukovets I., Snider M.D. Translation mediated by the internal ribosome entry site of the cat-1 mRNA is regulated by glucose availability in a PERK kinase-dependent manner. Journal of Biological Chemistry. 2002;277:11780–11787. doi: 10.1074/jbc.M110778200. [DOI] [PubMed] [Google Scholar]
- Fernandez J., Yaman I., Merrick W.C., Koromilas A., Wek R.C., Sood R. Regulation of internal ribosome entry site-mediated translation by eukaryotic initiation factor-2alpha phosphorylation and translation of a small upstream open reading frame. Journal of Biological Chemistry. 2002;277:2050–2058. doi: 10.1074/jbc.M109199200. [DOI] [PubMed] [Google Scholar]
- Fernandez J., Yaman I., Mishra R., Merrick W.C., Snider M.D., Lamers W.H. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. Journal of Biological Chemistry. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
- Frost G.I., Mohapatra G., Wong T.M., Csoka A.B., Gray J.W., Stern R. HYAL1LUCA-1, a candidate tumor suppressor gene on chromosome 3p21.3, is inactivated in head and neck squamous cell carcinomas by aberrant splicing of pre-mRNA. Oncogene. 2000;19:870–877. doi: 10.1038/sj.onc.1203317. [DOI] [PubMed] [Google Scholar]
- Gardner L.B. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Molecular and Cellular Biology. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi N., Skoda R.C. A single-base deletion in the thrombopoietin (TPO) gene causes familial essential thrombocythemia through a mechanism of more efficient translation of TPO mRNA. Blood. 1999;94:1480–1482. [PubMed] [Google Scholar]
- Ghilardi N., Wiestner A., Kikuchi M., Ohsaka A., Skoda R.C. Hereditary thrombocythaemia in a Japanese family is caused by a novel point mutation in the thrombopoietin gene. British Journal of Haematology. 1999;107:310–316. doi: 10.1046/j.1365-2141.1999.01710.x. [DOI] [PubMed] [Google Scholar]
- Hill J.R., Morris D.R. Cell-specific translation of S-adenosylmethionine decarboxylase mRNA. Regulation by the 5′ transcript leader. Journal of Biological Chemistry. 1992;267:21886–21893. [PubMed] [Google Scholar]
- Hill J.R., Morris D.R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. Journal of Biological Chemistry. 1993;268:726–731. [PubMed] [Google Scholar]
- Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annual Review of Microbiology. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. eIF3: a versatile scaffold for translation initiation complexes. Trends in Biochemical Sciences. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Hurtaud C., Gelly C., Bouillaud F., Levi-Meyrueis C. Translation control of UCP2 synthesis by the upstream open reading frame. Cellular and Molecular Life Sciences. 2006;63:1780–1789. doi: 10.1007/s00018-006-6129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono M., Mignone F., Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Inacio A., Silva A.L., Pinto J., Ji X., Morgado A., Almeida F. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. Journal of Biological Chemistry. 2004;279:32170–32180. doi: 10.1074/jbc.M405024200. [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Lareau L.F., Weissman J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Reviews Molecular Cell Biology. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C., Bruhat A., Carraro V., Urano F., Ferrara M., Ron D. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Research. 2001;29:4341–4351. doi: 10.1093/nar/29.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.M., Cho H., Kim Y.K. The upstream open reading frame of cyclin-dependent kinase inhibitor 1A mRNA negatively regulates translation of the downstream main open reading frame. Biochemical and Biophysical Research Communications. 2012;424:469–475. doi: 10.1016/j.bbrc.2012.06.135. [DOI] [PubMed] [Google Scholar]
- Kondo T., Okabe M., Sanada M., Kurosawa M., Suzuki S., Kobayashi M. Familial essential thrombocythemia associated with one-base deletion in the 5′-untranslated region of the thrombopoietin gene. Blood. 1998;92:1091–1096. [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Research. 1984;12:3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Research. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Molecular and Cellular Biology. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Molecular and Cellular Biology. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. Journal of Cell Biology. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. Journal of Biological Chemistry. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Research. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law G.L., Raney A., Heusner C., Morris D.R. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. Journal of Biological Chemistry. 2001;276:38036–38043. doi: 10.1074/jbc.M105944200. [DOI] [PubMed] [Google Scholar]
- Lee Y.Y., Cevallos R.C., Jan E. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. Journal of Biological Chemistry. 2009;284:6661–6673. doi: 10.1074/jbc.M806735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln A.J., Monczak Y., Williams S.C., Johnson P.F. Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames. Journal of Biological Chemistry. 1998;273:9552–9560. doi: 10.1074/jbc.273.16.9552. [DOI] [PubMed] [Google Scholar]
- Liu L., Dilworth D., Gao L., Monzon J., Summers A., Lassam N. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nature Genetics. 1999;21:128–132. doi: 10.1038/5082. [DOI] [PubMed] [Google Scholar]
- Luukkonen B.G., Tan W., Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. Journal of Virology. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M., Yachie N., Okada Y., Saito R., Tomita M. Bioinformatic analysis of post-transcriptional regulation by uORF in human and mouse. FEBS Letters. 2007;581:4184–4188. doi: 10.1016/j.febslet.2007.07.057. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature Genetics. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Mize G.J., Ruan H., Low J.J., Morris D.R. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. Journal of Biological Chemistry. 1998;273:32500–32505. doi: 10.1074/jbc.273.49.32500. [DOI] [PubMed] [Google Scholar]
- Moreno J.A., Radford H., Peretti D., Steinert J.R., Verity N., Martin M.G. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzarova V., Panek J., Gunisova S., Danyi I., Szamecz B., Valasek L.S. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genetics. 2011;7:e1002137. doi: 10.1371/journal.pgen.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesler B., Flohr T., Nothen M.M., Fischer C., Rietschel M., Franzek E. Association between the 5′ UTR variant C178T of the serotonin receptor gene HTR3A and bipolar affective disorder. Pharmacogenetics. 2001;11:471–475. doi: 10.1097/00008571-200108000-00002. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kashiwagi K., Matsuda Y., Janne O.A., Igarashi K. Gene structure and chromosomal localization of mouse S-adenosylmethionine decarboxylase. Gene. 1999;238:343–350. doi: 10.1016/s0378-1119(99)00355-8. [DOI] [PubMed] [Google Scholar]
- Oyama M., Itagaki C., Hata H., Suzuki Y., Izumi T., Natsume T. Analysis of small human proteins reveals the translation of upstream open reading frames of mRNAs. Genome Research. 2004;14:2048–2052. doi: 10.1101/gr.2384604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M., Kozuka-Hata H., Suzuki Y., Semba K., Yamamoto T., Sugano S. Diversity of translation start sites may define increased complexity of the human short ORFeome. Molecular and Cellular Proteomics. 2007;6:1000–1006. doi: 10.1074/mcp.M600297-MCP200. [DOI] [PubMed] [Google Scholar]
- Palam L.R., Baird T.D., Wek R.C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. Journal of Biological Chemistry. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola A.L., Kobilka B.K. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. Journal of Biological Chemistry. 1994;269:4497–4505. [PubMed] [Google Scholar]
- Peabody D.S. Translation initiation at an ACG triplet in mammalian cells. Journal of Biological Chemistry. 1987;262:11847–11851. [PubMed] [Google Scholar]
- Peabody D.S. Translation initiation at non-AUG triplets in mammalian cells. Journal of Biological Chemistry. 1989;264:5031–5035. [PubMed] [Google Scholar]
- Pecqueur C., Alves-Guerra M.C., Gelly C., Levi-Meyrueis C., Couplan E., Collins S. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress, and evidence for translational regulation. Journal of Biological Chemistry. 2001;276:8705–8712. doi: 10.1074/jbc.M006938200. [DOI] [PubMed] [Google Scholar]
- Pendleton L.C., Goodwin B.L., Flam B.R., Solomonson L.P., Eichler D.C. Endothelial argininosuccinate synthase mRNA 5′-untranslated region diversity. Infrastructure for tissue-specific expression. Journal of Biological Chemistry. 2002;277:25363–25369. doi: 10.1074/jbc.M111677200. [DOI] [PubMed] [Google Scholar]
- Pendleton L.C., Goodwin B.L., Solomonson L.P., Eichler D.C. Regulation of endothelial argininosuccinate synthase expression and NO production by an upstream open reading frame. Journal of Biological Chemistry. 2005;280:24252–24260. doi: 10.1074/jbc.M500106200. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes and Development. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon X., Wilson L.A., Stoneley M., Bastide A., King H.A., Somers J. RNA binding protein/RNA element interactions and the control of translation. Current Protein and Peptide Science. 2012;13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Kolupaeva V.G., Pisareva V.P., Merrick W.C., Hellen C.U., Pestova T.V. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes and Development. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley I.R., Kondrashov A., Young L.A., Dobbyn H.C., Hill K., Cannell I.G. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes and Development. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pöyry T.A., Kaminski A., Jackson R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes and Development. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabadan-Diehl C., Martinez A., Volpi S., Subburaju S., Aguilera G. Inhibition of vasopressin V1b receptor translation by upstream open reading frames in the 5′-untranslated region. Journal of Neuroendocrinology. 2007;19:309–319. doi: 10.1111/j.1365-2826.2007.01533.x. [DOI] [PubMed] [Google Scholar]
- Raney A., Baron A.C., Mize G.J., Law G.L., Morris D.R. In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. Journal of Biological Chemistry. 2000;275:24444–24450. doi: 10.1074/jbc.M003364200. [DOI] [PubMed] [Google Scholar]
- Raveh-Amit H., Maissel A., Poller J., Marom L., Elroy-Stein O., Shapira M. Translational control of protein kinase Ceta by two upstream open reading frames. Molecular and Cellular Biology. 2009;29:6140–6148. doi: 10.1128/MCB.01044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I., Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Current Opinion in Cell Biology. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Resch A.M., Ogurtsov A.Y., Rogozin I.B., Shabalina S.A., Koonin E.V. Evolution of alternative and constitutive regions of mammalian 5′UTRs. BMC Genomics. 2009;10:162. doi: 10.1186/1471-2164-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L.O., Jopling C.L., Jackson R.J., Willis A.E. Viral strategies to subvert the mammalian translation machinery. Progress in Molecular Biology and Translational Science. 2009;90:313–367. doi: 10.1016/S1877-1173(09)90009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.L., Pereira F.J., Morgado A., Kong J., Martins R., Faustino P. The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context. RNA. 2006;12:2160–2170. doi: 10.1261/rna.201406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nature Chemical Biology. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K., Krzyzosiak W.J. Structural determinants of BRCA1 translational regulation. Journal of Biological Chemistry. 2002;277:17349–17358. doi: 10.1074/jbc.M109162200. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs K.A., Bushell M., Willis A.E. Translational regulation of gene expression during conditions of cell stress. Molecular Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Stockklausner C., Breit S., Neu-Yilik G., Echner N., Hentze M.W., Kulozik A.E. The uORF-containing thrombopoietin mRNA escapes nonsense-mediated decay (NMD) Nucleic Acids Research. 2006;34:2355–2363. doi: 10.1093/nar/gkl277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B., Rutkai E., Cuchalova L., Munzarova V., Herrmannova A., Nielsen K.H. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes and Development. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A.C. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biology of the Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J.E. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Research. 1998;26:1150–1159. doi: 10.1093/nar/26.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Liu Y., Xu Y., Zhao Y., Hua R., Wang K. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nature Genetics. 2009;41:228–233. doi: 10.1038/ng.276. [DOI] [PubMed] [Google Scholar]
- Wiestner A., Schlemper R.J., van der Maas A.P., Skoda R.C. An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nature Genetics. 1998;18:49–52. doi: 10.1038/ng0198-49. [DOI] [PubMed] [Google Scholar]
- Yaman I., Fernandez J., Liu H., Caprara M., Komar A.A., Koromilas A.E. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- Zhao C., Datta S., Mandal P., Xu S., Hamilton T. Stress-sensitive regulation of IFRD1 mRNA decay is mediated by an upstream open reading frame. Journal of Biological Chemistry. 2010;285:8552–8562. doi: 10.1074/jbc.M109.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Palam L.R., Jiang L., Narasimhan J., Staschke K.A., Wek R.C. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. Journal of Biological Chemistry. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]