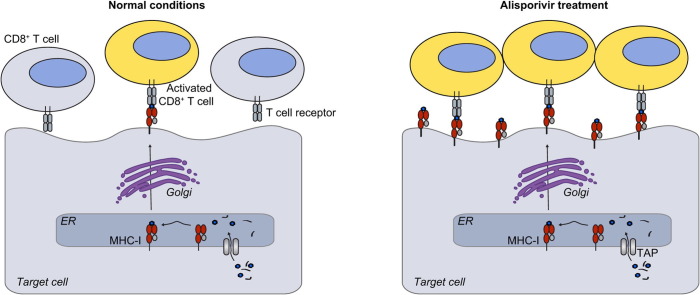
Graphical abstract
Abbreviations: HCV, hepatitis C virus; CsA, cyclosporine A; MHC, major histocompatibility complex; Cyp, cyclophilin; E:T ratio, effector-to-target ratio; NS5BEM, epitope-matched HCV non-structural protein 5B; IFN, interferon; b2m, beta-2 microglobulin; GLuc, Gaussia-luciferase; TFR, transferrin receptor; ERAP1, endoplasmic reticulum aminopeptidase 1; SFA, sanglifehrin A; CsH, cyclosporine H; TCR, T cell receptor; GeoMFI, geometric mean fluorescence intensity
Keywords: Cyclosporine A, Cyclophilin, HCV, MHC-I, Debio025, Antigen presentation
Abstract
Background & Aims
Cyclophilin-inhibitors have potent antiviral activity against Hepatitis C virus (HCV) and are promising candidates for broad-spectrum antiviral therapy. Cyclosporine A (CsA) acts immunosuppressive by blocking T cell activation and antigen presentation. Alisporivir, a non-immunosuppressive CsA analog in clinical development, does not inhibit T cell activation. In this study we explored the impact of alisporivir on antigen presentation.
Methods
Hepatoma cells endogenously expressing the epitope-restricting major histocompatibility complex-class I (MHC-I) allele HLA-A2 and constitutively expressing a viral antigen were established to study the impact of cyclophilin-inhibitors on antigen presentation. Antigen-specific CD8+ T cell activation and MHC-I surface expression were measured to quantify antigen presentation.
Results
Our work establishes a novel cell culture model to study antigen presentation in liver-derived cells. Authentic regulation of antigen presentation was ensured by the action of pro- and anti-inflammatory cytokines. Alisporivir pretreatment stimulated antigen presentation by hepatoma target cells, leading to enhancement of antigen-specific CD8+ T cell activation by 40%. Alisporivir, as well as a panel of other cyclophilin-inhibitors, induced an increase of MHC-I and beta-2 microglobulin on the surface of several cell lines. The drug neither enhanced MHC-I transcript or protein levels nor affected surface expression of other proteins or protein trafficking in general. Proteasome-inhibitors completely blocked the alisporivir-directed enhancement of surface MHC-I, suggesting an influence of the drug on peptide-availability.
Conclusions
Alisporivir stimulates antigen presentation by inducing enhanced MHC-I surface expression, thereby promoting antigen-specific CD8+ T cell activation. This immunostimulatory function might further contribute to the antiviral activity of non-immunosuppressive cyclophilin-inhibitors.
Introduction
Hepatitis C Virus (HCV) establishes persistence in a high rate of patients, resulting in more than 160 million chronic infections worldwide. In recent years, direct acting antivirals and host targeting antivirals have been identified, allowing new opportunities for antiviral therapy of patients chronically infected with HCV [1]. Characteristics of host targeting antivirals include their broad genotype coverage and their high barrier to resistance [1]. One such compound is the cyclophilin (Cyp)-inhibitor alisporivir, which targets the cellular protein and HCV host factor cyclophilin A (CypA), and has been in clinical development for HCV therapy [2], [3], [4]. Furthermore, due to involvement of cyclophilins in the lifecycle of a broad range of viruses like human immunodeficiency virus (HIV), Hepatitis B Virus (HBV) or several coronaviruses, alisporivir represents a promising candidate for pan-viral therapy [5]. The drug is an analog of the cyclic undecapeptide cyclosporine A (CsA), which blocks T cell activation by forming a ternary complex together with CypA and the phosphatase calcineurin [6]. In addition, CsA suppresses major histocompatibility complex-class I (MHC-I) dependent antigen presentation by a poorly defined mechanism [7]. The structure of alisporivir differs in two amino acid residues from the immunosuppressant CsA, abrogating the inhibitory effect on T cell activation [8]. However, its effect on antigen presentation has not been clarified so far.
We recently established an immunological model to characterize antiviral responses of HCV-specific CD8+ T cells activated by hepatoma cells harboring a self-replicating HCV replicon [9]. While this assay served to analyze the antiviral efficiency of T cell responses, its use to gain insights into MHC-I antigen presentation was limited due to ectopic expression of MHC-I.
We have now engineered liver cells endogenously expressing the epitope-restricting MHC-I allele and ectopically expressing an epitope-matched HCV protein and thereby adapted our initial model to quantify antigen presentation. Alisporivir and further non-immunosuppressive Cyp-inhibitors stimulated antigen presentation by upregulation of MHC-I surface expression, resulting in induction of enhanced antigen-specific CD8+ T cell responses. These findings point to additional therapeutic benefits for application of Cyp-inhibitors as antivirals mediated by enhanced immune responses.
Material and methods
Cell culture
Cell lines of hepatic origin: Huh7_Lunet, HepG2, Huh6.1 and cell lines of non-hepatic origin: HEK 293T, HeLa, 5637, LS180, CaCo2 and A549 were cultured in Dulbeccós modified essential medium (DMEM; Life Technologies, Darmstadt, Germany) supplemented with 2 mM L-glutamine, 0.1 mM nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin (all Life Technologies) and 10% fetal calf serum (FCS). Huh7_Lunet cells have been described previously [10]. Huh6.1 cells are a sub-clone of the human hepatoma cell line Huh6 and were generated by curing Huh6 cells harboring a persistent GT1b Con-1 HCV replicon with CsA and daclatasvir [11], [12]. HepaRG cells were cultured in Williams E medium (Life Technologies) supplemented with 5 μg/ml of insulin, 50 μM hydrocortisone hemisuccinate (both Sigma Aldrich, Steinheim, Germany), 100 U/ml of penicillin, 100 U/ml of streptomycin and 10% FCS. NS5B2594-2602 (NS5BEM)-specific CD8+ T cell clones were generated by limited dilution after sorting of tet+ CD8+ T cells as previously described [9]. All cells were incubated in a 37 °C incubator at 95% humidity and 5% CO2. Alisporivir (Novartis, Basel, Switzerland) was dissolved to 20 mg/ml in DMSO. Detailed information on other inhibitors can be found in [13] and Supplementary materials and methods.
T cell co-cultivation
A detailed description of the T cell co-cultivation assay has been published earlier [9]. In brief, 1 × 105 T cells were co-cultured with 1 × 105 Huh6.1 or HepG2 target cells in one well of a 96 well dish. Target cells were washed extensively before co-culture to remove residual drug amounts. Handling of NS5B2594 peptide (ALYDVVSKL; Biosynthan, Berlin, Germany) was described previously [14]. Upon 5 h of co-culture at an effector:target (E:T) ratio of 1:1, T cells were stained for intracellular IFN-γ and CD8 surface expression and analyzed on a BD FACSCanto II (Becton Dickinson, New Jersey, USA). Data were analyzed using FlowJo Software (Tree Star, USA).
Flow cytometry
For staining of surface proteins, 1 × 105 cells were seeded for drug treatment and detached on day 3 after seeding (resulting in ∼5 × 105 cells). Cells were resuspended in ice-cold PBS with 3% bovine serum albumin (PBS 3% BSA) containing a primary antibody at the individual working dilution. After 1 h incubation at 4 °C, cells were washed twice and taken up in ice-cold PBS 3% BSA containing a goat-anti-mouse antibody labeled with phycoerythrin (PE) (Santa Cruz, Dallas, USA). After incubation for 1 h at 4 °C in the dark, cells were washed twice, resuspended in ice-cold PBS and analyzed on a BD FACSCalibur (BD biosciences, New Jersey, USA). All washing steps were performed with ice-cold PBS. Data were analyzed using FlowJo Software (Tree Star, USA). Primary antibodies and respective concentrations used during this study were anti-IFN-γR1 (1 μg/ml; R&D systems, Minneapolis, Canada), anti-CD13 (1 μg/ml; abcam), anti-HLA-A∗02 (1 μg/ml; abcam, Cambridge, UK), anti-β2 microglobulin (1 μg/ml) and anti-HLA-A, B, C (W6/32; 4 μg/ml) (both Biolegend, London, UK). Isotype control antibodies were mouse IgG2bκ (Biolegend) and mouse IgG1 (R&D systems, Wiesbaden, Germany). For each condition data of at least 10,000 cells were collected. Staining for intracellular IFN-γ and surface CD8 was performed as described previously [9].
Statistical analysis
For statistical analysis two-tailed paired t test was performed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA). Further details on statistical tests are indicated in the figure legends; ∗p <0.05; ∗∗p <0.01; ∗∗∗p <0.001.
Transcriptome data
Transcriptome data are accessible via GSE68927.
Results
An in vitro immunological model to measure functional antigen presentation
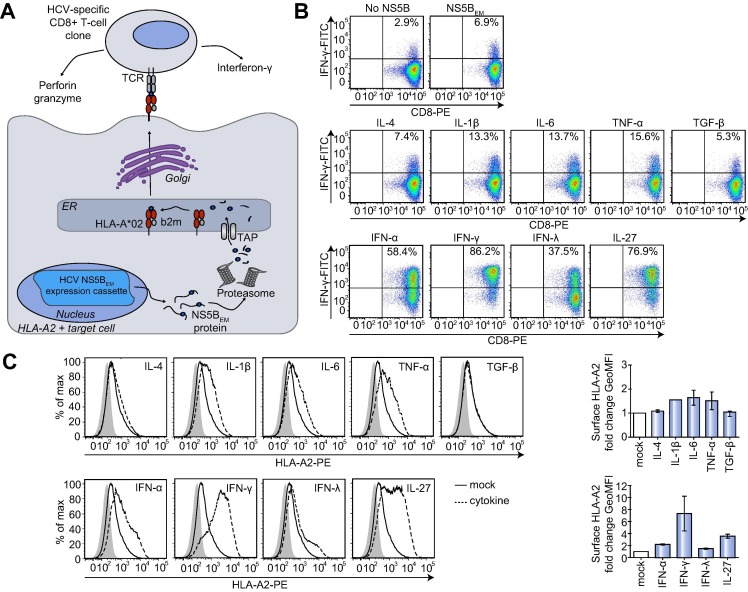
The first aim of our study was the establishment of a cell culture model allowing a quantitative assessment of functional antigen presentation by hepatocytes. To this end, we modified a previously established assay employing co-cultivation of HCV-specific HLA-A∗02-restricted CD8+ T cells and Huh7 hepatoma cells ectopically expressing the MHC-I allele HLA-A∗02 [9]. To generate a more physiological environment, we used hepatoma cell lines endogenously expressing HLA-A∗02 and stably expressing an epitope-matched HCV-NS5B protein (NS5BEM; EM = epitope-matched) (Fig. 1 A). NS5B-derived peptides are presented on the cellular surface via HLA-A∗02, resulting in antigen-specific T cell activation during co-culture of HLA-A∗02+ NS5BEM transduced hepatoma cells with NS5B-specific CD8+ T cells and concomitant production of various effector molecules (Fig. 1A). Importantly, this co-culture assay encompasses the whole antigen presentation process, from physiological regulation of MHC-I expression and processing of an endogenous antigen to surface presentation via MHC-I. Therefore, modifications at any step of this pathway significantly impacting on the efficiency of functional antigen presentation should be detectable as changes in CD8+ T cell activation.
Fig. 1.
An immunological model to measure antigen presentation. (A) Illustration of the co-cultivation assay as modified from Jo et al.[9]. Hepatoma target cells endogenously expressing HLA-A∗02 and stably expressing an epitope-matched HCV NS5B protein (NS5BEM) were co-cultured with HLA-A∗02-restricted NS5BEM-specific CD8+ T cell clones. Antigen-specific CD8+ T cell activation induced by binding of the T cell receptor (TCR) to the NS5BEM-peptide-MHC-I complex stimulates IFN-γ-production which is measured as indirect readout for the antigen presentation capacity of target cells. (B) Various cytokines stimulate antigen presentation by Huh6.1 cells as measured by a co-cultivation assay. Huh6.1 cells expressing NS5BEM, were pretreated with the indicated cytokines and co-cultured with CD8+ T cells. CD8+ T cells were stained for intracellular IFN-γ and surface CD8. The upper right quadrant includes the percentage of activated IFN-γ-producing CD8+ T cells. Depicted are results of one representative experiment (n = 3). (C) Cytokines which stimulate antigen presentation enhance MHC-I surface expression. Huh6.1 cells were treated with the indicated cytokines and stained for HLA-A∗02. Data are presented as histograms. Shaded area, IgG2bκ isotype control; Diagrams on the right present the mean fold change of geometric mean fluorescence intensities (GeoMFIs) of two independent experiments (n = 2). ER, endoplasmic reticulum; HLA, human leukocyte antigen; b2m, β-2 microglobulin; TAP, transporter associated with antigen processing; TCR, T cell receptor.
We tested two HLA-A∗02+ hepatoma cell lines, characterized by either high (HepG2) or low (Huh6.1) HLA-A∗02 expression levels (Supplementary Fig. 1A). More than 90% of CD8+ T cells were activated upon co-culture with HepG2_NS5BEM cells, which could not be further enhanced by pretreatment of HepG2 cells with IFN-γ, a well-known stimulant of MHC-I antigen presentation (Supplementary Fig. 1B) [15]. Therefore, HepG2 cells were not suitable for our model, since any condition enhancing antigen presentation would be missed. In contrast, Huh6.1_NS5BEM cells gave rise to a significant portion of activated CD8+ T cells, which was lower than for HepG2 (Fig. 1B), possibly due to lower MHC-I expression levels (Supplementary Fig. 1A). The basic CD8+ T cell activation rate in co-cultivation assays varied between 6% and 42% in different experiments (compare Figs. 1B, 2 C, D), due to passage number and pre-activation status of CD8+ T cells. Still, modulatory effects of cytokines remained the same, independent from baseline activation levels (data not shown). Upon confirming cytokine receptor expression on Huh6 Con1 cells (Supplementary Table 1), the founder cell line of Huh6.1, we used several pro-inflammatory cytokines (IL-1β, IL-6, TNF-α), anti-inflammatory cytokines (TGF-β and IL-4) and type I, II and III interferons (IFN-α, IFN-γ, IFN-λ) to evaluate physiological regulation of antigen presentation in our system and to define its dynamical range. Pretreatment of Huh6.1 cells with IL-1β, IL-6 and TNF-α led to an enhancement of CD8+ T cell activation, while IL-4 and TGF-β, as expected, did not have a stimulating effect (Fig. 1B). However, type I and II IFNs, well-known stimulants of antigen presentation [15] as well as a type III IFN and IL-27, which was reported to have IFN-γ-like effects on hepatocytes [16] stimulated functional antigen presentation in Huh6.1 cells even stronger (Fig. 1B). Furthermore, alterations in HLA-A∗02 surface expression of Huh6.1 target cells upon treatment with different cytokines correlated with changes in their antigen presentation potency (Fig. 1C).
Fig. 2.
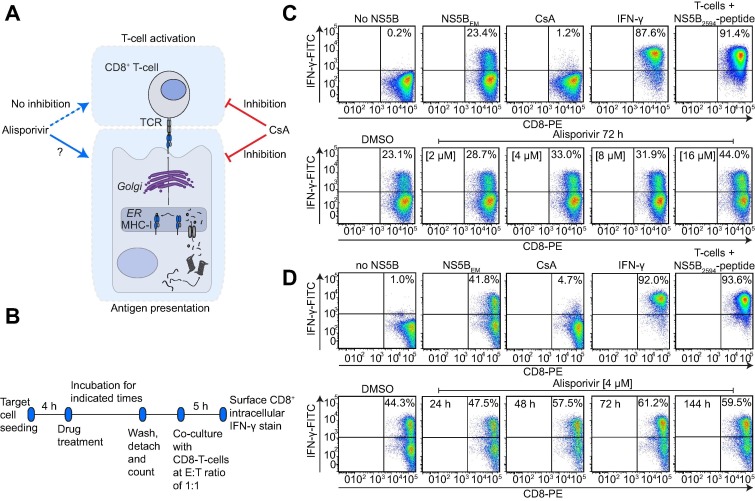
The cyclophilin-inhibitor Alisporivir stimulates antigen presentation. (A) Illustration of the inhibition profile of Cyp-inhibitor CsA and its analog alisporivir. CsA acts immunosuppressive, blocking T cell activation as well as antigen presentation, whereas alisporivir does not inhibit T cell activation, while its effect on antigen presentation is unclear. (B) Assay-setup applied to explore effects of alisporivir on antigen presentation. Note that prior to co-culture cells were washed extensively to avoid drug carryover. (C) Huh6.1 cells were treated for 72 h with the indicated alisporivir concentrations, DMSO, CsA or IFN-γ. Huh6.1 cells not expressing the epitope-matched NS5B (no NS5B) or CD8+ T cells exogenously stimulated with the epitope containing peptide (NS5B2594-peptide) served as technical controls. Depicted are data derived from one representative experiment (n = 2). (D) After incubation of Huh6.1 for the indicated times, co-culture with CD8+ T cells was performed according to description in (B) and (C). Data are derived from one representative experiment (n = 2).
In conclusion, endogenously processed peptides were presented by Huh6.1 cells and gave rise to an HLA-A∗02-restricted antigen-specific CD8+ T cell activation. HLA-A∗02 surface expression of Huh6.1 cells allowed physiological modulation of functional antigen presentation by cytokines, likely reflecting low MHC-I expression levels in human liver [17]. Overall, the Huh6.1 model therefore appeared suitable to detect immunomodulatory effects on antigen presentation.
Alisporivir stimulates functional antigen presentation
While CsA and FK506 have been reported to block T cell activation by inhibition of calcineurin and to antagonize antigen presentation [6], [7], the CsA analog alisporivir does not suppress T cell activation [8]. However, whether alisporivir impairs antigen presentation, has not been investigated (Fig. 2A). Therefore, we pretreated Huh6.1 target cells with different concentrations and for different durations with alisporivir and determined the impact on CD8+ T cell activation as a measure of functional antigen presentation. Before co-culture with HCV-specific CD8+ T cells, target cells were detached and washed extensively to avoid drug carryover into co-culture (Fig. 2B).
IFN-γ pretreatment of Huh6.1 cells and preloading of T cells with a NS5B2594 peptide served as positive controls [14], [15] (Fig. 2C), both resulting in strong activation of CD8+ T cells. As expected, pretreatment with CsA abolished antigen presentation by Huh6.1 cells (Fig. 2C). In striking contrast, alisporivir pretreatment of Huh6.1 target cells dose-dependently increased the percentage of activated CD8+ T cells, suggesting a stimulation of the antigen presentation capacity of Huh6.1 by alisporivir (Fig. 2C). We used 2 or 4 μM alisporivir in all subsequent experiments since this concentration range was regarded as physiologically relevant, based on its efficacy to inhibit HCV replication in vitro and in vivo [18], [19]. Next, Huh6.1 cells were pretreated for different times with alisporivir to explore the kinetics of immunostimulation. A 24 h pretreatment only had minor effects, whereas 48 h after treatment, the increase of CD8+ T cell activation reached a plateau and remained stable for at least 6 days (Fig. 2D), arguing for a slow and accumulating process increasing antigen presentation by alisporivir. The percentage of activated CD8+ T cells was enhanced by approximately 40% upon alisporivir treatment during all experiments (compare Fig. 2C, D).
In summary, we demonstrated that alisporivir has an immunostimulatory effect on antigen presentation by human hepatoma cells, possibly supporting its antiviral activity in vivo.
Alisporivir enhances MHC-I surface expression on cell lines of hepatic and non-hepatic origin
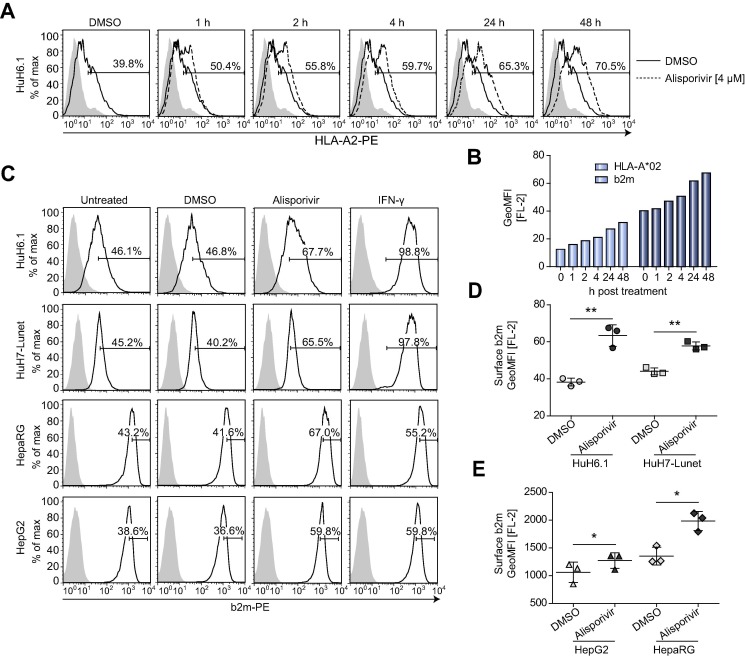
So far, we detected an immunostimulatory effect of alisporivir by using CD8+ T cell activation as an indirect readout for antigen presentation. Assuming that the amount of MHC-I/peptide on the cell surface is a limiting factor of antigen-specific T cell activation, we measured surface HLA-A∗02 on Huh6.1 cells after different times of alisporivir treatment (Fig. 3 A, B). Indeed, alisporivir slowly and continuously enhanced HLA-A∗02 and beta-2-microglobulin (b2m) surface expression over time (Fig. 3A, B). To assess whether this effect was restricted to Huh6.1 cells we analyzed further liver-derived cell lines (Fig. 3C). Since these cell lines, apart from HepG2, endogenously expressed HLA alleles differing from HLA-A∗02 we measured their b2m surface expression, which was indeed significantly upregulated in Huh6.1, Huh7_Lunet, HepG2 and HepaRG after 48 h of alisporivir treatment compared to DMSO controls (Fig. 3C–E). Analogous results were obtained with an MHC-I specific antibody (Supplementary Fig. 2A, B). Furthermore, alisporivir treatment significantly enhanced MHC-I surface expression on lung epithelial (A549) cells and a similar trend was observed for further cell lines of non-hepatic origin (Supplementary Fig. 2C, D).
Fig. 3.
Alisporivir enhances β-2 microglobulin (b2m) surface expression on various liver-derived cell lines. (A) Huh6.1 cells were treated with alisporivir for the indicated times and HLA-A∗02 surface expression was analyzed. Data are presented as histograms and include the percentage of cells with a positive signal for HLA-A∗02. Shaded area: IgG2bκ isotype control. Data are derived from one experiment which was repeated with an anti-b2m antibody. (B) Depicted are GeoMFIs derived from (A) and GeoMFIs for a b2m surface-stain on Huh6.1 cells treated as described in (A). (C) b2m surface-stain after treatment of various liver-derived cell lines with alisporivir. Huh6.1, Huh7_Lunet, HepG2 and HepaRG cells were treated as indicated. Data are presented as histograms including the percentage of cells showing a high expression of b2m. Shaded area: IgG1 isotype control. Depicted are data from one representative experiment (n = 3). (D, E) GeoMFIs of b2m surface-stains of various liver-derived cell lines performed according to (C). Data are derived from three independent experiments. Mean and SD are depicted as scatter plot. Data were analyzed for statistical significance by paired t test (n = 3). SD; standard deviation.
In summary, upregulation of MHC-I surface expression by alisporivir was not restricted to a particular cell line, but found in cells of various origins.
Alisporivir does not affect MHC-I mRNA or protein levels nor generally impact on protein secretion
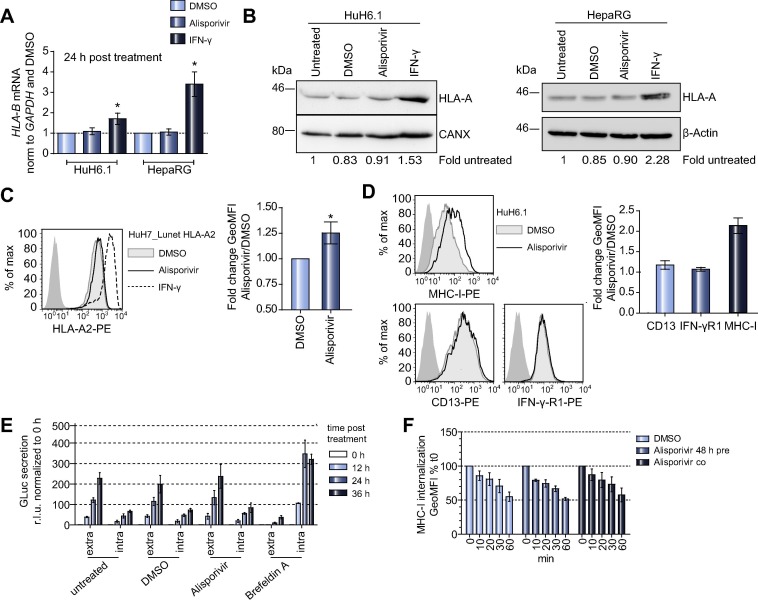
We next wanted to address, whether the alisporivir-induced enhancement of MHC-I surface expression was due to changes in MHC-I mRNA and protein expression levels. While IFN-γ, as expected, increased mRNA expression of both HLA-B and b2m, no effect was detected for alisporivir (Fig. 4 A; Supplementary Fig. 3A). In line with these data, MHC-I protein levels in Huh6.1 or HepaRG cells were not altered by alisporivir, in contrast to IFN-γ treatment (Fig. 4B). In addition, we found a slight but significant upregulation of HLA-A∗02 surface expression on Huh7_Lunet cells ectopically expressing HLA-A∗02 under control of a heterologous promoter (Huh7_Lunet HLA-A∗02) [9], further arguing against a mechanism involving regulation of HLA gene expression (Fig. 4C). An enhancement of MHC-I surface expression by alisporivir could also be caused by a general effect on protein surface expression, in line with a previous study reporting an upregulation of pathways associated with protein trafficking by CsA analog NIM-811 [20]. However, neither surface expression of IFN-γ-receptor chain 1 (IFN-γR1), a type I transmembrane protein like MHC-I α-chain nor CD13, which is like MHC-I co-translationally glycosylated inside the endoplasmic reticulum and transported through the Golgi to the cell surface [21], was affected by alisporivir treatment (Fig. 4D). A general effect on cellular transport was further excluded, because alisporivir did not affect Gaussia-luciferase (GLuc) secretion, transferrin receptor (TFR) recycling dynamics (Fig. 4E; Supplementary Fig. 3B) or internalization of MHC-I/b2m (Fig. 4F; Supplementary Fig. 3C). Finally, alisporivir did not induce enhanced MHC-I passage through the Golgi (Supplementary Fig. 3D, E), nor cause obvious alterations of intracellular MHC-I distribution (Supplementary Fig. 4).
Fig. 4.
Alisporivir does not enhance MHC-I transcript or protein expression nor affect overall protein secretion. (A) HLA-B mRNA levels in Huh6.1 and HepaRG cells were determined by quantitative PCR after indicated treatments. Results are normalized to GAPDH as internal reference and DMSO as vehicle control. Shown are mean and SD derived from triplicate values of three independent experiments (n = 3). Data were analyzed for statistical significance by paired t test. Please note that HepaRG cells do not express HLA-A2, therefore HLA-B mRNA was quantified instead, since primers detecting all HLA-B alleles have been published [23] and transcriptional regulation is comparable to HLA-A. (B) Western blot analyses of HLA-A protein levels in Huh6.1 and HepaRG cells after 48 h of DMSO, alisporivir or IFN-γ treatment. Calnexin (CANX) and β-Actin served as loading controls. Values below indicate expression signals normalized to loading control and to untreated. Data are derived from one representative experiment (n = 2). (C) Huh7_Lunet cells expressing HLA-A∗02 under control of a constitutive (EF1α) promoter were treated as indicated and stained with an antibody specific for HLA-A∗02 or an IgG2bκ isotype control antibody (dark grey area). Left: Data of one representative experiment are depicted as histogram. Right: Mean and SD of GeoMFIs of four independent experiments. Data were analyzed for statistical significance by paired t test (n = 4). (D) Surface expression of MHC-I, CD13 and IFN-γ receptor 1 (IFN-γR1) on Huh6.1 cells after treatment with alisporivir. Left: Data are depicted as histograms. Dark shaded area, goat-anti-mouse-PE antibody (MHC-I) or IgG1 isotype control antibody (CD13, IFN-γR1). Right: Depicted are the mean fold changes and SD of GeoMFIs of Alisporivir treated cells normalized to DMSO. Data are derived from two independent experiments (n = 2). (E) Gaussia-luciferase (GLuc) secretion over time in Huh6.1 cells upon transfection of a GLuc expression plasmid into Huh6.1 cells. Data were normalized to the 0 h value. Depicted are means and SD of two independent experiments (n = 2). (F) MHC-I internalization over time in Huh6.1 cells. Huh6.1 cells were either pretreated for 48 h or treated during the time of the experiment with alisporivir. GeoMFIs were normalized to t0 and plotted against time (min) to monitor MHC-I internalization. Data show mean and SD derived from two independent experiments (n = 2), which were repeated using identical settings and a b2m-specific antibody (Supplementary Fig. 3C).
In summary, our data suggested a specific enhancement of MHC-I and b2m surface expression upon alisporivir treatment, which was neither caused by an induction of basal MHC-I expression nor by general effects on cellular protein trafficking. We also found no evidence for a strong impact of alisporivir on MHC-I internalization or its passage through the Golgi.
Alisporivir enhances surface expression of peptide-loaded MHC-I molecules
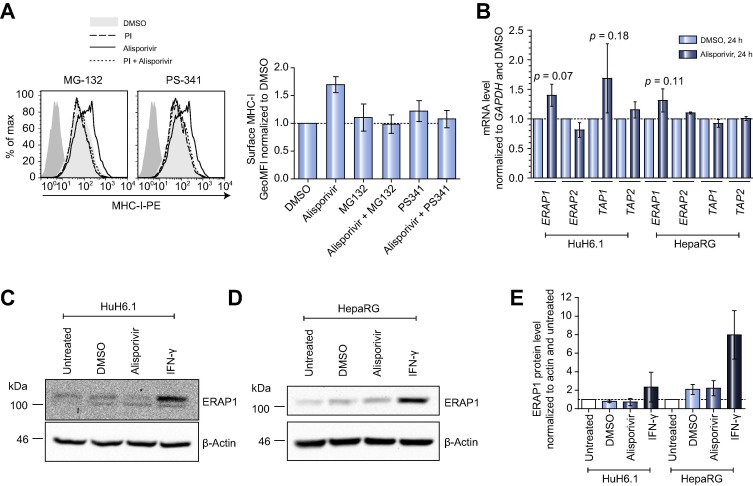
We were wondering if alisporivir induced effects depended on peptide availability, since peptide amount is a limiting factor for MHC-I antigen presentation [22]. Indeed, we observed a complete inhibition of MHC-I upregulation if alisporivir was combined with proteasome-inhibitors (MG-132 or PS-341), whereas basal MHC-I surface expression was not affected under these conditions (Fig. 5 A). These results argued for enhanced surface expression primarily of peptide-loaded MHC-I and for a critical role of peptide-availability, suggesting peptide-generation as potential target of alisporivir. Interestingly, a recent study observed upregulation of surface MHC-I upon p53 activation mediated by enhanced expression of endoplasmic reticulum aminopeptidase 1 (ERAP1) and transporter associated with antigen processing 1 (TAP1) [23]. We indeed detected a minor but insignificant upregulation of ERAP1 mRNA by alisporivir (Fig. 5B) but found no changes in ERAP1 protein levels or p53 activation status (Fig. 5C–E and data not shown).
Fig. 5.
Alisporivir induces enhanced surface expression of peptide-loaded MHC-I. (A) Proteasome-inhibitors (PI) block an alisporivir-induced upregulation of surface MHC-I. Left: Huh6.1 cells were treated with DMSO as vehicle control, alisporivir, a PI; left: MG-132; right: PS-341) or a combination of alisporivir and PI. MHC-I surface expression was detected with W6/32 antibody. Depicted is one representative out of two independent experiments. Dark shaded area, goat-anti-mouse-PE antibody. Right: Depicted are mean and SD of two independent experiments. GeoMFIs were normalized to DMSO control (n = 2). (B) Transcript levels of ERAP and TAP after alisporivir treatment. Huh6.1 or HepaRG cells were treated with DMSO or alisporivir. 24 h later total RNA was prepared and transcript levels were determined in triplicates by quantitative PCR. Data were normalized to GAPDH and DMSO and are given as mean and SD of three independent experiments (n = 3). Statistical data analysis was performed using paired t test. (C, D) Western blot analyses of ERAP1 expression in Huh6.1 (C) and HepaRG (D) cells. Cells were left untreated or treated with DMSO, alisporivir or IFN-γ. Presented is one representative out of two independent experiments for each cell line (n = 2). (E) Densitometric analysis of Western blots against ERAP1 using Quantity One 1-D software (BioRad). Data were normalized to β-Actin and to untreated and are given as mean and SD of two independent experiments (n = 2).
In conclusion, alisporivir led to upregulation of peptide-loaded MHC-I complexes, possibly due to enhanced peptide-availability by a yet to be defined mechanism.
Cyclophilin-inhibitors and analogs thereof induce upregulation of MHC-I surface expression
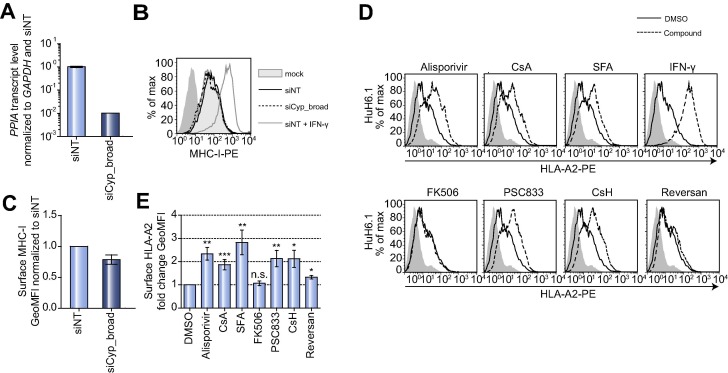
In search for the molecular target of alisporivir we first focused on CypA and CypB, which are the most common and best studied of the Cyp family [20] and like other members, show comparable expression in Huh6.1 and Huh7 cells (Supplementary Table 2). However, simultaneous knockdown of CypA and B did not significantly affect MHC-I surface expression of untreated or alisporivir treated Huh6.1 cells (Fig. 6 A–C and data not shown), suggesting that these most abundant Cyps were not the main determinants mediating surface accumulation of MHC-I. Cyp-inhibitors cover a broad range of cellular targets, including adenosine triphosphate binding cassette (ABC)-transporters, Na+-taurocholate co-transporting polypeptide (SLC10A1, NTCP) or calcineurin [24], [25]. Therefore, we tested a panel of compounds with different inhibition profiles [13] for their effect on MHC-I surface expression, using non-cytotoxic concentrations (Supplementary Fig. 5C). CsA and the structurally unrelated Cyp-inhibitor sanglifehrin A (SFA) stimulated a significant increase of HLA-A∗02 and MHC-I surface expression to an extent similar to alisporivir (Fig. 6D, E; Supplementary Fig. 5A, B), arguing for Cyp-inhibition as underlying cause. In agreement with this assumption, ABC-transporter inhibitors reversan and piperine both did not strongly impact on the amount of surface HLA-A∗02 or MHC-I, nor did FK506, which inhibits a different group of peptidyl-prolyl cis trans isomerases (Fig. 6D, E; Supplementary Fig. 5A, B) [5]. In contrast, the cyclic undecapeptides PSC833 and cyclosporine H (CsH), both with reduced affinity at least for the most common Cyps [20], [26] induced a significant upregulation of HLA-A∗02 and MHC-I surface expression to a similar degree as alisporivir (Fig. 6D, E; Supplementary Fig. 5A, B). In summary, cyclophilins are most likely the targets responsible for upregulation of MHC-I surface expression.
Fig. 6.
Cyclophilin-inhibitors and analogs thereof promote an upregulation of MHC-I surface expression. (A) CypA (PPIA) knockdown efficiency in Huh6.1 cells. Cells were transfected with siCyp_broad, targeting expression of CypA (PPIA) and CypB (PPIB). PPIA mRNA levels were measured in triplicates by quantitative PCR at 72 h post transfection and normalized to GAPDH and a non-targeting control siRNA (siNT). Presented are mean and SD of triplicate values of two independent experiments in logarithmic scale (n = 2). (B) Detection of MHC-I surface expression 72 h after Cyp silencing in Huh6.1 using a W6/32 antibody. Depicted is one representative out of two independent experiments (n = 2). Dark shaded area, goat-anti-mouse PE secondary antibody. (C) Mean and SD of GeoMFIs of MHC-I surface expression normalized to siNT. Data are derived from two independent experiments (n = 2). (D) Cyp-inhibitors enhance HLA-A∗02 surface expression. Huh6.1 cells were treated with Alisporivir (2 μM), CsA (2 μM), Sanglifehrin A (SFA; 4 μM), IFN-γ (10 ng/ml), FK506 (2 μM), PSC833 (2 μM), CsH (2 μM) or Reversan (2 μM). MHC-I was detected with a HLA-A∗02-specific antibody. Data are derived from one representative experiment of at least three independent experiments for each inhibitor (n ⩾3). Dark shaded area, IgG2bκ isotype control. (E) Depicted are mean fold changes of GeoMFIs normalized to DMSO controls of at least three independent experiments performed as described in (D) (n ⩾3). For statistical analysis paired t test was performed.
Discussion
For our study of Cyp-inhibitor mediated effects on antigen presentation, we modified our previously developed immunological model [9] by using Huh6.1 cells, intrinsically expressing HLA-A∗02 and with constitutive ectopic expression of HCV-NS5B. Huh6.1 cells (HLA-A∗02low) activated CD8+ T cells and provided a dynamic range for up and downregulatory effects. In addition, pro-inflammatory cytokines and IFNs enhanced MHC-I surface expression and stimulation of CD8+ T cells during co-culture, arguing for physiological regulation of functional antigen presentation in Huh6.1 cells. Using this model, we were able to validate the proposed IFN-γ-like effect of IL-27 on hepatocytes [16].
CsA has been shown to inhibit antigen presentation [7], however, we now found that the CsA analog alisporivir exerted an immunostimulatory effect, mediated by slow enhancement of MHC-I surface expression over time, accompanied by a higher percentage of activated HCV-specific CD8+ T cells. Interestingly, CsA also increased MHC-I surface expression, but still entirely blocked functional antigen presentation to T cells by a yet to be defined mechanism [7]. Upregulation of MHC-I surface expression was confirmed in a panel of cell lines of diverse origins and not due to changes in total MHC-I expression levels. Furthermore, alisporivir specifically upregulated MHC-I since neither expression of other surface proteins (CD13, IFN-γR1) nor overall protein secretion was modified. We also could not find evidence for an impact of alisporivir on MHC-I trafficking, endocytosis or degradation, although we cannot exclude that we missed subtle effects in MHC-I transport dynamics, due to limitations of available assays. Still, we favor a mechanism mediated by enhancing peptide amounts available for MHC-I loading, since the stimulatory effect of alisporivir, but not regular MHC-I surface expression, was completely abrogated by proteasome-inhibitors. We hypothesize, that inhibition of the chaperone-like functions of Cyps by alisporivir might enhance peptide pools available for presentation on MHC-I by increasing the amount of unfolded proteins [24]. Still, further studies are required to clarify the mechanism how alisporivir stimulates MHC-I surface expression.
Cyps are a group of peptidyl-prolyl cis trans isomerases, including at least 17 family members in humans, most of which are poorly studied regarding their inhibition by Cyp-inhibitors and their molecular function [20], [27]. Most Cyp-inhibitors are not restricted to Cyps, but also act against additional targets, like ABC-transporters or Calcineurin [24]. An involvement of Cyps in MHC-I surface upregulation was supported by the fact that all Cyp-inhibitors tested increased MHC-I surface expression, in contrast to FK506, an inhibitor of a different class of peptidyl-prolyl cis trans isomerases [5] and two ABC-transporter inhibitors, reversan and piperine [13]. However, PSC833 and CsH, CsA analogs with reduced affinity for the best studied Cyps [20], [26] induced the same phenotype as observed for CsA, alisporivir and SFA, whereas siRNA-mediated knockdown of CypA and CypB, the most common Cyps, did not affect MHC-I surface expression, as well as individual knockdown of several other Cyps (data not shown). Altogether, our results either point to a potential role of less studied Cyps, which can still be blocked by all compounds [27] or to a redundant role of many Cyps. Due to the variety of potential targets, the possible involvement of several Cyps with analogous functions, we did not attempt to further define specific Cyps potentially involved in regulation of MHC-I surface expression. For the same reasons and due to high abundance of many Cyps, we neither carried out Cyp-overexpression. Alisporivir, as well as other non-immunosuppressive Cyp-inhibitors like SCY-635 have been in clinical development for antiviral therapy of chronically HCV-infected patients [2], [3], [4] and represent potential broad-spectrum antiviral drugs since many viruses depend on CypA as host factor [5], [28]. Based on our in vitro data, we speculate that alisporivir exerts an immunostimulatory effect on hepatocyte antigen presentation during therapy of chronically HCV-infected patients which might complement its antiviral properties by supporting an efficient adaptive immune response. Stimulation of the antigen presentation capacity of hepatocytes, which normally express only low levels of MHC-I [17] might promote recognition of infected cells by CD8+ T cells and finally contribute to their clearance. As previously reported, IFN-free therapy of HCV-infected patients with direct acting antivirals, but not IFN-based treatment, has the potential, to restore functional T cell responses presumably by lowering high antigen levels, which may support T cell exhaustion [29]. Along these lines, it seems plausible, that alisporivir, if applied as an IFN-free regimen, could restore T cell responses even more efficiently and additionally promote clearance of infected cells by CD8+ T cells via enhancing functional antigen presentation. Indeed, immunomodulatory activities on peripheral blood mononuclear cells of HCV-infected patients have been observed in clinical trials of the Cyp-inhibitor SCY-635 [30]. To gain insights into the contribution of these immunostimulatory effects of Cyp-inhibitors to the cure of HCV infections, future clinical studies should also monitor the development of T cell responses during therapy. Alternatively, testing the drug in suitable animal models allowing thorough quantification of T cell responses could further shed light on the impact of alisporivir on adaptive immune responses in various organs in vivo. In conclusion, we could demonstrate that non-immunosuppressive Cyp-inhibitors, in striking contrast to CsA, increase functional antigen presentation by upregulation of MHC-I surface expression. Importantly, the enhanced antigen presentation upon alisporivir treatment might also support clearance of other viruses which are sensitive to Cyp-inhibitors.
Financial support
This project was funded by grants from the Deutsche Forschungsgemeinschaft (VL: FOR1202, TP3, RT: FOR1202 TP2; CNH: FOR1202 TP8).
Conflict of interest
The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
Authors’ contributions
VL, KEN, CNH and RT designed experiments and interpreted data, VL and KEN drafted the manuscript, KEN acquired data, JS and KN generated and provided CD8+ T cell clones, KN, CNH and RT critically revised manuscript.
Acknowledgements
The authors especially thank Rahel Klein and Ulrike Herian for excellent technical assistance and Novartis for alisporivir and sanglifehrin A.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jhep.2016.02.027.
Supplementary data
References
Author names in bold designate shared co-first authorship
- 1.Rupp D., Bartenschlager R. Targets for antiviral therapy of hepatitis C. Semin Liver Dis. 2014;34:9–21. doi: 10.1055/s-0034-1371006. [DOI] [PubMed] [Google Scholar]
- 2.Flisiak R., Feinman S.V., Jablkowski M., Horban A., Kryczka W., Pawlowska M. The cyclophilin inhibitor Debio 025 combined with PEG IFNalpha2a significantly reduces viral load in treatment-naive hepatitis C patients. Hepatology. 2009;49:1460–1468. doi: 10.1002/hep.22835. [DOI] [PubMed] [Google Scholar]
- 3.Pawlotsky J.M., Flisiak R., Sarin S.K., Rasenack J., Piratvisuth T., Chuang W.L. Alisporivir plus ribavirin, interferon free or in combination with pegylated interferon, for hepatitis C virus genotype 2 or 3 infection. Hepatology. 2015;62:1013–1023. doi: 10.1002/hep.27960. [DOI] [PubMed] [Google Scholar]
- 4.Zeuzem S., Flisiak R., Vierling J.M., Mazur W., Mazzella G., Thongsawat S. Randomised clinical trial: alisporivir combined with peginterferon and ribavirin in treatment-naive patients with chronic HCV genotype 1 infection (ESSENTIAL II) Aliment Pharmacol Ther. 2015;42:829–844. doi: 10.1111/apt.13342. [DOI] [PubMed] [Google Scholar]
- 5.Frausto S.D., Lee E., Tang H. Cyclophilins as modulators of viral replication. Viruses. 2013;5:1684–1701. doi: 10.3390/v5071684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Farmer J.D., Jr., Lane W.S., Friedman J., Weissman I., Schreiber S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y.R., Yang I.H., Lee Y.H., Im S.A., Song S., Li H. Cyclosporin A and tacrolimus, but not rapamycin, inhibit MHC-restricted antigen presentation pathways in dendritic cells. Blood. 2005;105:3951–3955. doi: 10.1182/blood-2004-10-3927. [DOI] [PubMed] [Google Scholar]
- 8.Landrieu I., Hanoulle X., Bonachera F., Hamel A., Sibille N., Yin Y. Structural basis for the non-immunosuppressive character of the cyclosporin A analogue Debio 025. Biochemistry. 2010;49:4679–4686. doi: 10.1021/bi1003266. [DOI] [PubMed] [Google Scholar]
- 9.Jo J., Aichele U., Kersting N., Klein R., Aichele P., Bisse E. Analysis of CD8+ T cell-mediated inhibition of hepatitis C virus replication using a novel immunological model. Gastroenterology. 2009;136:1391–1401. doi: 10.1053/j.gastro.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 10.Esser-Nobis K., Romero-Brey I., Ganten T.M., Gouttenoire J., Harak C., Klein R. Analysis of hepatitis C virus resistance to silibinin in vitro and in vivo points to a novel mechanism involving nonstructural protein 4B. Hepatology. 2013;57:953–963. doi: 10.1002/hep.26260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grünvogel O., Esser-Nobis K., Reustle A., Schult P., Müller B., Metz P. DDX60L is an interferon-stimulated gene product restricting hepatitis C virus replication in cell culture. J Virol. 2015;89:10548–10568. doi: 10.1128/JVI.01297-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Windisch M.P., Frese M., Kaul A., Trippler M., Lohmann V., Bartenschlager R. Dissecting the interferon-induced inhibition of hepatitis C virus replication by using a novel host cell line. J Virol. 2005;79:13778–13793. doi: 10.1128/JVI.79.21.13778-13793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esser-Nobis K., Harak C., Schult P., Kusov Y., Lohmann V. Novel perspectives for hepatitis A virus therapy revealed by comparative analysis of hepatitis C virus and hepatitis A virus RNA replication. Hepatology. 2015;62:397–408. doi: 10.1002/hep.27847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bengsch B., Seigel B., Ruhl M., Timm J., Kuntz M., Blum H.E. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou F. Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol. 2009;28:239–260. doi: 10.1080/08830180902978120. [DOI] [PubMed] [Google Scholar]
- 16.Bender H., Wiesinger M.Y., Nordhoff C., Schoenherr C., Haan C., Ludwig S. Interleukin-27 displays interferon-gamma-like functions in human hepatoma cells and hepatocytes. Hepatology. 2009;50:585–591. doi: 10.1002/hep.22988. [DOI] [PubMed] [Google Scholar]
- 17.Thomson A.W., Knolle P.A. Antigen-presenting cell function in the tolerogenic liver environment. Nat Rev Immunol. 2010;10:753–766. doi: 10.1038/nri2858. [DOI] [PubMed] [Google Scholar]
- 18.Paeshuyse J., Kaul A., Clercq De., Rosenwirth B., Dumont J.M., Scalfaro P. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen T.H., Mentre F., Levi M., Yu J., Guedj J. A pharmacokinetic-viral kinetic model describes the effect of alisporivir as monotherapy or in combination with peg-IFN on hepatitis C virologic response. Clin Pharmacol Ther. 2014;96:599–608. doi: 10.1038/clpt.2014.173. [DOI] [PubMed] [Google Scholar]
- 20.Gaither L.A., Borawski J., Anderson L.J., Balabanis K.A., Devay P., Joberty G. Multiple cyclophilins involved in different cellular pathways mediate HCV replication. Virology. 2010;397:43–55. doi: 10.1016/j.virol.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 21.Riemann D., Kehlen A., Langner J. CD13–not just a marker in leukemia typing. Immunol Today. 1999;20:83–88. doi: 10.1016/S0167-5699(98)01398-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neefjes J., Jongsma M.L., Paul P., Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 23.Wang B., Niu D., Lai L., Ren E.C. P53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat Commun. 2013;4:2359. doi: 10.1038/ncomms3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galat A., Bua J. Molecular aspects of cyclophilins mediating therapeutic actions of their ligands. Cell Mol Life Sci. 2010;67:3467–3488. doi: 10.1007/s00018-010-0437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nkongolo S., Ni Y., Lempp F.A., Kaufman C., Lindner T., Esser-Nobis K. Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol. 2014;60:723–731. doi: 10.1016/j.jhep.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 26.de Paulis A., Ciccarelli A., de Crescenzo G., Cirillo R., Patella V., Marone G. Cyclosporin H is a potent and selective competitive antagonist of human basophil activation by N-formyl-methionyl-leucyl-phenylalanine. J Allergy Clin Immunol. 1996;98:152–164. doi: 10.1016/s0091-6749(96)70237-3. [DOI] [PubMed] [Google Scholar]
- 27.Naoumov N.V. Cyclophilin inhibition as potential therapy for liver diseases. J Hepatol. 2014;61:1166–1174. doi: 10.1016/j.jhep.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Bekerman E., Einav S. Infectious disease. Combating emerging viral threats. Science. 2015;348:282–283. doi: 10.1126/science.aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin B., Hennecke N., Lohmann V., Kayser A., Neumann-Haefelin C., Kukolj G. Restoration of HCV-specific CD8+ T cell function by interferon-free therapy. J Hepatol. 2014;61:538–543. doi: 10.1016/j.jhep.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 30.Probst P., Mosier S., Scorneaux B., Sluder A., Heuman D., Borroto-Esoda K. The cyclophilin inhibitor SCY-635 restores the innate recognition of HCV by peripheral blood mononuclear cells (PBMC) from HCV positive subjects. J Hepatol. 2013;58:476. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.