Abstract
We recently characterized the interaction between the intraviral domains of envelope glycoproteins (Gn and Gc) and ribonucleoprotein (RNP) of Puumala and Tula hantaviruses (genus Hantavirus, family Bunyaviridae). Herein we report a direct interaction between spike-forming glycoprotein and nucleic acid. We show that the envelope glycoprotein Gn of hantaviruses binds genomic RNA through its cytoplasmic tail (CT). The nucleic acid binding of Gn-CT is unspecific, as demonstrated by interactions with unrelated RNA and with single-stranded DNA. Peptide scan and protein deletions of Gn-CT mapped the nucleic acid binding to regions that overlap with the previously characterized N protein binding sites and demonstrated the carboxyl-terminal part of Gn-CT to be the most potent nucleic acid-binding site. We conclude that recognition of the RNP complex by the Gn-CT could be mediated by interactions with both genomic RNA and the N protein. This would provide the required selectivity for the genome packaging of hantaviruses.
Keywords: Hantavirus, Glycoprotein, Cytoplasmic tail, Genomic RNA, Ribonucleoprotein, Assembly, Packaging, Transciption
Highlights
► Genomic RNA binding activity of hantavirus Gn. ► Gn cytoplasmic tails of Puumala and Tula hantaviruses bind RNA and DNA. ► No evidence for specificity. ► Nucleic acid-binding activity of Gn tail is mainly located to its C-terminal part. ► Possible role in virus assembly and/or transcription.
Introduction
Rodent- and insectivore-borne hantaviruses (genus Hantavirus) are enveloped viruses that together with four other genera (Orthobunya-, Nairo-, Phlebo- and Tospovirus) constitute the family Bunyaviridae (Elliott et al., 2000, Nichol et al., 2005). The asymptomatic carrier rodents and insectivores spread hantaviruses in their excreta, which is considered the main route for transmission to humans (Jonsson et al., 2010, Kariwa et al., 2007, Vaheri et al., 2011). When transmitted to man hantaviruses cause two severe diseases: hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS). Old World hantaviruses include causative agents of HFRS, whereas the hantaviruses found in the New World have been associated with HCPS (Mir, 2010; C. Schmaljohn and Hjelle, 1997). Additionally hantaviruses cause nephropathia epidemica, a mild form of HFRS, that occurs mainly in Northern Europe (Vaheri et al., 2008, Vapalahti et al., 2003).
The single-stranded, negative-sense RNA genome of hantaviruses is segmented into small (S), middle (M) and large (L) segments encoding nucleocapsid (N) protein, glycoproteins (Gn and Gc) and the RNA-dependent RNA polymerase (L protein), respectively (Plyusnin et al., 1996). The RNA genome is encapsidated via oligomerization of N protein to form RNP complexes (Alfadhli et al., 2001, Alfadhli et al., 2002, Alminaite et al., 2006, Alminaite et al., 2008, Kaukinen et al., 2005, Wang et al., 2008). The RNPs interact with the spike complex, formed of four units of both Gn and Gc glycoproteins (Battisti et al., 2010, Hepojoki et al., 2010a, Huiskonen et al., 2010), by binding to the cytoplasmic tails (CTs) of both Gn and Gc (Hepojoki et al., 2010b, Wang et al., 2010).
For enveloped viruses it is common that the contacts between RNP and envelope proteins are mediated by a matrix protein (Timmins et al., 2004). However, members of the family Bunyaviridae do not contain a separate matrix protein and therefore interactions between the spike complex and RNPs are assumed to initiate assembly of the virions. Of the two hantavirus glycoprotein CTs, Gn-CT is considerably larger and capable of forming domain structures (~ 110 amino acids) while Gc-CT represents an intraviral insertion of a transmembrane anchor (~ 10 amino acids). The Gn-CT harbors a tandem zinc finger (ZF) fold, the structure of which was resolved by nuclear-magnetic resonance (NMR) (Estrada et al., 2009). Gn-CT carries also a conserved endocytosis signal YxxL, functionally characterized in many cell surface proteins (Owen and Evans, 1998), but its role in hantaviruses remains elusive. This motif resembles a class of late domain (L-domain) motifs (Bieniasz, 2006, Groseth et al., 2010), that are commonly utilized by matrix proteins to drive the budding process of viruses. Taken together, the Gn-CT is assumed to operate as a surrogate matrix protein of hantaviruses.
We previously reported that both Gn-CT and Gc-CT are able to bind isolated RNP, and interact with purified recombinant N protein (Hepojoki et al., 2010b, Wang et al., 2010). We showed that the Gn-CT is fundamental to the RNP-spike interaction in the case of native proteins. Additionally we found that there are at least three binding sites in the Gn-CT towards N protein which are located on either side adjacent to the ZF domain and on the very C-terminal part of the protein. Recent studies suggest that Gn-CTs of Rift Valley Fever virus (RVFV, genus Phlebovirus) and Crimean-Congo hemorrhagic fever virus (CCHFV, genus Nairovirus) are able to interact with genomic RNA (Estrada and De Guzman, 2011, Piper et al., 2011). For CCHFV this interaction was shown to involve a ZF domain of Gn-CT that closely resembles the one found in hantaviruses. However, in the same study the ZF domain of hantaviruses could not bind RNA when expressed without the flanking regions. In this report we aimed to investigate the ability of the full Gn-CT of hantaviruses to interact with genomic RNA. We show an interaction between PUUV Gn (but not Gc) and exogenous 32P-labeled genomic RNA by cross-linking and co-immunoprecipitation experiments. The nucleic acid binding site is pin-pointed to Gn-CT using GST-fused recombinant proteins and synthetic peptides. The results indicate that the binding ability of Gn-CT towards nucleic acids is rather unspecific since in addition to genomic RNA, also unrelated RNA as well as single-stranded DNA was able to interact with Gn-CTs of PUUV and TULV. The nucleic acid binding activity of hantavirus Gn-CT resided predominantly at the C-terminal part of the protein. In conclusion we hypothesize the RNA–Gn-CT interaction to be linked to the assembly process of hantaviruses.
Results
Cross-linking of RNA to PUUV and TULV Gn
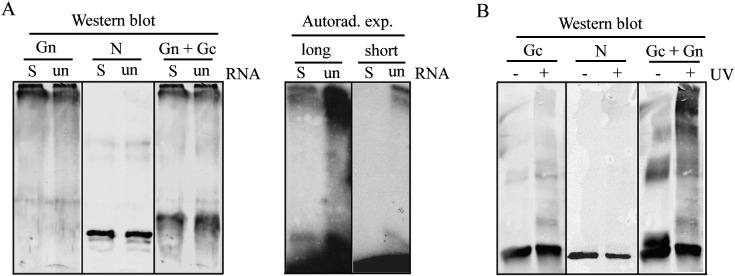
We assessed biochemically, whether one or both of the glycoproteins of PUUV would directly bind genomic RNA. Purified, lysed and micrococcal nuclease (MNase)-treated virus preparation and in vitro transcribed and radiolabeled genomic PUUV S segment were allowed to form RNA–protein complexes prior to UV cross-linking. Samples were subsequently treated with RNase A in order to facilitate separation of proteins according to their molecular size in SDS-PAGE. Proteins were detected by Western blotting using N, Gn and Gc antibodies. The RNA binding to viral proteins was studied in parallel by separating identically treated sample in SDS-PAGE and using autoradiography for detection. Firstly, the successful transcriptions of the S segment (~ 2 kilobases {kB}) together with an unrelated marker RNA (unRNA of 2 kB) were verified on a TBE-Urea gel (Fig. 1A). From Fig. 1B it is evident that the bands migrating above 250 kDa and at 50 kDa remained radiolabeled after RNase A-treatment, indicating cross-linking of RNA to proteins of the indicated sizes. Interestingly, the most intense radioactivity (above 250 kDa) overlapped with the band recognized by anti-Gn immunoblot which suggested that the genomic RNA interacts with Gn. The band detected by autoradiography at 50 kDa overlapped with the band recognized by N protein immunoblot. This was expected since the RNA-binding properties of N protein are well-characterized (Gott et al., 1993, Mir and Panganiban, 2004, Severson et al., 1999).
Fig. 1.
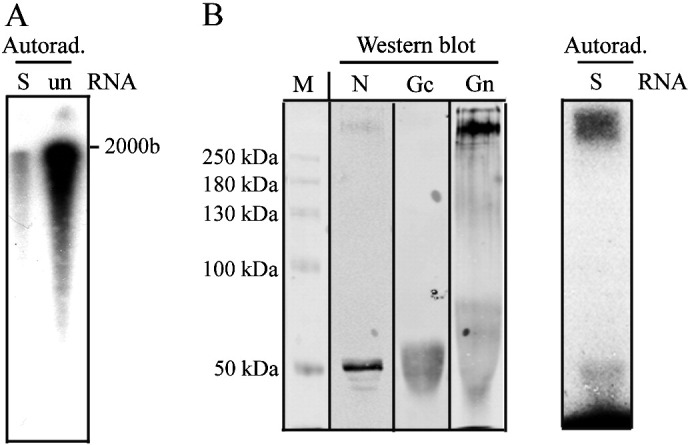
Cross-linking of exogenous genomic 32P-RNA to PUUV proteins. Expression of in vitro transcribed 32P-labeled PUUV S segment RNA (32P-S-RNA) and an unrelated RNA (32P-unRNA) of similar size (2 kB) are shown in urea-PAGE (A). The S-RNA (500 CPM) was incubated with purified MNase-treated PUUV lysate, UV cross-linked, treated with RNase A and the proteins were separated in 6% SDS-PAGE (B). As indicated, the PUUV proteins were detected by immunoblotting using Gn- or Gc-specific PAb and N-specific MAb 5E1, a combination of appropriate IRDye-conjugated secondary antibodies and Odyssey infrared imaging. Protein-bound nucleotides were detected by autoradiography. M indicates the marker lane.
Next we performed similar experiments as before using TULV as the source of viral proteins (Fig. 2A). In addition to the genomic S segment RNA of PUUV, we used an excessive amount of radiolabeled unrelated RNA (unRNA) as a non-genomic RNA substrate to analyze the RNA-binding specificity of viral proteins. Similarly to what was observed using PUUV, the radioactivity after RNase A-treatment was retained in the bands that by migration represent Gn and N proteins. The same pattern of RNA-bound proteins was observed with exogenous hantavirus S segment and the unRNA. The capacity of both N and Gn protein to bind both viral and unRNA suggests that the RNA-binding activity of these proteins did not absolutely require a specific sequence or structure of the RNA substrate. However, our experimental set-up was not optimized to study the binding affinity of proteins to different RNA sequences since the amounts of added RNAs differed and thus we cannot rule out the possibility that viral proteins would prefer genomic RNA over unrelated. We further observed that while N and Gc proteins migrated mainly as monomers, the mobility of PUUV and TULV Gn were drastically reduced after RNA-binding and UV-induced cross-linking (Fig. 1, Fig. 2). To assess whether this phenomenon is due to cross-linking alone we treated TULV as before but without the addition of exogenous RNA (Fig. 2B). The mobility of Gn was clearly reduced by UV cross-linking thereby showing that the exogenously applied RNA is probably not responsible for the reduced mobility of Gn in SDS-PAGE.
Fig. 2.
Cross-linking of exogenous 32P-RNA to TULV proteins. TULV RNA-binding proteins were detected by Western blotting (using Gn- or Gc-specific PAb and N-specific MAb 3C11) and autoradiography as described in Fig. 1B (A). Two different autoradiography exposures (long and short) emphasize the difference in the concentrations of the applied exogenous S-RNA vs. unRNA (500 vs. 2500 CPM; specific activities of the probes were similar). Reduced mobility of Gn in SDS-PAGE due to UV-induced cross-linking is shown (B). TULV preparations were treated and immunoblotted as described in Fig. 1B but without exogenous RNA.
Co-immunoprecipitation of RNA with Gn
Next we wanted to confirm the ability of Gn to bind RNA and chose to use co-immunoprecipitation with the well-described PUUV-specific monoclonal antibodies (MAbs) 5A2 (Gn-specific), 4G2 (Gc-specific) and 5E1 (N-specific) in parallel with rabbit polyclonal antibodies (PAbs) directed to PUUV Gn, Gc or N. Since these PAbs are known to cross-react with TULV proteins in Western blot, we firstly wanted to verify their ability also to immunoprecipitate the envelope proteins of TULV. For this purpose we used metabolically radiolabeled lysates of both PUUV and TULV as the source of viral proteins in immunoprecipitation with Gn- and Gc-specific PAbs. Immunoprecipitated proteins were separated in SDS-PAGE and detected by autoradiography according to their typical migration pattern (Fig. 3A). The results demonstrated that the Gn- and Gc-specific PAbs cross-react between PUUV and TULV and in both cases precipitate, in addition to their respective target antigens, also a complex formed of Gn and Gc. Unexpectedly the PUUV Gc PAb seemed to precipitate TULV Gc more efficiently than its native antigen. This suggests differences in the accessibility between Gc of PUUV and TULV towards this PAb. Both PAbs favor their target antigens over the Gn–Gc complex which is in contrast to the PUUV Gn- and Gc-specific MAbs 5A2 and 4G2 that both precipitate close to equimolar amounts of Gn and Gc (Hepojoki et al., 2010a, Hepojoki et al., 2010b, Strandin et al., 2011).
Fig. 3.
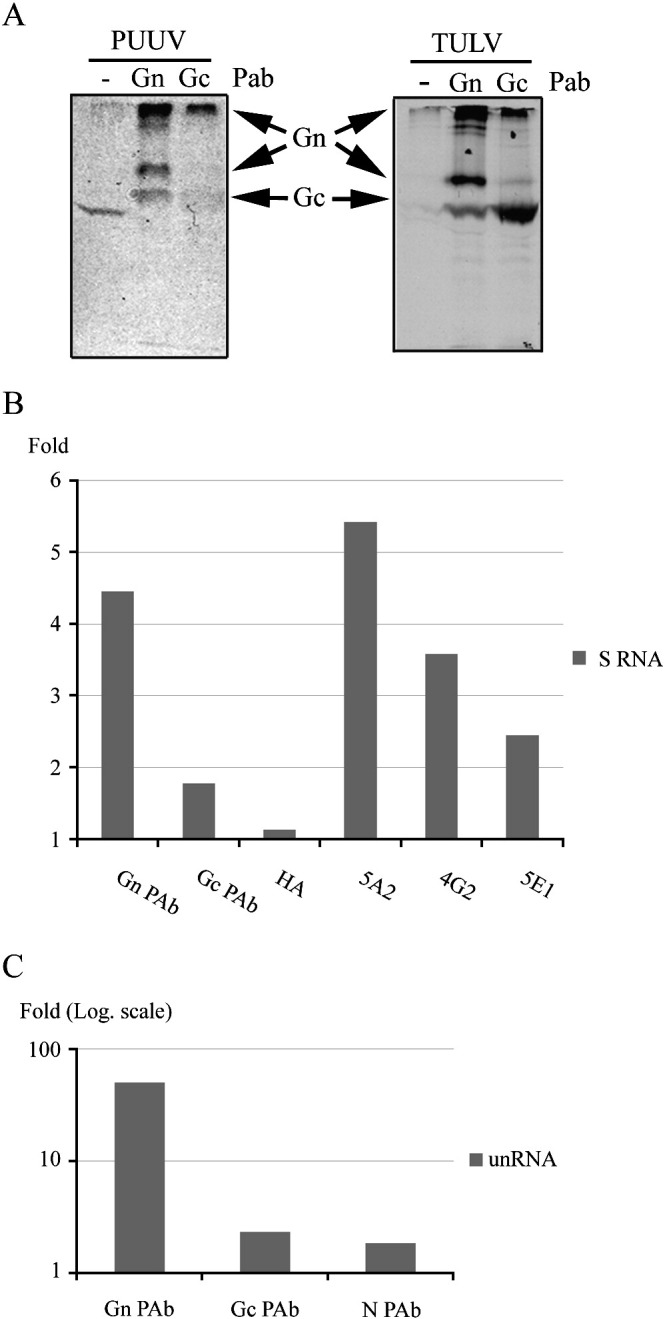
Immunoprecipitation of exogenous 32P-RNA by hantavirus proteins. Lysates of metabolically 35S-Cys and 35S-Met labeled and purified PUUV or TULV were subjected to immunoprecipitation by Gn- or Gc-specific PAbs, proteins separated in 10% SDS-PAGE and detected by autoradiography (A). Negative serum was used as a control (−). Viral proteins, according to their typical migration in SDS-PAGE, are indicated. Co-immunoprecipitation of 500 CPM 32P-S-RNA (B) or 2500 CPM 32P-unRNA (C) with purified, lysed and MNase-treated PUUV or TULV proteins, respectively, are shown. The radioactivity retained on protein G Sepharose-beads after immunoprecipitation was measured by scintillation counting and the fold of change as compared to negative serum is indicated. The MAbs 5A2, 4G2 and 5E1 are specific for PUUV Gn, PUUV Gc and PUUV N, respectively. The MAb indicated HA is specific for hemagglutinin-tag and used as a negative control.
The capability of structural proteins to bind RNA was analyzed by incubating purified, lysed and micrococcal nuclease (MNase)-treated virus preparations with radioactive PUUV S segment RNA in the case of PUUV (Fig. 3B) and unrelated RNA in the case of TULV (Fig. 3C) as was done in the cross-linking experiments (Figs. 1B and 2A). The samples were immunoprecipitated with MAbs or PAbs specific to N, Gn or Gc and retained radioactivity on protein G beads after washing was measured by scintillation counting. The amount of precipitated RNA is reported as a fold of increase compared to the value obtained with negative PAb. The Gn-specific antibodies precipitated the highest amount of RNA in the case of both PUUV and TULV, and thus the RNA-binding activity of Gn was further solidified. The substantially higher amount of RNA precipitated by TULV Gn compared to PUUV Gn is at least partially explained by the different amounts of applied RNA (approximately 5 times more of unRNA than S segment RNA). The amount of viral proteins in precipitates of PUUV and TULV was nearly the same (Fig. 3A) suggesting that the maximum RNA-binding capacity of PUUV Gn could be considerably higher than detected with the amount of exogenous S segment RNA used in this experiment.
Surprisingly, the antibodies against N protein precipitated exogenous RNA–N protein complexes very weakly if at all. The likely explanation to this is a nuclease-induced aggregation or modification of the N protein, thus rendering it unrecognizable by antibodies. Another possibility could be inefficiency of nucleases to degrade the endogenous N protein-bound genomic RNA. This would also explain the fairly low amount of exogenous RNA cross-linked to N protein as compared to Gn in the cross-linking experiments (Fig. 1, Fig. 2). The fact that Gc-specific MAb 4G2 precipitated more RNA than Gc-specific PAb is probably due to effective co-immunoprecipitation of the Gn–Gc complex by this MAb, as mentioned earlier.
The nucleic acid binding activity of PUUV and TULV Gn is retained in their cytoplasmic tails
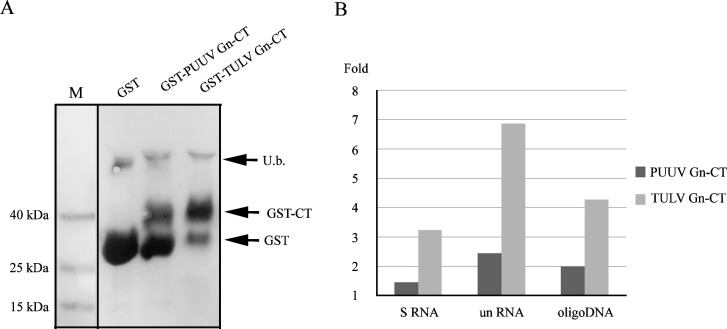
Based on our results using native viral proteins, the Gn of both PUUV and TULV can bind RNA. In the native virion or in infected cells the Gn protein can only encounter nucleic acids via its cytoplasmic tail and therefore the RNA-binding activity should reside in this part of the protein to be biologically relevant. The ability of Gn-CT of PUUV and TULV to bind nucleic acids was studied with GST-fused Gn-CTs purified using reduced glutathione (GSH)-beads. The expression of the recombinant proteins eluted from GSH-beads was analyzed by Ponceau S staining (Fig. 4A). The RNA-binding of bead-bound Gn-CTs of either PUUV S segment or unrelated RNA, which were applied in similar radiolabeled amounts, was measured by scintillation counting. In parallel, we also tested with single-stranded 42-mer DNA whether the Gn-CTs would bind DNA. The sequence of this IRdye800-labeled DNA was derived from the 3′-end of the PUUV genomic S segment and the binding of this DNA was detected by Odyssey infrared imaging. We compared the amounts of Gn-CT bound nucleic acids to plain GST and report the results as fold change (Fig. 4B). The results indicated that the Gn-CTs of PUUV and TULV bind to both RNA and DNA. The observed difference in nucleic acid binding between Gn-CT of TULV and Gn-CT of PUUV is likely due to a higher level of expression of TULV Gn-CT as compared to PUUV Gn-CT as demonstrated in Fig. 4A. It seems that there is also more plain GST in the PUUV Gn-CT preparation as compared to TULV Gn-CT. However, since plain GST as negative control did not show significant binding to nucleic acids, this should not influence the obtained results. The fact that the unrelated RNA bound both CTs more efficiently than the genomic RNA and the ability of Gn-CT to bind also relative short DNAs further solidifies the unspecific nature of the nucleic acid recognition of Gn-CT. However, due to the different detection methods used, we could not directly compare the binding strength of RNA vs. DNA.
Fig. 4.
Pull-down of nucleic acids by GST-fused hantavirus Gn cytoplasmic tails. PUUV and TULV Gn cytoplasmic tails (CT) were expressed as GST-fusion proteins in parallel with plain GST, separated in 6% SDS/Tricine-PAGE, blotted onto nitrocellulose and visualized by Ponceau S staining (A). M indicates a marker lane. Glutathione beads loaded with PUUV and TULV Gn-CT were used to pull-down a similar amount of PUUV 32P-S-RNA (400 CPM) or 32P-unRNA (500 CPM) (B). The bead-retained radioactivity was measured by scintillation counting. In comparison IRdye800-conjugated DNA, instead of RNA, was used in pull-down. The 42-mer single stranded DNA (sequence derived from PUUV S-segment 3′-end) was bound to GST-Gn-CTs at 100 nM, eluted from glutathione beads by Laemmli sample buffer, cross-linked onto a nitrocellulose membrane and quantified by Odyssey infrared imaging. Results of RNA and DNA pull-downs are reported as a fold increase as compared to plain GST.
Peptide mapping of nucleic acid binding sites in PUUV Gn-CT
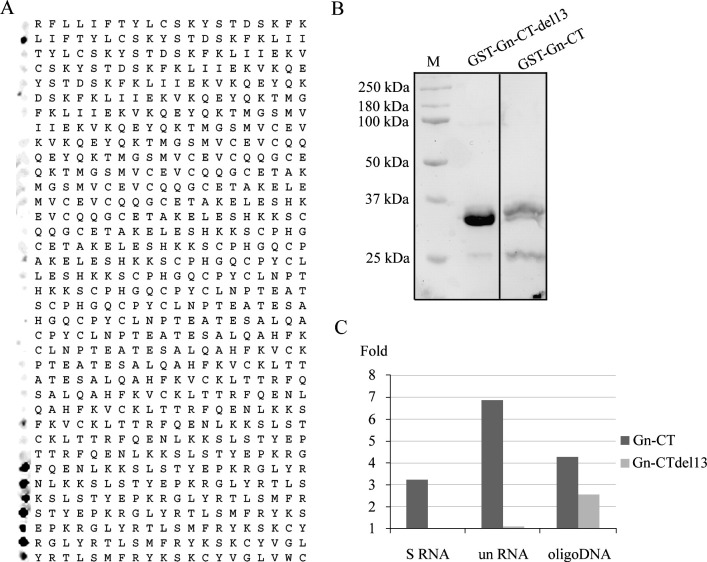
To define the regions in Gn-CT that bind nucleic acids, we synthesized the Gn-CT of PUUV as overlapping 20-mer peptides in a 3 residue shift in CelluSpot™ format. The CelluSpot™ array was probed with the IRdye800-conjugated DNA shown to interact with the GST-tagged Gn-CT of PUUV (Fig. 4B). The mapping indicated that the same peptides that were previously shown to interact with RNP and N protein (Hepojoki et al., 2010b) were capable of binding also nucleic acids (Fig. 5A). Next we wanted to study the nucleic acid binding capacity of the three binding sites individually using biotinylated soluble peptides GnN, GnM and GnC, which represent the N-binding sites 1, 2 and 3 of PUUV Gn-CT as described earlier (Hepojoki et al., 2010b). Schematic presentation of the peptides with respect to the primary sequence of PUUV Gn-CT is shown in Fig. 6 . We also used the synthetic peptide corresponding to Gc-CT, which in the earlier study was also shown to bind N protein (Hepojoki et al., 2010b). The peptides were pre-bound to avidin beads through biotin and analyzed for RNA- or DNA-binding activity similarly as was done for GST-fused proteins (Fig. 4). The GnC showed the strongest binding of the analyzed peptides irrespective of the applied nucleic acids (Fig. 5B). Also GnM showed significant RNA-binding capacity but only very little DNA-binding activity. The binding levels of GnN and Gc-CT were above control (biotin-bound avidin beads) but clearly lower than those of GnM or GnC. In addition, in the case of Gc-CT, this observed interaction might not have any biological relevance since we did not detect any cross-linking of RNA to native Gc (Fig. 1, Fig. 2).
Fig. 5.
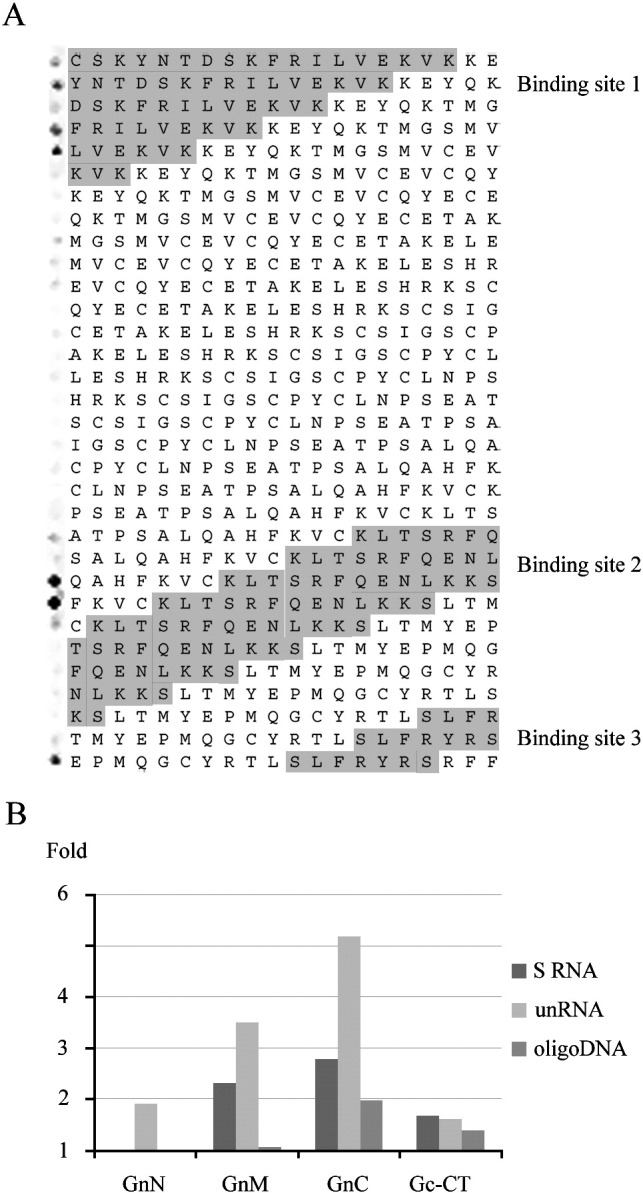
Mapping of the nucleic acid binding sites in the Gn-CT of PUUV. The Gn-CT (amino acids 525–638) was synthesized in CelluSpot™ format as partially overlapping 20-residue peptides with a 3-residue shift. The CelluSpotTM slide was probed with the IRdye800-conjugated 42-mer single-stranded DNA (A). The signal detected in Odyssey infrared imaging for each individual peptide is shown on the left side next to the amino acid sequence of the respective peptide. The indicated binding sites 1–3 were determined for N protein using SPOT peptide assay as previously described (Hepojoki et al., 2010b, Wang et al., 2010), and are colored grey. PUUV Gn-CT peptides were used to pull-down nucleic acids (B). Monomeric avidin beads preloaded to saturation by the biotinylated peptides GnN, GnM, GnC and Gc-CT were used to pull-down RNA (32P-S-RNA in 400 CPM and 32P-unRNA in 500 CPM) or IRdye800-conjugated DNA (IRdye800-conjugated 42mer 3′-end of hantavirus S-segment, 100 nM) and detections were done by scintillation counting and Odyssey infrared imaging, respectively. Results are indicated as a fold increase in respect to biotin-saturated avidin beads.
Fig. 6.
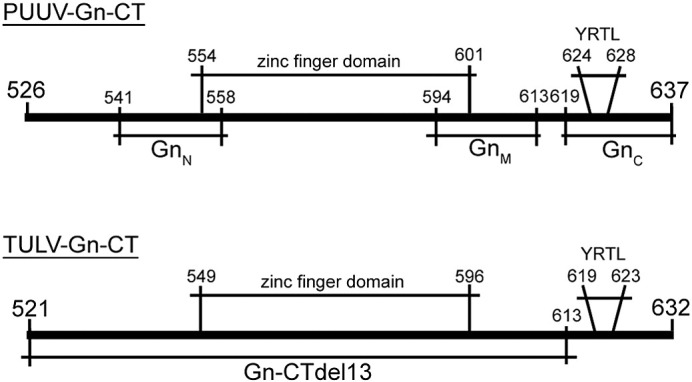
Scheme of Gn-CT peptides and proteins used to map nucleic acid-binding sites in PUUV and TULV. The full length cytoplasmic tails of PUUV and TULV Gn (111 amino acids) and TULV Gn-CT with 13 residue C-terminal deletion (Gn-CTdel13) were expressed as N-terminal GST-fusion proteins (Wang et al., 2010). Biotinylated peptides (GnN, GnM and GnC) derived from PUUV Gn-CT was synthesized as described previously (Hepojoki et al., 2010b). The amino acid numbers in the image refer to the PUUV or TULV glycoprotein precursors.
Mapping of the nucleic acid binding site in TULV Gn-CT
Finally we were interested in mapping the nucleic acid binding sites of TULV Gn-CT and thus performed a similar CelluSpot™ assay as with PUUV Gn-CT, shown in Fig. 5A. The strongest DNA-binding peptides of TULV Gn-CT (Fig. 7A) located to the C-terminal part of the protein and overlapped with the corresponding sequence of PUUV GnC peptide. This result suggested that the main nucleic acid binding site in Gn is conserved among hantaviruses and locates to the C-terminal part of the Gn-CT which contains the putative late domain motif YRTL and is adjacent to the membrane-spanning signal sequence for Gc. To confirm the significance of this region to nucleic acid binding we used a previously described (Wang et al., 2010) truncated GST-fusion protein of TULV Gn-CT lacking the C-terminal DNA-binding sequence (Fig. 6, Fig. 7B). When comparing the RNA- and DNA-binding activities of this mutant (Gn-CTdel13) to the full-length Gn-CT of TULV in a similar GST pull-down as before, we observed that the RNA-binding capacity of the mutant was completely abolished (Fig. 7C). However, the DNA-binding activity was seen only to diminish due to the deletion of the C-terminal part, and thus we conclude that while the whole Gn-CT containing the ZF domain might be important in regulating nucleic acid binding it is the C-terminal part that is mainly responsible for this activity.
Fig. 7.
Mapping of the nucleic acid binding sites in the Gn-CT of TULV. The Gn-CT (amino acids 525–638) peptides as 20-mers in a 3-residue shift were synthesized and printed on glass slides in CelluSpot™ format. The IRdye800-conjugated 42-mer single-stranded DNA overlay assay and detection in Odyssey imaging system were performed as for PUUV (A). The detected signal for each individual peptide is shown on the left side next to the amino acid sequence of the respective 20-mer peptide. TULV Gn-CT (wt) and its 13 amino acid C-terminal deletion (del13) expressed as GST-fusion proteins were separated in 10% SDS- PAGE, blotted onto nitrocellulose and visualized by Ponceau S staining (B). M indicates a marker lane. Nucleic acid binding of Gn-CT and Gn-CTdel13 of TULV were compared (C). GST-fused TULV Gn-CT and TULV Gn-CTdel13 were bound on glutathione beads and analyzed for nucleic acid binding of RNA (32P-S-RNA in 400 CPM and 32P-unRNA in 500 CPM) and DNA (IRdye800-conjugated 42mer 3′-end of hantavirus S-segment, 100 nM) as in original comparison of GST-Gn-CTs of PUUV and TULV (Fig. 4). The fold of change to GST control is indicated.
Discussion
In this report we show using several techniques that the Gn protein of hantaviruses is able to bind nucleic acids and map this interaction to its CT. Furthermore, by using peptide array we were able to pin-point this interaction to the very C-terminus of Gn-CT. In our previous study we demonstrated that the CTs of both Gn and Gc interact with the native RNP consisting of N protein and N protein-encapsidated genomic RNA (Hepojoki et al., 2010b, Wang et al., 2010). We also showed recombinant N protein to be able to mediate binding to Gn- and Gc-CT; however, we were unable to rule out the involvement of nucleic acids in these interactions. The results in this report show that the N protein encapsidated genomic RNA could be directly involved in the interaction between glycoprotein CTs and RNP. This is supported by the fact that the binding sites of nucleic acids and N protein either overlap or are contiguous in the primary sequence of Gn-CT. Thus amino acid residues of N protein and nucleotides from genomic RNA would form the interaction surface in RNP that mediates the binding to glycoprotein CTs. Our results corroborate recent findings suggesting that Gn-CTs of other bunyaviruses could bind genomic RNA. For RVFV of phleboviruses it was demonstrated that genomic RNA is required for the efficient release of infectious VLPs and the authors hypothesized that this would occur through direct recognition of viral RNA by Gn-CT (Piper et al., 2011). Furthermore a ZF domain in Gn-CT of CCHFV (nairovirus), closely resembling the one found in hantavirus Gn-CT (Estrada et al., 2009), was found to bind viral RNA and again a role for this interaction in virus assembly was suggested (Estrada and De Guzman, 2011).
Specific recognition of the RNP structure is a crucial step in the egress of enveloped viruses. During infection the cells may contain various types of nucleic acids encapsidated by the viral nucleoproteins; however, mainly the genomic RNA is incorporated into virions and this requires accentuated regulation of genome packaging. Several enveloped viruses encode a matrix protein that bridges between the RNP and the viral envelope. Hantaviruses do not possess an individual matrix protein and based on recent evidence, instead, the relatively large Gn-CT substitutes for this activity (Battisti et al., 2010, Hepojoki et al., 2010a, Huiskonen et al., 2010, Wang et al., 2010). The matrix protein of both family Bornaviridae (Neumann et al., 2009) and Orthomyxoviridae (Elster et al., 1997, Wakefield and Brownlee, 1989, Ye et al., 1989) viruses have been shown to bind, in addition to the viral core proteins, also the genomic RNA. Thus our results suggest by analogy the Gn-CT of hantaviruses to be equivalent of a matrix protein in the family Bunyaviridae. Prior to recent reports for CCHFV and RVFV (Estrada and De Guzman, 2011, Piper et al., 2011), interaction involving a viral envelope protein and its genomic RNA has only been shown for the family Coronaviridae, where the envelope protein M recognizes the viral RNA packaging signal and incorporates itself in to the nascent virions independently of the core protein (Narayanan et al., 2003). However, the M protein of coronaviruses possesses only a tiny extracellular domain, thus differing from the spike-forming glycoprotein Gn of hantaviruses.
The segmented, negative-sense RNA viruses are divided in three families: Orthomyxoviridae, Bunyaviridae and Arenaviridae. In the case of influenza viruses of the family Orthomyxoviridae, the eight-segmented genome is recognized via packaging signals formed of secondary structures in RNA (Hutchinson et al., 2010). Although the exact mechanism of genome incorporation in to virions is not known, it has been shown that the matrix protein M1 interacts with RNP (Baudin et al., 2001, Elster et al., 1997, Noton et al., 2007, Ye et al., 1999). The M1 protein harbors a ZF and a nuclear localization signal with RNA-binding abilities (Elster et al., 1997, Wakefield and Brownlee, 1989, Ye et al., 1999, Ye et al., 1989). Interestingly, the influenza M1 has been shown to repress endogenous viral transcription through its RNA-binding domains (Perez and Donis, 1998, Watanabe et al., 1996, Ye et al., 1989). This activity could be biologically relevant in the late stages of infection where it would halt viral mRNA production and thereby trigger the packaging of viral genome eventually leading to the egress of virions. Similarly to influenza M1, the matrix protein Z of arenaviruses also acts as a transcriptional repressor (Cornu and de la Torre, 2001, Cornu and de la Torre, 2002, Lopez et al., 2001), and a ZF domain is required for this activity. Curiously, most bunyavirus Gn-CTs also contain a ZF domain (Estrada et al., 2009) the functions of which are currently unknown. Unlike the ZF domain of CCHFV, the ZF-domain of hantavirus Gn-CT is unable to bind RNA directly (Estrada and De Guzman, 2011). However, according to the results of the present study there are other motifs in Gn-CT capable of binding RNA and thus a regulatory role in viral transcription for the hantavirus ZF cannot be ruled out. Since the matrix proteins of some non-segmented negative-sense RNA viruses have also been described to regulate transcription through interaction with the RNP (Ghildyal et al., 2002, Iwasaki et al., 2009, Suryanarayana et al., 1994), it is intriguing to speculate that hantavirus Gn-CT would, through binding to RNP, mediate regulatory functions in gene expression and in virion packaging. Further studies regarding the functions of Gn-CT in hantavirus assembly or replication would greatly benefit from reverse genetics or infectious virus-like particle generation systems that are unfortunately still lacking today for these viruses.
Materials and methods
Hantavirus cultivation, radiolabeling and purification
PUUV Sotkamo strain and TULV Moravia strain 5302 were propagated in Vero E6 cells (green monkey kidney epithelial cell line; ATCC: CRL-1586) in which they have been isolated and to which they are adapted producing titers of 104 to 108 FFU/ml in to conditioned medium (Schmaljohn et al., 1985, Strandin et al., 2008, Vapalahti et al., 1996). Vero E6 cells, grown at 37 °C in a humidified atmosphere containing 5% CO2, were propagated in minimal essential medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, 100 IU/ml of penicillin and 100 μg/ml of streptomycin. Cell cultures in 75-cm2 flasks were inoculated for 1 h at 37 °C with virus suspension and conditioned medium collected 7–10 days post infection (d.p.i.) for TULV and 12–21 d.p.i. for PUUV. Conditioned medium containing virus (stored in aliquots at − 70 °C) was used as an inoculum. For radiolabeling of viral proteins infected Vero E6 cell cultures were starved for 1 h at 37 °C with medium depleted of methionine and cysteine, and propagated with a 1 mCi mixture of 35S-cysteine and 35S-methionine (Wallac Perkin-Elmer) for 3 days at 37 °C. For purification and concentration of viruses, cell culture medium, passed through a 0.22-μm filter (Millipore), was concentrated by pelleting through a 30% (w/v) sucrose cushion (Beckman SW28 rotor, 27,000 rpm, 2 h, 4 °C) and suspended in 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.4; 150 mM NaCl (HBS).
Antibodies and reagents
Neutralizing bank vole monoclonal antibodies (MAbs) directed to PUUV Gn (5A2) and Gc (4G2) and to PUUV N (5E1) and TULV N (3C11) (Lundkvist and Niklasson, 1992, Lundkvist et al., 1993, Lundkvist et al., 1996a, Lundkvist et al., 1996b) were kindly provided by Prof. Åke Lundkvist (Swedish Institute for Infectious Disease Control). Polyclonal antisera (PAbs) raised against glutathione-S-transferase (GST) fusion proteins of PUUV Gn, Gc and N have been described previously (Vapalahti et al., 1995). The anti-hemagglutinin (HA) MAb was from Abcam. The IRdye800- and AlexaFluor680-conjugated secondary antibodies were from Li-COR and Invitrogen, respectively. Micrococcal nuclease (MNase) and RNase A were from MBI Fermentas.
Nucleic acids
The genomic PUUV S segment was in vitro transcribed (TranscriptAid™ T7 High Yield Transcription Kit; MBI Fermentas) from a pGEM-T plasmid containing the respective sequence information under a T7 promoter. The PUUV S segment together with an unrelated RNA (unRNA) of similar size (included in TranscriptAid™ T7 High Yield Transcription Kit; MBI Fermentas) were radioactively labeled with 32P-UTP (Wallac PerkinElmer) during transcription and purified using Tripure (Roche). The specific activity of these RNAs was assumed similar since both contained approximately 1/3 of uridine residues. Successful RNA synthesis was verified on a denaturing polyacrylamide gel electrophoresis (PAGE) in 5% gel containing 8 M urea in TBE buffer (22.5 mM Tris base, 22.5 mM boric acid, 2.5 mM EDTA). The gel was dried and RNA visualized by autoradiography. The 42-mer single-stranded DNA represented the 3′-end of the genomic PUUV S segment: 5′-TCCAGACTTTCTCGTAGTAGCTTTTCAAGGAGTCTACTACTA-3′. An IRdye800-moiety was conjugated to the 5′-end of the oligonucleotide which was synthesized by Oligomer Oy, Finland.
Cross-linking
The purified PUUV or TULV, lysed in 0.5% TritonX-100, were treated with 0.1 U/μl MNase in 50 mM Tris–HCl pH 7.5; 5 mM CaCl2 for 1 h at 37 °C. MNase was inactivated with 10 mM ethylene glycol tetra-acetic acid (EGTA) for 15 min at room temperature prior to addition of 32P-labeled RNA, that was added in amounts of 500 counts per minute (CPM) of PUUV S segment to both PUUV and TULV extracts or 2500 CPM of unrelated RNA (unRNA) to TULV. After 2-h incubation at RT the RNAs were cross-linked to proteins under a UV lamp for 30 min at RT and RNA cleaved with 10 μg RNase A for 1 h at 37 °C. The enzyme was inactivated by addition of Laemmli sample buffer, samples boiled and proteins separated on 6% SDS-PAGE. To visualize the RNA-interacting proteins, the gel was either subjected to autoradiography or proteins were transferred to nitrocellulose and immunoblotted with Gn- or Gc-specific PAbs and N-specific 3C11 or 5E1 MAbs. After probing with infrared dye-conjugated secondary antibodies the results were visualized by Odyssey infrared detection system (Li-COR).
Immunoprecipitation
The PUUV or TULV preparations, incubated with radioactive RNA prior to cross-linking as described earlier, were subjected to immunoprecipitation in binding buffer (BB; 50 mM Tris–HCl, 150 mM NaCl, 0.1% Triton X-100). Firstly, 10 μg of MAbs (5A2, 4G2, 5E1 or HA) or 20 μl of PAbs (Gn-, Gc-, N-specific or negative serum) were incubated with the samples for 1 h at RT. Then 10 μl of Protein G Sepharose beads (GE Healthcare) were added and incubation continued for 1 h under end-over-end rotation in Eppendorf tubes at RT. The beads were then washed two times with BB and liquid removed prior to scintillation counting (Cerenkov-32P; Wallac 140 MicroBeta Trilux liquid scintillation counter). The results were calculated as a fold change in respect to a negative control sample. The efficiency of Gn-, Gc- or negative PAb to bind viral proteins was monitored with metabolically radiolabeled virion lysates of PUUV or TULV by immunoprecipitation similarly as described earlier. The protein G bound protein complexes were eluted to Laemmli sample buffer, boiled and separated in 10% SDS-PAGE. The gel was dried and proteins visualized by autoradiography.
Recombinant protein production
The Gn-CTs of PUUV and TULV were expressed as glutathione S-transferase (GST) fusion proteins in E. coli according to protocols of the manufacturer (GST Gene Fusion System Handbook, Amersham Bioscience 18-1157-58 Edition AA). The expression plasmids for GST-tagged Gn-CTs were constructed by inserting respective Gn-CT fragments (residues 526–637 of PUUV and 521–632 of TULV) into the pGEX-4T-3 vector (GE Healthcare). Protein production was induced by addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) to cultures at 0.5 OD600, protein expression was performed under vigorous shaking over 4 h at 30 °C, bacteria were lysed and the expressed GST and GST-fusion proteins were purified by glutathione (GSH) Sepharose 4B beads (Amersham Pharmacia, GE Healthcare) using HBS as washing buffer. The GST-fusion protein with GST as the negative control (eluted from 10 μl of GSH beads to reducing Laemmli sample buffer) was separated in 6% SDS-PAGE in Tris/tricine/SDS running buffer (Bio-Rad) or in conventional 12% SDS-PAGE, and the proteins were transferred to nitrocellulose for recording of the relative protein amounts by Ponceau S staining.
Pull-downs
The synthesis of biotin-conjugated peptides GnN, GnM, GnC and Gc-CT has been described earlier (Hepojoki et al., 2010b) and contained the following amino acid sequences: GnN = Biotin-KVKKEYQKTMGSMVCEVC-OH, GnM = Biotin-QAHFKVCKLTSRFQENLKK-OH, GnC = Biotin-EPMQGCYRTLSLFRYRS-OH and Gc-CT = Biotin-CPRRPSYKKDHKP-OH. These peptides or biotin as a negative control were bound on monomeric avidin beads (Pierce) at saturating amounts. The GST-fusion protein coated on GSH and biotinylated peptide coated on avidin beads were used in the pull-down experiments of the radiolabeled S segment RNA (400 CPM) or unRNA (500 CPM). After 1 h incubation under shaking in BB at RT the beads were washed twice with BB and the amount of bound RNA detected by scintillation counting. For the binding of DNA, the IRdye800-conjugated single-stranded oligonucleotide (100 nM) was incubated for 1 h under shaking at RT and the bound DNA was eluted after two washes with BB by boiling in Laemmli sample buffer. 1 μl of the LSB eluate (corresponding to 1 μl bead volume) was dot-blotted on to nitrocellulose, cross-linked by UV and the amount of IRdye800-conjugated oligonucleotide quantified using Odyssey. The results are presented as a fold change as compared to negative control.
CelluSpot™ peptide arrays
Gn-CTs of TULV and PUUV were synthesized in CelluSpot™ format as 20-residue long peptides on MultiPep synthesizer (Intavis Ag) and the slides printed with SlideSpotter (Intavis Ag), all according to the manufacturer's instructions as described (Beutling et al., 2008). Prior to probing, the peptide array slides were rinsed with ethanol and incubated in 10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) and 0.0005% Tween-20 (TENT) for 5 min. The 5′-IRdye800-conjugated oligonucleotide was applied in blocking buffer for 1 h at RT, unbound DNA washed from slide with TENT and bound DNA detected with Odyssey.
Acknowledgments
This work was supported by the Helsinki University Nanoscience Training and Research Programme (to H.L.), The Finnish Funding Agency for Technology and Innovation, (TEKES 40264/07 to H.L.), Finnish Cultural Foundation (to T.S.), Magnus Ehrnrooth Foundation (to T.S. and J.H.), Paulo Foundation (to T.S.), Maud Kuistila Memorial Foundation (to T.S.), Otto Malm Donation Foundation (to T.S.), Instrumentarium Science Foundation (to J.H.), EU grant (QLK2-CT-2002-01358 to A.V.), and Sigrid Jusélius Foundation (to A.V.). We thank Irina Suomalainen and Kirsi Aaltonen for expert technical assistance.
References
- Alfadhli A., Love Z., Arvidson B., Seeds J., Willey J., Barklis E. Hantavirus nucleocapsid protein oligomerization. J. Virol. 2001;75:2019–2023. doi: 10.1128/JVI.75.4.2019-2023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A., Steel E., Finlay L., Bachinger H.P., Barklis E. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 2002;277:27103–27108. doi: 10.1074/jbc.M203395200. [DOI] [PubMed] [Google Scholar]
- Alminaite A., Backstrom V., Vaheri A., Plyusnin A. Oligomerization of hantaviral nucleocapsid protein: charged residues in the N-terminal coiled-coil domain contribute to intermolecular interactions. J. Gen. Virol. 2008;89:2167–2174. doi: 10.1099/vir.0.2008/004044-0. [DOI] [PubMed] [Google Scholar]
- Alminaite A., Halttunen V., Kumar V., Vaheri A., Holm L., Plyusnin A. Oligomerization of hantavirus nucleocapsid protein: analysis of the N-terminal coiled-coil domain. J. Virol. 2006;80:9073–9081. doi: 10.1128/JVI.00515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti A.J., Chu Y.K., Chipman P.R., Kaufmann B., Jonsson C.B., Rossmann M.G. Structural studies of hantaan virus. J. Virol. 2010;85:835–841. doi: 10.1128/JVI.01847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin F., Petit I., Weissenhorn W., Ruigrok R.W. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology. 2001;281:102–108. doi: 10.1006/viro.2000.0804. [DOI] [PubMed] [Google Scholar]
- Beutling U., Stading K., Stradal T., Frank R. Large-scale analysis of protein–protein interactions using cellulose-bound peptide arrays. Adv. Biochem. Eng. Biotechnol. 2008;110:115–152. doi: 10.1007/10_2008_096. [DOI] [PubMed] [Google Scholar]
- Bieniasz P.D. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Cornu T.I., de la Torre J.C. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 2001;75:9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu T.I., de la Torre J.C. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J. Virol. 2002;76:6678–6688. doi: 10.1128/JVI.76.13.6678-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.M., Bouloy M., Calisher C.H., Goldbach R., Moyer J.T., Nichol S.T. Virus taxonomy: the classification and nomenclature of viruses. In: Van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., editors. Bunyaviridae. Academic Press; San Diego: 2000. [Google Scholar]
- Elster C., Larsen K., Gagnon J., Ruigrok R.W., Baudin F. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J. Gen. Virol. 1997;78(Pt 7):1589–1596. doi: 10.1099/0022-1317-78-7-1589. [DOI] [PubMed] [Google Scholar]
- Estrada D.F., Boudreaux D.M., Zhong D., St Jeor S.C., De Guzman R.N. The hantavirus glycoprotein G1 tail contains a dual CCHC-type classical zinc fingers. J. Biol. Chem. 2009;284:8654–8660. doi: 10.1074/jbc.M808081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada D.F., De Guzman R.N. Structural characterization of the crimean-congo hemorrhagic fever virus gn tail provides insight into virus assembly. J. Biol. Chem. 2011;286:21678–21686. doi: 10.1074/jbc.M110.216515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildyal R., Mills J., Murray M., Vardaxis N., Meanger J. Respiratory syncytial virus matrix protein associates with nucleocapsids in infected cells. J. Gen. Virol. 2002;83:753–757. doi: 10.1099/0022-1317-83-4-753. [DOI] [PubMed] [Google Scholar]
- Gott P., Stohwasser R., Schnitzler P., Darai G., Bautz E.K. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology. 1993;194:332–337. doi: 10.1006/viro.1993.1263. [DOI] [PubMed] [Google Scholar]
- Groseth A., Wolff S., Strecker T., Hoenen T., Becker S. Efficient budding of the tacaribe virus matrix protein z requires the nucleoprotein. J. Virol. 2010;84:3603–3611. doi: 10.1128/JVI.02429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepojoki J., Strandin T., Vaheri A., Lankinen H. Interactions and oligomerization of hantavirus glycoproteins. J. Virol. 2010;84:227–242. doi: 10.1128/JVI.00481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepojoki J.M., Strandin T., Wang H., Vapalahti O., Vaheri A., Lankinen H. The cytoplasmic tails of hantavirus glycoproteins interact with the nucleocapsid protein. J. Gen. Virol. 2010;91:2341–2350. doi: 10.1099/vir.0.021006-0. [DOI] [PubMed] [Google Scholar]
- Huiskonen J.T., Hepojoki J., Laurinmaki P., Vaheri A., Lankinen H., Butcher S.J. Electron cryotomography of tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 2010;84:4889–4897. doi: 10.1128/JVI.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson E.C., von Kirchbach J.C., Gog J.R., Digard P. Genome packaging in influenza A virus. J. Gen. Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Takeda M., Shirogane Y., Nakatsu Y., Nakamura T., Yanagi Y. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J. Virol. 2009;83:10374–10383. doi: 10.1128/JVI.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson C.B., Figueiredo L.T., Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariwa H., Yoshimatsu K., Arikawa J. Hantavirus infection in east asia. Comp. Immunol. Microbiol. Infect. Dis. 2007;30:341–356. doi: 10.1016/j.cimid.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Kaukinen P., Vaheri A., Plyusnin A. Hantavirus nucleocapsid protein: a multifunctional molecule with both housekeeping and ambassadorial duties. Arch. Virol. 2005;150:1693–1713. doi: 10.1007/s00705-005-0555-4. [DOI] [PubMed] [Google Scholar]
- Lopez N., Jacamo R., Franze-Fernandez M.T. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 2001;75:12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundkvist A., Horling J., Athlin L., Rosen A., Niklasson B. Neutralizing human monoclonal antibodies against puumala virus, causative agent of nephropathia epidemica: a novel method using antigen-coated magnetic beads for specific B cell isolation. J. Gen. Virol. 1993;74(Pt 7):1303–1310. doi: 10.1099/0022-1317-74-7-1303. [DOI] [PubMed] [Google Scholar]
- Lundkvist A., Kallio-Kokko H., Sjolander K.B., Lankinen H., Niklasson B., Vaheri A. Characterization of puumala virus nucleocapsid protein: identification of B-cell epitopes and domains involved in protective immunity. Virology. 1996;216:397–406. doi: 10.1006/viro.1996.0075. [DOI] [PubMed] [Google Scholar]
- Lundkvist A., Niklasson B. Bank vole monoclonal antibodies against puumala virus envelope glycoproteins: identification of epitopes involved in neutralization. Arch. Virol. 1992;126:93–105. doi: 10.1007/BF01309687. [DOI] [PubMed] [Google Scholar]
- Lundkvist A., Vapalahti O., Plyusnin A., Sjolander K.B., Niklasson B., Vaheri A. Characterization of tula virus antigenic determinants defined by monoclonal antibodies raised against baculovirus-expressed nucleocapsid protein. Virus Res. 1996;45:29–44. doi: 10.1016/0168-1702(96)01360-3. [DOI] [PubMed] [Google Scholar]
- Mir M.A. Hantaviruses. Clin. Lab. Med. 2010;30:67–91. doi: 10.1016/j.cll.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 2004;78:8281–8288. doi: 10.1128/JVI.78.15.8281-8288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., Lieber D., Meyer S., Dautel P., Kerth A., Kraus I. Crystal structure of the borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3710–3715. doi: 10.1073/pnas.0808101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol S.T., Beaty B.J., Elliott R.M. Bunyaviridae. 2005:695–716. [Google Scholar]
- Noton S.L., Medcalf E., Fisher D., Mullin A.E., Elton D., Digard P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.J., Evans P.R. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez D.R., Donis R.O. The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology. 1998;249:52–61. doi: 10.1006/viro.1998.9318. [DOI] [PubMed] [Google Scholar]
- Piper M.E., Sorenson D.R., Gerrard S.R. Efficient cellular release of rift valley fever virus requires genomic RNA. PLoS One. 2011;6:e18070. doi: 10.1371/journal.pone.0018070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A., Vapalahti O., Vaheri A. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 1996;77(Pt 11):2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- Schmaljohn C., Hjelle B. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn C.S., Hasty S.E., Dalrymple J.M., LeDuc J.W., Lee H.W., von Bonsdorff C.H. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science. 1985;227:1041–1044. doi: 10.1126/science.2858126. [DOI] [PubMed] [Google Scholar]
- Severson W., Partin L., Schmaljohn C.S., Jonsson C.B. Characterization of the hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 1999;274:33732–33739. doi: 10.1074/jbc.274.47.33732. [DOI] [PubMed] [Google Scholar]
- Strandin T., Hepojoki J., Wang H., Vaheri A., Lankinen H. Hantaviruses and TNF-alpha act synergistically to induce ERK1/2 inactivation in vero E6 cells. Virol. J. 2008;5:110. doi: 10.1186/1743-422X-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandin T.M., Hepojoki J.M., Wang H., Vaheri A., Lankinen H.M. Inactivation of hantaviruses by N-ethylmaleimide preserves virion integrity. J. Gen. Virol. 2011;92:1189–1198. doi: 10.1099/vir.0.027896-0. [DOI] [PubMed] [Google Scholar]
- Suryanarayana K., Baczko K., ter Meulen V., Wagner R.R. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J. Virol. 1994;68:1532–1543. doi: 10.1128/jvi.68.3.1532-1543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins J., Ruigrok R.W., Weissenhorn W. Structural studies on the ebola virus matrix protein VP40 indicate that matrix proteins of enveloped RNA viruses are analogues but not homologues. FEMS Microbiol. Lett. 2004;233:179–186. doi: 10.1111/j.1574-6968.2004.tb09480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaheri A., Mills J.N., Spiropoulou C.F., Hjelle B. Zoonoses — biology, clinical practice and public health”. In: Palmer S.R., Soulsby L., Brown D., Torgerson P., editors. Hantaviruses. Oxford University Press; Oxford. U.K.: 2011. pp. 307–322. [Google Scholar]
- Vaheri A., Vapalahti O., Plyusnin A. How to diagnose hantavirus infections and detect them in rodents and insectivores. Rev. Med. Virol. 2008;18:277–288. doi: 10.1002/rmv.581. [DOI] [PubMed] [Google Scholar]
- Vapalahti O., Kallio-Kokko H., Narvanen A., Julkunen I., Lundkvist A., Plyusnin A. Human B-cell epitopes of puumala virus nucleocapsid protein, the major antigen in early serological response. J. Med. Virol. 1995;46:293–303. doi: 10.1002/jmv.1890460402. [DOI] [PubMed] [Google Scholar]
- Vapalahti O., Lundkvist A., Kukkonen S.K., Cheng Y., Gilljam M., Kanerva M. Isolation and characterization of tula virus, a distinct serotype in the genus hantavirus, family bunyaviridae. J. Gen. Virol. 1996;77(Pt 12):3063–3067. doi: 10.1099/0022-1317-77-12-3063. [DOI] [PubMed] [Google Scholar]
- Vapalahti O., Mustonen J., Lundkvist A., Henttonen H., Plyusnin A., Vaheri A. Hantavirus infections in Europe. Lancet Infect. Dis. 2003;3:653–661. doi: 10.1016/s1473-3099(03)00774-6. [DOI] [PubMed] [Google Scholar]
- Wakefield L., Brownlee G.G. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 1989;17:8569–8580. doi: 10.1093/nar/17.21.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Alminaite A., Vaheri A., Plyusnin A. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein gn. Virus Res. 2010;151:205–212. doi: 10.1016/j.virusres.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Wang Y., Boudreaux D.M., Estrada D.F., Egan C.W., St Jeor S.C., De Guzman R.N. NMR structure of the N-terminal coiled coil domain of the Andes hantavirus nucleocapsid protein. J. Biol. Chem. 2008;283:28297–28304. doi: 10.1074/jbc.M804869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Handa H., Mizumoto K., Nagata K. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J. Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Liu T., Offringa D.P., McInnis J., Levandowski R.A. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.P., Baylor N.W., Wagner R.R. Transcription-inhibition and RNA-binding domains of influenza A virus matrix protein mapped with anti-idiotypic antibodies and synthetic peptides. J. Virol. 1989;63:3586–3594. doi: 10.1128/jvi.63.9.3586-3594.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]