Abstract
Background & Aims
Cyclophilins are host factors required for hepatitis C virus replication. Cyclophilin inhibitors such as alisporivir have shown strong anti–hepatitis C virus activity in vitro and in clinical studies. However, little is known about whether hepatocyte cyclophilins are involved in the hepatitis B virus (HBV) life cycle. We investigated the effects of 2 cyclophilin inhibitors (alisporivir and NIM811) on HBV replication and hepatitis B surface antigen (HBsAg) production in cell lines.
Methods
Liver-derived cell lines producing full-length HBV and HBsAg particles, owing to stable (HepG2215) or transient (HuH-7) transfection, or infected with HBV (HepaRG cells; Invitrogen [Carlsbad, CA]), were incubated with alisporivir or NIM811 alone, or alisporivir in combination with a direct antiviral (telbivudine). The roles of individual cyclophilins in drug response was evaluated by small interfering RNA knockdown of cyclophilin (CYP)A, CYPC, or CYPD in HepG2215 cells, or CYPA knockdown in HuH-7 cells. The kinetics of antiviral activity were assessed based on levels of HBV DNA and HBsAg and Southern blot analysis.
Results
In HepG2215, HuH-7, and HepaRG cells, alisporivir reduced intracellular and secreted HBV DNA, in a dose-dependent manner. Knockdown of CYPA, CYPC, or CYPD (reduced by 80%) significantly reduced levels of HBV DNA and secreted HBsAg. Knockdown of CYPA significantly reduced secretion of HBsAg, leading to accumulation of intracellular HBsAg; the addition of alisporivir greatly reduced levels of HBsAg in these cells. The combination of alisporivir and telbivudine had greater antiviral effects than those of telbivudine or alisporivir alone.
Conclusions
Alisporivir inhibition of cyclophilins in hepatocyte cell lines reduces replication of HBV DNA and HBsAg production and secretion. These effects are potentiated in combination with direct antiviral agents that target HBV-DNA polymerase.
Keywords: Cyclophilin Inhibition, Direct-Acting Antiviral Agent, Peptidyl-Prolyl Isomerase, Viral Replication
Abbreviations used in this paper: ALV, alisporivir; BL, baseline; CYP, cyclophilin; DMEM, Dulbecco’s modified Eagle medium; DMSO, dimethyl sulfoxide; ELISA, enzyme-linked immunosorbent assay; FCS, fetal calf serum; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; HCV, hepatitis C virus; iHBsAg, intracellular hepatitis B surface antigen; KD, knockdown; LDH, lactate dehydrogenase; mRNA, messenger RNA; PCR, polymerase chain reaction; sHBsAg, secreted hepatitis B surface antigen; siRNA, small interfering RNA
The cyclophilins are a group of cellular proteins with peptidyl-prolyl isomerase enzymatic activity, which catalyze the cis to trans conversion of proline-containing peptides and facilitate protein folding.1, 2 There are 7 main cyclophilins in human beings: cyclophilin A (CYP)A, CYPB, CYPC, CYPD, CYPE, CYP40, and CYP natural killer (NK),1 which are localized in different cellular compartments. For example, CYPA and CYP40 are present in the cytosol, CYPB and CYPC reside in the lumen of the endoplasmic reticulum, CYPD is present in the mitochondria, and CYPE is localized in the nucleus.
Cyclophilins are involved in the life cycle of a wide range of viruses including hepatitis C virus (HCV), human immunodeficiency virus, vaccinia virus, coronaviruses, and polyomavirus BK, acting as host co-factors essential for virus replication.3, 4, 5, 6, 7, 8 CYPA is the main cyclophilin that is involved directly in the life cycle of HCV2, 3, 4 and inhibition of its peptidyl-prolyl isomerase activity with cyclophilin inhibitors was shown to interfere at multiple sites of the HCV life cycle in hepatocytes, affecting not only replication but also the HCV secretion from infected cells.9, 10, 11, 12, 13 The role of cellular cyclophilins in the hepatitis B virus (HBV) life cycle, however, is poorly understood.
In the present study we investigated whether hepatocyte cyclophilins are involved in HBV replication, hepatitis B surface antigen (HBsAg) production and secretion, and the effects of nonimmunosuppressive cyclophilin inhibitors alone and in combination with a direct antiviral agent targeting HBV polymerase.
Materials and Methods
Cell Lines Supporting HBV Replication
Four human hepatoma cell lines that produce full HBV virions and HBsAg subviral particles were used in this study: (1) HuH-7 cells (Japan Health Science Research Resources Bank, Osaka, Japan), and (2) HuH-7 with stable knockdown (KD) of CYPA using a short hairpin,11 both cell lines were transfected with HBV DNA; (3) HepG2215 cells, which are stably transfected with HBV DNA; and (4) HepaRG cells (Invitrogen, Carlsbad, CA), which were infected with HBV. The HuH-7 and HepG2215 cells were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Paisley, UK) with 10% fetal calf serum (FCS) (Life Technologies), as described previously.14 The HuH-7 CYPA KD cells were cultured in DMEM/10% FCS plus 1× nonessential amino acids and 1 μg/mL of the selection marker puromycin (Life Technologies). HepaRG cells are terminally differentiated and were purchased from Invitrogen (Carlsbad, CA). The cells were cultured according to the manufacturer’s instructions (HepaRG Cell User Guide; Invitrogen) on collagen I–coated plates. Initially, cells were grown in William’s Medium E with HepaRG Thaw, Plate&General Purpose Medium Supplement and GlutaMAX (Invitrogen), and prepared in a standardized 2-step process. After 7 days, cells were maintained in William’s Medium E with HepaRG Maintenance Medium Supplement plus GlutaMAX.
HuH-7 and HuH-7 CYPA KD cells were transfected with a HBV plasmid, pSM2 (kindly provided by Professor Hans Will), which contained a head-to-tail HBV-DNA dimer.15 Briefly, HuH-7 cells, at a density of 1.5 × 105/well, were seeded onto 24-well plates and cultured for 48 hours at 37°C to reach confluency. The cells then were transfected with 0.5 μg of pSM2 with Fugene 6 (Roche, Burgess Hill, UK) according to the manufacturer’s instructions. The transfection efficiency was determined using a β-gal staining kit (Life Technologies). HepaRG cells were infected with HBV derived from 5-day culture supernatants of AD38 cells, as described.16 HepG2215 and HBV-infected HepaRG cells were seeded at 1 and 0.3 ×106 cells, respectively, onto 24-well plates and maintained at 37°C. The cell lines subsequently were treated as described later.
Small Interfering RNA Transfection for Silencing Cellular Cyclophilin Expression
HepG2215 cells were transfected with CYPA, B, C, or D small interfering RNA (siRNA) (siGENOME SMART pool; Thermo Scientific Dharmacon, Epsom, UK). To optimize the knockdown effect of cyclophilin expression, preliminary experiments with a range of siRNA concentrations, specific for each cyclophilin, were conducted by testing 30, 60, 100, and 150 nmol/L over 96 hours (see the Supplementary Materials and Methods section). The cyclophilin messenger RNA (mRNA) expression at baseline and at different time points after siRNA transfection was assessed by real-time polymerase chain reaction (PCR) with primers specific for each CYPA, B, C, and D. After these optimization experiments, siRNA stock solutions of 20 μmol/L were diluted in 50 μL Opti-MEM medium (Life Technologies) for a final concentration of 60 nmol/L for CYPA and CYPD siRNA, and 100 nmol/L for CYPC. Lipofectamine 2000 (Invitrogen, Life Technologies) was diluted into 50 μL Opti-MEM medium and incubated for 5 minutes at room temperature. Combined diluted siRNA and diluted Lipofectamine were incubated for 30 minutes at room temperature.
HepG2215 cells (1 × 105 cells per well) then were treated with the siRNA–Lipofectamine complex and incubated for 48 hours at 37°C. Nontargeting scrambled siRNA was used as a negative control; glyceraldehyde-3-phosphate dehydrogenase siRNA and siGLO (Thermo Scientific Dharmacon) Green were used as positive controls for siRNA delivery and transfection, respectively. The specificity of siRNA silencing was cross-checked using real-time PCR and primers specific for CYPA, CYPC, CYPD mRNA with HepG2215 cells transfected with CYPA, CYPC, or CYPD siRNA, respectively. This confirmed that each siRNA selectively decreased only targeted cyclophilin mRNA (by approximately 80%), whereas the other cyclophilin (CYPA, CYPC, or CYPD) mRNA expression was comparable with the control (scramble siRNA). The knock-down effect for CYPA protein was tested further by Western blotting11 and by enzyme-linked immunosorbent assay (ELISA), as described later.
Compounds and Cell Treatment
Alisporivir and NIM811 are nonimmunosuppressive cyclophilin inhibitors9; telbivudine, a nucleoside analog, is a potent inhibitor of HBV-DNA polymerase.17 All compounds were provided by Novartis (Basel, Switzerland).
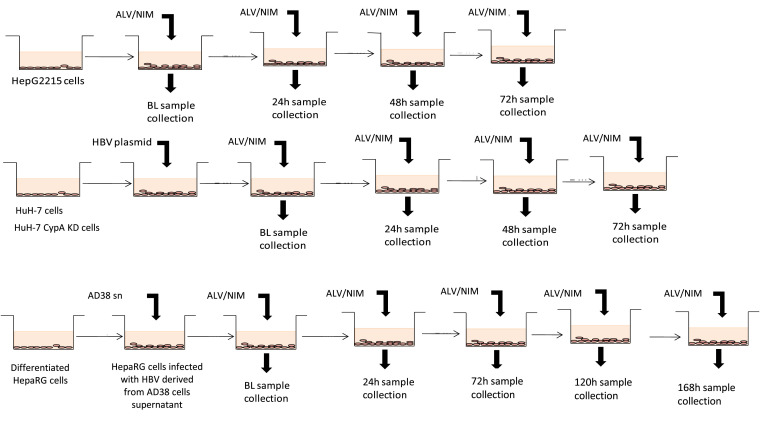
In a series of experiments, the cells used (stably transfected HepG2215, transiently transfected HuH-7, HuH-7 CYPA KD cells, and HBV-infected HepaRG cells) were cultured alone and with several concentrations of alisporivir or NIM811. Alisporivir and NIM811 stock solutions were prepared as 2000× stocks in 100% dimethyl sulfoxide (DMSO) and the final working concentrations of alisporivir and NIM811 (0.25, 1, 5, and 20 μg/mL) were prepared daily in DMEM/10% FCS with a final DMSO concentration of 0.05%. The control also contained 0.05% DMSO. The culture media were replaced every 24 hours with 1.5 mL of fresh alisporivir-containing or NIM-containing medium. Supernatants and cells were collected at baseline (BL), and at 24, 48, and 72 hours. HBV-infected HepaRG cells were treated with alisporivir or NIM811 for 7 days, cells and supernatants were collected at BL, and at 24, 72, 120, and 168 hours (Supplementary Figure 1). For all conditions and time points, HepG2215 and HuH-7 cells were tested in at least 3 independent experiments with each condition run in triplicate wells. HepaRG and HuH-7 CYPA KD cells were tested in duplicates. Because the culture medium was replaced every 24 hours, the graphic representations of the results for viral particles secreted in supernatants are shown with each time point starting from zero after each medium change.
Cytotoxicity was assessed by microscopic observation of the cells, trypan blue exclusion, and/or by lactate dehydrogenase (LDH) release in supernatants using the LDH-cytotoxicity assay kit (Biovision Research Products, Mountain View, CA).
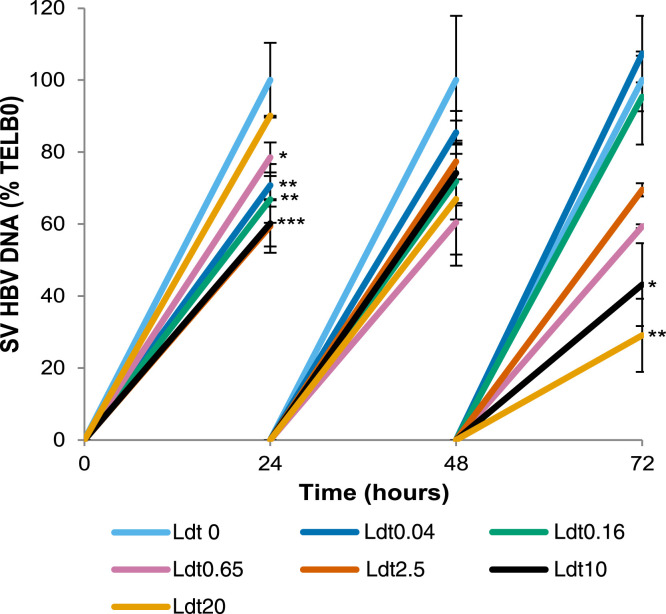
Before treating HepG2215 cells with the combination of alisporivir (ALV) and telbivudine, we first tested the antiviral activity of a range of telbivudine concentrations in the current model. Stock solutions with 2000× telbivudine in 100% DMSO were used to prepare 6 working concentrations daily, ranging from 0.04 to 20 μmol/L telbivudine (Supplementary Figure 2). The cell culture medium containing these working concentrations was replaced daily and samples were collected at BL, and at 24, 48, and 72 hours. Based on this experiment, in the subsequent combination experiments telbivudine was used at 10 μmol/L (2.42 μg/mL), which corresponds to the maximum concentration levels observed in clinical studies (see Supplementary Materials and Methods section). To test the antiviral activity of ALV in combination with telbivudine, HepG2215 cells were treated for 72 hours with the following: ALV 0.25 μg/mL/telbivudine 10 μmol/L, ALV 1 μg/mL/telbivudine 10 μmol/L, ALV 5 μg/mL/telbivudine 10 μmol/L, and ALV 20 μg/mL/telbivudine 10 μmol/L.
Evaluation of HBV Replication: Quantitative Real-Time Polymerase Chain Reaction and Southern Blotting
Intracellular, nucleocapsid-associated viral DNA was extracted from transfected HuH-7 and 2215 cells or HBV-infected HepaRG cells, as described previously.14, 18 The culture supernatants were treated with DNase I for 1 hour at 37°C, 10 minutes at 75°C, and then placed on ice for 1 minute. The supernatants were centrifuged at 10,000 rpm and the pellets were discarded. The DNA was extracted from the supernatants using the QiAamp DNA mini Qiagen kit (Qiagen, Sussex, UK). HBV DNA was quantitated by TaqMan real-time polymerase chain reaction (Applied Biosystems 7500; Applied Biosystems, Paisley, UK) using the EuroHep HBV standard and HBV-DNA plasmid.14, 18
For Southern blot analysis, 20 μg of HinD III–treated intracellular DNA was analyzed on a 1% agarose gel and transferred to a Hybond-XL (GE Healthcare Life Sciences, Pittsburgh, PA) membrane, as described.19 Radioactive HBV-specific probes were generated from a Sac I–HinD III fragment derived from pSP65-ayw-1.3 by nick translation and purified by spin column chromatography. Prehybridization, hybridization, and washes were as described previously.20
Quantitation of HBsAg, CypA, and CypB by ELISA
HBsAg secreted during cell culture (sHBsAg) and intracellular HBsAg (iHBsAg) were quantitated using a commercially available HBsAg ELISA (Abazyme, Needham, MA). HBsAg levels were quantitated in triplicates of the culture supernatants and cytoplasmic extracts of stably and transiently transfected cells. A standard curve was established by using a serial dilution of a known quantity of purified HBsAg protein (American Research Products, Belmont, MA).
The intracellular CYPA and CYPB levels in supernatants were quantitated by ELISA, as described previously.21
Statistics
Statistical analyses were performed using the SPSS package (IBM, New York, NY). One-way analysis of variance was used, followed by Dunnett's post hoc test to compare one treatment group with the nontreated group and the Tukey post hoc test was used for multiple comparisons between groups.22 A P value less than .05 was considered statistically significant.
Results
Alisporivir and NIM811 Reduce HBV Replication
Alisporivir treatment of HepG2215 cells resulted in a progressive reduction of secreted and intracellular, nucleocapsid-associated HBV DNA dependent on both drug concentration and time of drug exposure (Figure 1 A and B). The secreted viral DNA was reduced by 52% after 72 hours of treatment with alisporivir 20 μg/mL, compared with cells incubated with medium only (P < .01). The level of intracellular HBV DNA also was reduced by approximately 60% after 72 hours of treatment with 5 and 20 μg/mL of alisporivir (P < .01). NIM811 treatment of HepG2215 and of HepaRG cells also reduced the secreted and intracellular nucleocapsid-associated HBV DNA, however, its antiviral effect was lower than alisporivir (Supplementary Figure 3). The difference between NIM811 and alisporivir in reducing HBV-DNA level was particularly apparent with HepG2215 cells at 5 μg/mL: 6% vs 34% HBV-DNA reduction at 48 hours.
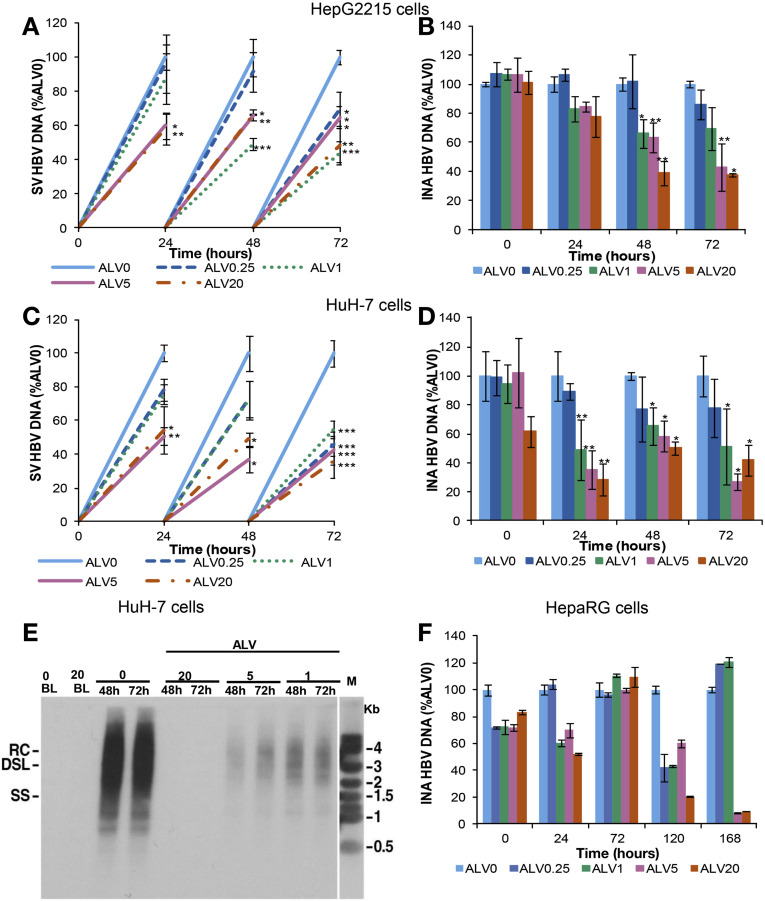
Figure 1.
ALV suppresses HBV replication in liver-derived cell lines. HepG2215, HuH-7, and HepaRG cells were treated with 0.25, 1, 5, and 20 μg/mL of ALV and HBV DNA was quantitated by real-time PCR. Cells and supernatants were collected every 24 hours. HepG2215 cells: (A) secreted-virus (SV) HBV DNA and (B) intracellular nucleocapsid-associated (INA) HBV DNA. HuH-7 cells: (C) SV HBV DNA and (D) INA HBV DNA. (E) Southern blot analysis of intracellular HBV DNA from HuH-7 cells. The positions of relaxed circular (RC), double-strand linear (DSL), and single-strand (SS) DNA intermediates are indicated. (F) HepaRG: INA HBV DNA during 7 days (168 hours) of treatment. Mean values and the SD of 3 independent experiments are shown. *P values less than .05, **P values less than .01, and ***P values less than .001 for ALV-treated vs nontreated cells.
In HuH-7 cell cultures, the control experiments for transfection efficiency (β-gal staining) showed HBV replication in 30%–40% of cells. Alisporivir treatment of HBV-transfected HuH-7 cells also resulted in a dose-dependent reduction of secreted and intracellular HBV DNA. The most profound reduction was observed after 72 hours of treatment with alisporivir at 5 μg/mL (58% and 73%, respectively) and 20 μg/mL (64% and 58%, respectively) (Figure 1 C and D). Overall, the alisporivir antiviral activity was greater in transiently transfected HuH-7 cells in comparison with stably transfected HepG2215 cells (P = .038 for secreted HBV DNA and P < .001 for cytoplasmic HBV DNA). As stated earlier, alisporivir at 5 μg/mL reduced intracellular HBV-DNA levels by 73% and 58% in HuH-7 and HepG2215 cells, respectively, after 72 hours of treatment (Figure 1 B and D). This difference was even more apparent at earlier time points. After 24 hours, the intracellular HBV-DNA reduction in HepG2215 cells and HuH-7 cells was 16% and 64%, respectively (Figure 1 B and D). Importantly, the Southern blot analysis of intracellular DNA supports the quantitative real-time PCR results outlined earlier because alisporivir reduced intracellular HBV DNA in a dose-dependent manner (Figure 1 E).
HBV-infected HepaRG cells were treated with alisporivir or NIM811 for 7 days, starting 16 hours after HBV inoculation (Supplementary Figure 1). Also in this model, the cyclophilin inhibitors reduced HBV-DNA level (Figure 1 F and Supplementary Figure 3). After 120 and 168 hours of treatment with the highest concentration of alisporivir, intracellular nucleocapsid-associated HBV DNA was reduced by 80% and 90%, respectively, similar to the effect of NIM811: 70% and 90% reduction, respectively.
Alisporivir and NIM811 Inhibit HBsAg Production and Secretion
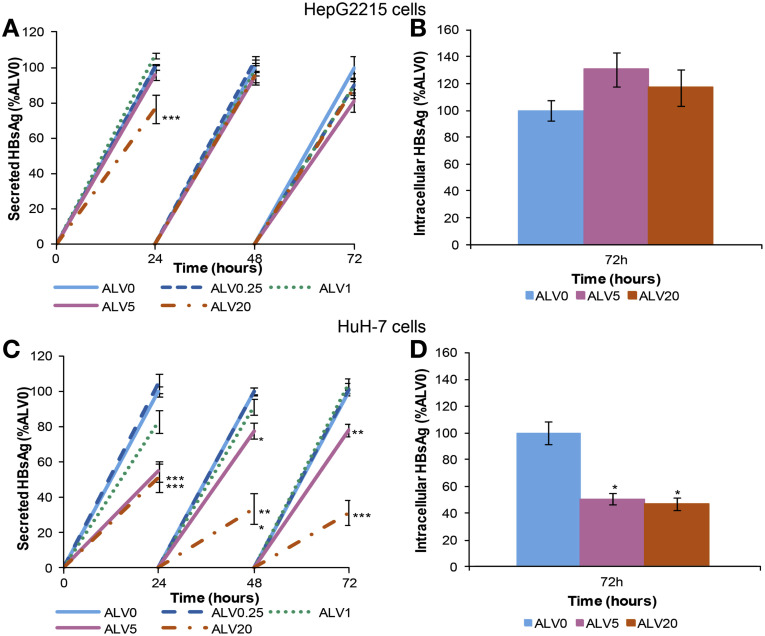
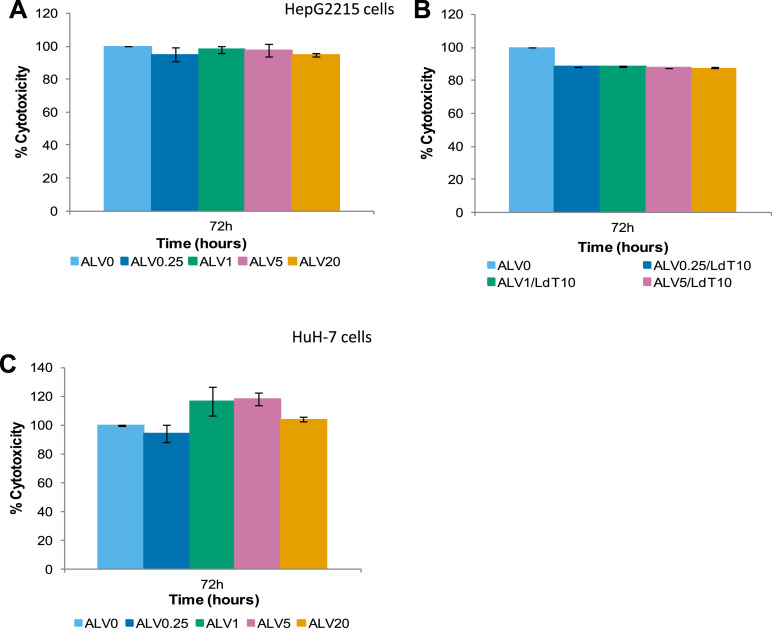
In HepG2215 cells, there were minimal changes in sHBsAg or iHBsAg levels (Figure 2 A and B). The magnitude of reduction of sHBsAg by both alisporivir and NIM811 was lower than the reduction of HBV-DNA level (Supplementary Figures 3 and 4). In HuH-7 cells, significant reductions in both sHBsAg and iHBsAg were observed with the two highest alisporivir concentrations (Figure 2 C and D). The most significant change was seen after 72 hours of treatment with 20 μg/mL alisporivir, in which sHBsAg was reduced by 69% and iHBsAg was reduced by 53%. In all experiments, alisporivir treatment was not cytotoxic for HepG2215 and HBV-transfected HuH-7 cells, as shown by LDH release assay (Supplementary Figure 5).
Figure 2.
ALV decreases HBsAg production and secretion. HepG2215 and HuH-7 cells were treated with 0.25, 1, 5, and 20 μg/mL of ALV over 72 hours. Cells and supernatants were collected every 24 hours and HBsAg levels were quantitated by ELISA. HepG2215 cells: (A) sHBsAg and (B) iHBsAg. HuH-7 cells: (C) sHBsAg and (D) iHBsAg. Mean values and the SD of 3 independent experiments are shown. *P values less than .05, **P values less than .01, and ***P values less than .001 for ALV-treated vs nontreated cells.
Silencing of Cyclophilin Expression Reduces HBV Replication
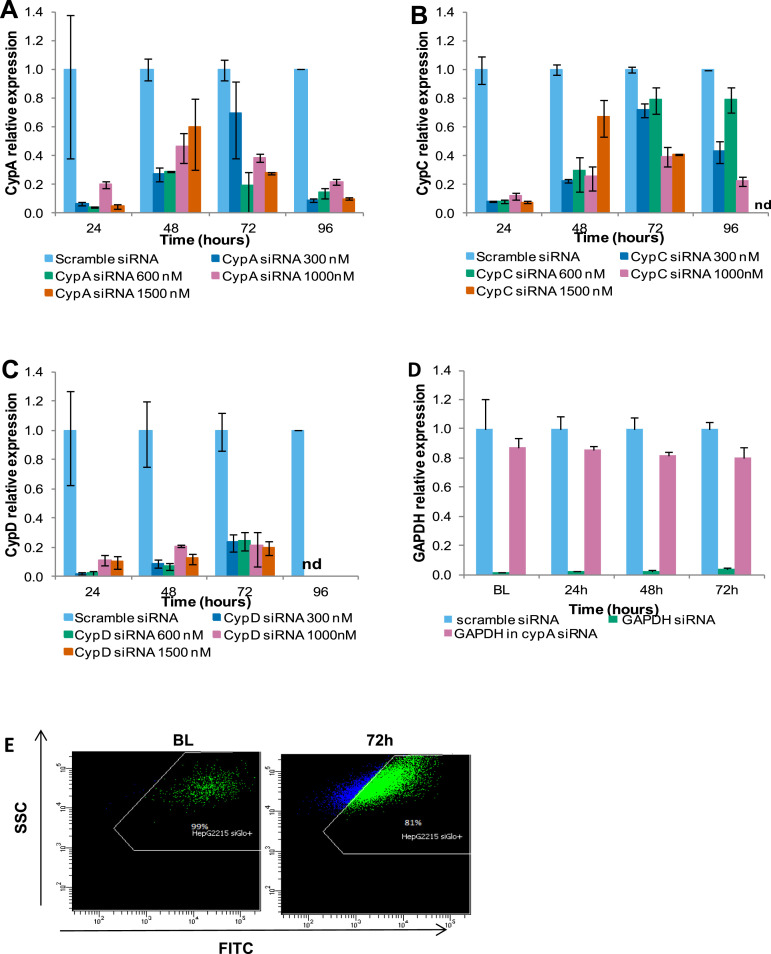
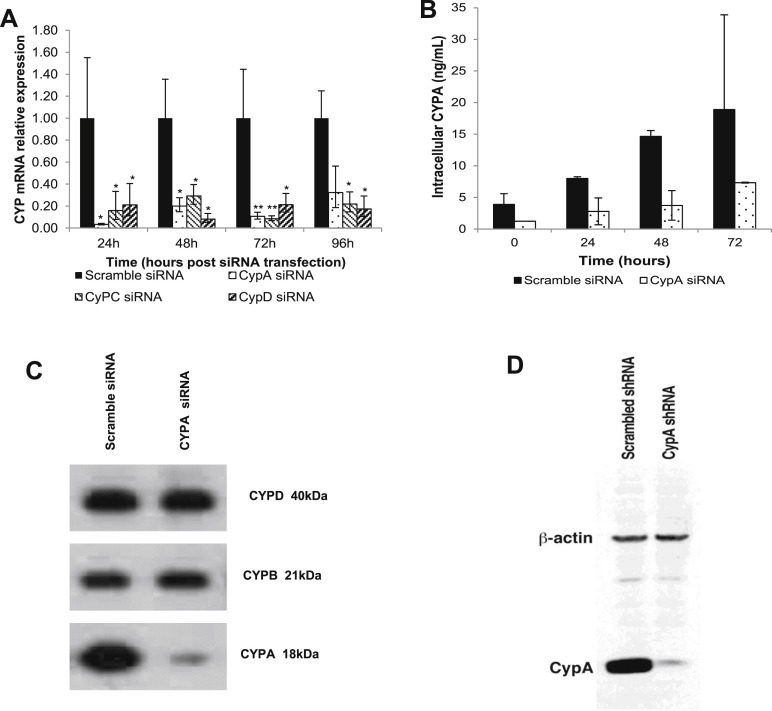
To determine the relative involvement of different cyclophilins in HBV replication, the expression of CYPA, B, C, and D were silenced selectively with a respective siRNA. The degree of gene silencing with a range of CYPA, B, C, or D siRNA concentrations was assessed over a 96-hour period by real-time PCR (Supplementary Figure 6). The efficiency of the transfection and the siRNA delivery were assessed with siGLO and glyceraldehyde-3-phosphate dehydrogenase, respectively (Supplementary Figure 6 D and E). The expression of CYPA, C, or D mRNA in HepG2215 cells was reduced markedly (by ≥80%) in comparison with the controls treated with nontargeting, scrambled siRNA for the duration of the experiment (Supplementary Figure 7 A). The CYPA gene silencing also efficiently reduced the intracellular CYPA protein when tested by ELISA (Supplementary Figure 7 B), or by Western blot (Supplementary Figure 7 C).
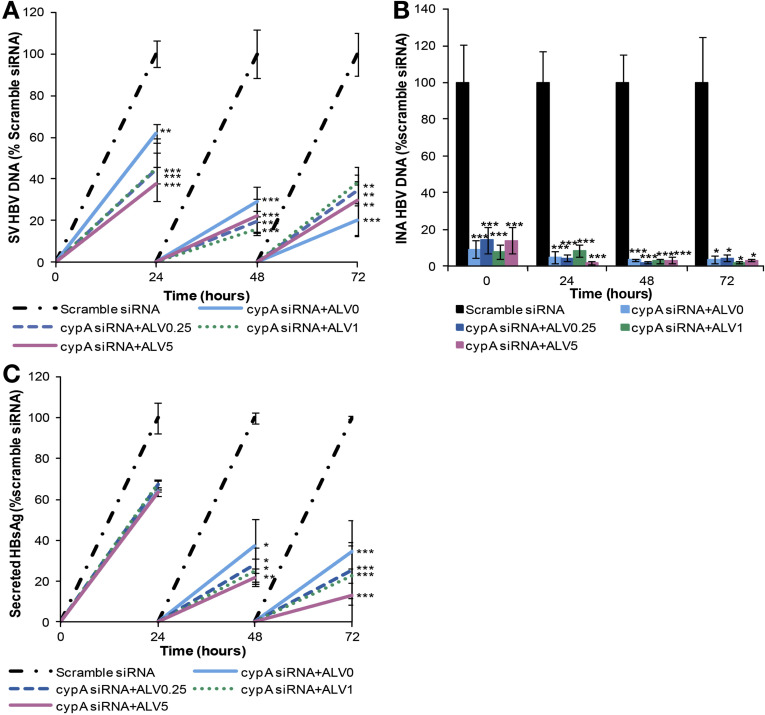
Both secreted and intracellular HBV-DNA levels were reduced by 80% and 97%, respectively, in CYPA-silenced cells (Figure 3 A and 3 B). Secreted HBsAg was reduced by 65% in the same cells (Figure 3 C). The CYPA-silenced cells were treated with alisporivir and compared with the CYPA-silenced cells without alisporivir (CYPA + no alisporivir) to determine whether alisporivir inhibition of other cellular cyclophilins would enhance the antiviral effect of silencing CYPA. The addition of ALV did not result in any further decrease in the levels of secreted and intracellular nucleocapsid-associated HBV DNA and sHBsAg (Figure 3).
Figure 3.
Silencing of CYPA suppresses HBV replication and HBsAg secretion. HepG2215 cells were transfected with CYPA siRNA and seeded in a culture plate for 48 hours and then treated with 0.25, 1, and 5 μg/mL of ALV over 72 hours. Cells and supernatants were collected every 24 hours. (A) Secreted-virus (SV) HBV DNA, (B) intracellular nucleocapsid-associated (INA) HBV were quantified by real time PCR. (C) sHBsAg was measured by ELISA. Mean values and the SD of 3 independent experiments are shown. *P values less than .05, **P values less than .01, and ***P values less than .001 for CYPA siRNA ± ALV-treated vs scrambled siRNA-treated cells.
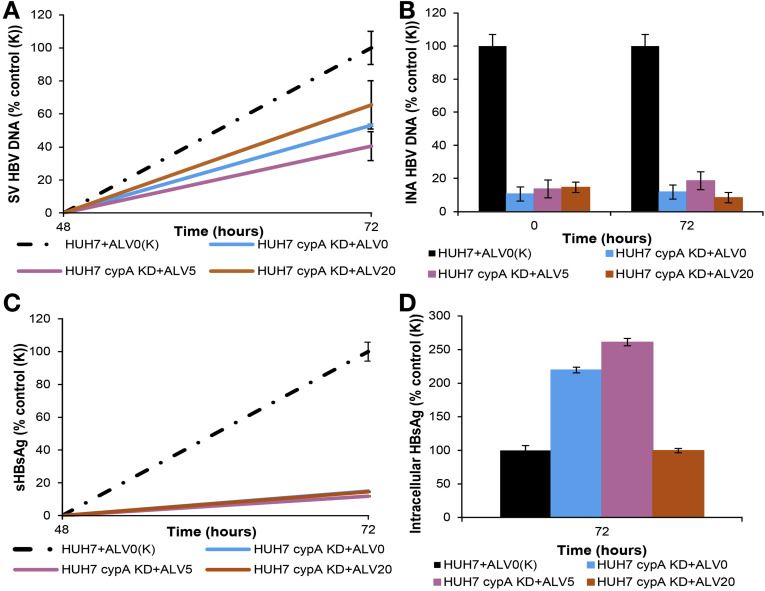
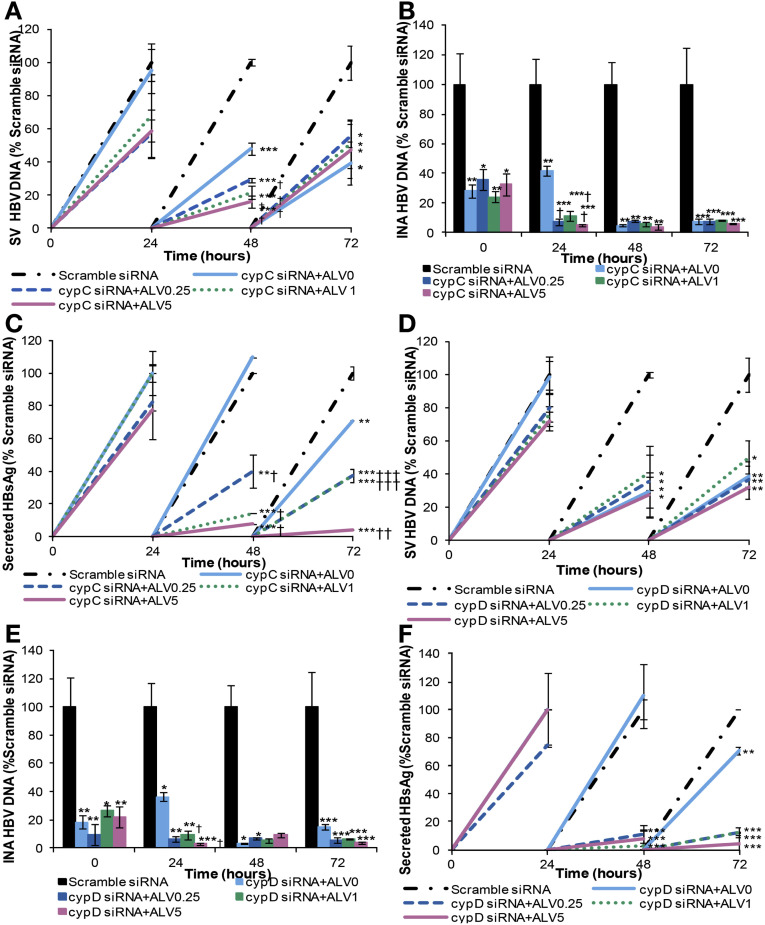
We also used HuH-7 cells with stable CYPA KD (Supplementary Figure 7 D) and assessed the impact on HBV replication and HBsAg. The secreted HBV DNA was reduced by 50% in these cells in comparison with normal HuH-7 cells (Figure 4 A), and adding alisporivir had only a marginal effect. Intracellular nucleocapsid-associated HBV DNA was affected even more in this cell line with a 90% reduction (Figure 4 B). Secreted HBsAg also was reduced significantly (90%) whereas intracellular HBsAg accumulated in these cells (Figure 4 C and D). Interestingly, treatment of the CYPA KD cells with alisporivir (20 μg/mL) profoundly decreased intracellular HBsAg levels. In CYPC-silenced cells, secreted and intracellular nucleocapsid-associated HBV DNA were reduced by 61% and 93%, respectively, compared with negative controls (Figure 5 A and B) and HBsAg levels were reduced by 30% (Figure 5 C). The result of silencing CYPD was similar to silencing CYPC-as secreted and intracellular nucleocapsid-associated HBV DNA and sHBsAg, which were reduced by 62%, 96%, and 30%, respectively (Figure 5 D–F). CYPA silencing with siRNA resulted in a greater reduction in secreted HBV DNA and sHBsAg compared with siRNA silencing of CYPC or CYPD. As stated earlier, at 72 hours treatment, the silencing of CYPA reduced secreted HBV DNA by 80% (Figure 3 A) compared with 60% in CYPC or CYPD-silenced cells (Figure 5 A and D). At the same time point, sHBsAg was reduced by 65% (Figure 3 C) compared with 30% in CYPC or CYPD silenced cells (Figures 5 C and F, respectively). Interestingly, adding alisporivir to CYPC and CYPD-silenced cells further reduced the 3 viral parameters measured (Figure 5). This increased effect in the presence of alisporivir was seen only when the levels of secreted and intracellular DNA as well as sHBsAg were still increased in the cyclophilin-silenced cells such as at 24 hours for intracellular nucleocapsid-associated DNA and 48 and 72 hours for sHBsAg. This additional effect of alisporivir, however, disappeared when the levels of secreted and intracellular nucleocapsid-associated HBV DNA became very low in cyclophilin-silenced cells.
Figure 4.
Stable KD of CypA decreases HBV replication and HBsAg secretion but not HBsAg production. CYPA KD HuH-7 cells were seeded in a culture plate and transfected with HBV DNA plasmid pSM2 and then treated with 5 and 20 μg/mL of ALV over 72 hours. Cells and supernatants were collected at BL and 72 hours. (A) Secreted-virus (SV) HBV DNA and (B) intracellular nucleocapsid-associated (INA) HBV DNA were quantified by real-time PCR. (C) SHBsAg and (D) iHBsAg were measured by ELISA.
Figure 5.
siRNA silencing of CYPC and CYPD decreases HBV replication and HBsAg secretion but to a lesser extent than CYPA. HepG2215 cells were transfected with CYPC and CYPD siRNA and seeded in a culture plate for 48 hours and then treated with 0.25, 1, and 5 μg/mL of ALV over 72 hours. Cells and supernatants were collected every 24 hours. (A and D) Secreted-virus (SV) HBV DNA and (B and E) intracellular nucleocapsid-associated (INA) HBV DNA were quantified by real-time PCR. (C and F) SHBsAg was measured by ELISA. Mean values and the SD of 3 independent experiments are shown. *P values less than .05, **P values less than .01, ***P values less than .001 for CYPC/D siRNA ± ALV-treated vs scramble siRNA-treated cells. †P values less than .05, ††P values less than .01, †††P values less than .001 for CYPC/D siRNA-treated vs CYPC/D siRNA+ALV-treated cells.
Cyclophilin Levels in Alisporivir-Treated Cells
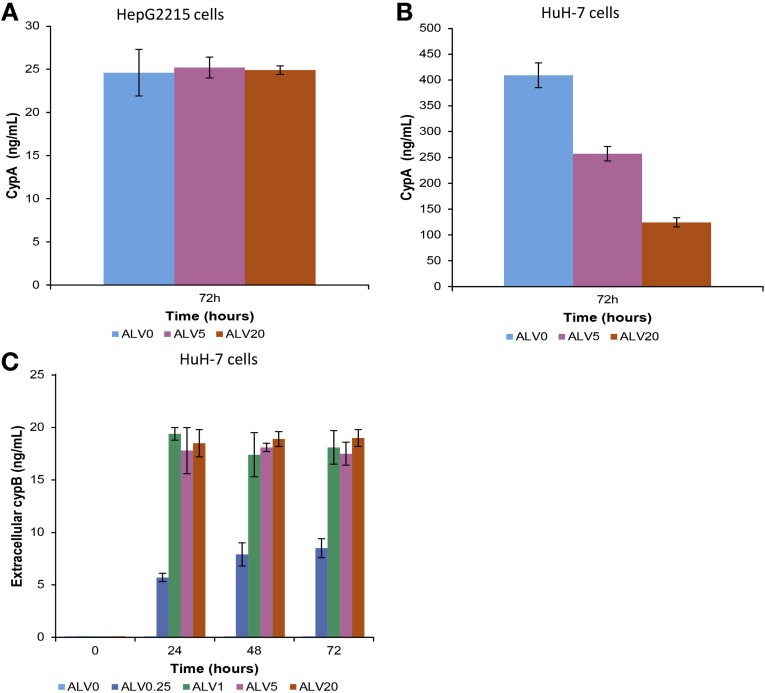
Because CYPA represents a key target for cyclophilin inhibitors, we analyzed the changes of intracellular CYPA levels in untreated and alisporivir-treated HepG2215 and HuH-7 cells. In HepG2215 cells, the CYPA levels were 16 times lower than in HuH-7 cells (Figure 6 A and B) and were not affected by the alisporivir treatment, whereas in HuH-7 cells the alisporivir treatment resulted in a dose-dependent decrease of intracellular CYPA levels. The corresponding levels of CYPB in culture supernatants of HuH-7 cells increased steadily, dependent on time and alisporivir concentration (Figure 6 C), in accordance with the previous findings of CYPB release from alisporivir-treated cells.21 In HepaRG cells, the CYPA and CYPB levels were similar to those in HuH-7 (data not shown).
Figure 6.
CYPA and CYPB levels in alisporivir-treated cells HepG2215 and HBV-transfected HuH-7 cells were treated with 0.25, 1, 5, and 20 μg/mL of alisporivir over 72 hours. Intracellular CYPA was quantitated by ELISA in (A) HepG2215 and (B) HuH-7 cells. (C) CYPB in supernatants was quantitated by ELISA in HuH-7 cells.
Combination of Alisporivir Plus a Direct Antiviral Agent Results in a Greater Reduction of HBV and HBsAg Levels
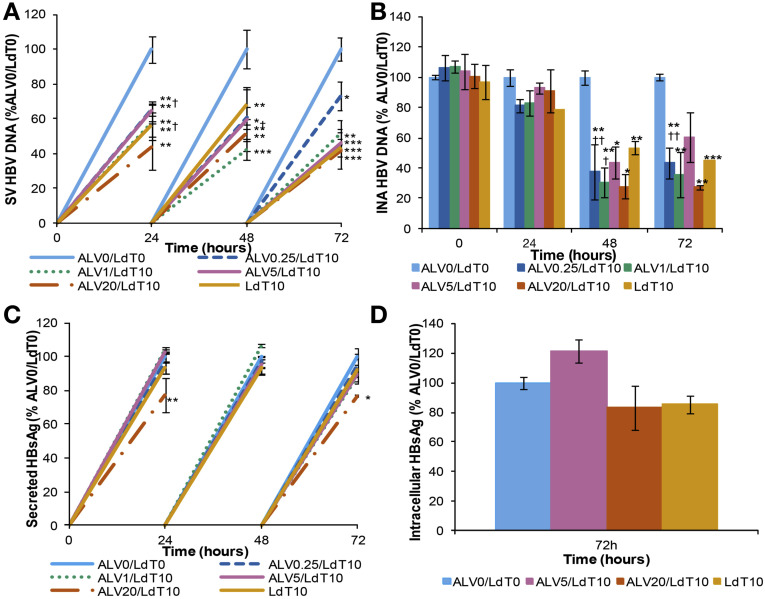
A significantly greater reduction of HBV replication was observed with alisporivir in combination with a direct antiviral agent, telbivudine, compared with alisporivir alone. This was more apparent at the lower concentrations of alisporivir: 0.25 and 1 μg/mL (Figure 7 ). Secreted viral DNA and intracellular nucleocapsid-associated HBV DNA were reduced by 39% and 62%, respectively, in cells treated with 0.25 μg/mL alisporivir plus 10 μmol/L telbivudine vs 9% and 0% in cells treated with alisporivir alone at 48 hours (Figure 7 A and B compared with Figure 1 A and B). Furthermore, at 48 hours, the reduction of secreted HBV DNA was more apparent with alisporivir plus telbivudine, compared with telbivudine alone (Figure 7 A). The combination of alisporivir plus telbivudine had a similar effect on sHBsAg and iHBsAg levels compared with alisporivir alone (Figure 7 C and D compared with Figure 2 A and B).
Figure 7.
Telbivudine (LdT) plus alisporivir combination has a greater antiviral effect than alisporivir or telbivudine alone. HepG2215 cells were treated with 0.25, 1, 5, and 20 μg/mL of alisporivir alone and in combination with 10 μmol/L of LdT or 10 μmol/L of LdT alone. Cells and supernatants were collected every 24 hours. (A) Secreted-virus (SV) HBV DNA and (B) intracellular nucleocapsid-associated (INA) HBV DNA were quantified by real-time PCR. (C) sHBsAg and (D) iHBsAg were quantified by ELISA. Mean values and the SD of 3 independent experiments are shown. *P values less than .05, **P values less than .01, ***P values less than .001 for ALV/LdT-treated vs nontreated cells. †P values less than .05, ††P values less than .01 for alisporivir-treated vs ALV/LdT-treated cells.
Discussion
HBV is the smallest human DNA virus with a unique genomic organization and replication mechanism.23 Viral replication takes place in the cytoplasm of infected hepatocytes with 3 subsets of HBV-RNA transcripts: pregenomic RNA, envelope preS/S RNAs, and HBx mRNA, encoding all structural and nonstructural viral proteins. Characteristic of HBV infection are the spheric and filamentous subviral particles composed exclusively of viral envelope proteins and host-derived lipids, which are produced 103- to 106-fold in excess to full virions and secreted from hepatocytes.23 By using 3 different in vitro HBV models: stably transfected (HepG2215), transiently transfected (HuH-7), and HBV-infected (HepaRG) cells, the present study shows that cellular cyclophilins have a major role in the HBV life cycle in hepatocytes. Blocking cyclophilins’ enzymatic activity with small molecules, alisporivir or NIM811, or the selective silencing of individual cyclophilins with siRNA, markedly reduced HBV-DNA replication, as well as HBsAg production and secretion from cells. CYPA is one of the most abundant cytosolic proteins (approximately 0.1% of cell proteins)2 and the present study shows the major utilization of cyclophilin A both in HBV replication and in HBV envelope protein secretion from hepatocytes. The experiments using HuH-7 cells with stable CYPA KD, and the findings that ALV enhanced HBV DNA and HBsAg reduction in CYPC and CYPD-silenced cells show that CYPA is important for the formation of intracellular, nucleocapsid-associated HBV DNA and viral secretion, as well as for HBsAg secretion from cells. The quantitation of intracellular CYPA showed a greater amount in HuH-7 than in HepG2215 cells, which is the likely explanation for the much greater effect of alisporivir on HBsAg secretion from HuH-7 compared with HepG2215 cells. Importantly, cyclophilin inhibitors, such as alisporivir and NIM811, block all cellular cyclophilins and in HepG2215 cells there was a dose-dependent reduction in HBV DNA. The data thus suggest that, in this model, the antiviral effect was primarily a result of alisporivir and NIM811 targeting other cyclophilins (eg, CYPD and/or CYPC), leading to the observed HBV-DNA reduction in these cells.
The results from the present study together with previously published data indicate that the mechanisms by which alisporivir and NIM811 impact HBV replication and HBsAg production are likely to involve interference at multiple sites of the HBV life cycle. First, the finding that cyclophilin inhibition reduces intracellular nucleocapsid-associated HBV DNA in the cytoplasm suggests an antiviral effect on the production of new virions and a reduction in the recirculation of HBV nucleocapsids from the cytoplasm to the nucleus, which is a key mechanism to replenish the viral template. We found a marked reduction of intracellular HBV DNA in cells with stable CYPA KD, along with intracellular HBsAg accumulation. Previous studies have shown the intracellular interaction between CYPA and HBsAg and their close association during secretion from liver cells.24 Thus, cyclophilin inhibitors will disrupt the CYPA/HBsAg complex and reduce envelope protein secretion. Second, Hepatitis B x Antigen (HBxAg), which is essential for HBV replication, associates with the outer membrane of mitochondria, and by regulating cytosolic calcium and signaling it drives HBV replication.25, 26, 27 NIM811 was shown to block cytosolic calcium signaling and the opening of mitochondrial permeability transition pores as a result of its inhibition of CYPD.28 Therefore, blocking CYPD interferes with HBxAg regulation of calcium and will impact HBV replication negatively. Third, CYPA is an important co-factor for lipids and apolipoprotein B (apoB) trafficking and NIM811 was shown to decrease apoB secretion, as well as the egress of HCV particles from JFH1–HCV–infected cells.13 Because cellular lipids are part of the HBV envelope proteins, the impact of cyclophilin inhibition on apoB secretion is likely to interfere with the intracellular formation and secretion of lipoproteins part of HBV viral and subviral particles. Fourth, recent findings have shown that the sodium taurocholate cotransporting polypeptide, a membrane bile acid transporter expressed in the liver, is a functional receptor for HBV and hepatitis Delta virus, via pre-S1 binding.29, 30 Moreover, cyclosporin A and nonimmunosuppressive analogues, such as alisporivir, recently were shown to inhibit HBV entry into cells by blocking the sodium taurocholate cotransporting polypeptide, which is a cyclophilin-independent mechanism.31, 32
The ultimate treatment goal of patients with chronic hepatitis B is to prevent the development of cirrhosis, liver failure, and hepatocellular carcinoma. HBsAg is the hallmark of HBV infection and HBsAg loss, with or without seroconversion to antibody to hepatitis B surface antigen, is the ideal treatment end point, is associated with improved long-term outcome,33, 34, 35 shows markedly enhanced antiviral T-cell reactivity,36 and it allows therapy to be stopped with a minimal risk of relapse. Although the nucleos(t)ide polymerase antivirals are potent inhibitors of HBV replication and result in a profound reduction of circulating HBV, they have little effect on HBsAg transcription/subviral particle production. HBsAg loss has been reported to occur in 2%–8% of HBeAg-positive patients and in 0%–5% of HBeAg-negative patients after 3–5 years of continuous treatment.33, 37 The clinical implication of the present study is that it provides in vitro evidence supporting a new therapeutic approach in chronic hepatitis B with a combination of a direct antiviral agent (such as the HBV-DNA polymerase inhibitors) with a host-targeting antiviral (such as alisporivir), which is likely to increase the rate of HBsAg clearance. Alisporivir is a host-targeting antiviral agent currently in development for the treatment of chronic hepatitis C. Analyses of the clinical database, involving more than 2000 patients, show that alisporivir is well tolerated and has a markedly better safety profile when given as interferon-free treatment than with interferon-containing treatment.38 A combination treatment with an HBV nucleos(t)ide polymerase inhibitor plus alisporivir could be beneficial because of the interference at multiple sites of the HBV life cycle, especially the additional effects in reducing HBsAg production and secretion, plus blocking HBV entry into noninfected hepatocytes. In the present study, the combination of alisporivir plus telbivudine showed greater impact on HBsAg and supports the potential utility of this combination, which deserves to be tested in clinical studies.
In conclusion, cyclophilins are involved in multiple steps of the HBV life cycle in infected hepatocytes and blocking their enzymatic activity using nonimmunosuppressive cyclophilin inhibitors reduces viral replication and HBV envelope protein production and secretion. The combination of alisporivir, a host-targeting agent, and an HBV polymerase inhibitor, was found to decrease HBsAg production markedly, as well as HBV replication, indicating a potential utility for a more effective therapy in chronic hepatitis B.
Acknowledgment
The authors thank Professor Hans Will (Hamburg, Germany) for providing the pSM2 plasmid and the HepG2215 cells.
Footnotes
Conflicts of interest These authors disclose the following: Shilpa Chokshi has received a research grant from Novartis, and Nikolai Naoumov is an employee of Novartis Pharma AG, Basel, Switzerland. The remaining authors disclose no conflicts.
Funding This work was supported in part by a research grant from Novartis Pharma AG, Basel, Switzerland.
Author names in bold designate shared co-first authors.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2014.10.004.
Supplementary Materials and Methods
Dose-Dependent Silencing of Cellular Cyclophilin Expression
The siRNA oligonucleotides for CypA, CypB, CypC, and CypD (M-004979-01-0005; M-004606-00-0005; M-008819-00-0005; M-009708-00-0005; siGENOME SMART pool; Thermo Scientific Dharmacon) were prepared in Opti-MEM medium at the following concentrations: 300, 600, 1000, and 1500 nmol/L. Lipofectamine was diluted in Opti-MEM medium at a ratio of 1:50 for 5 minutes. Each siRNA concentration was mixed with the Lipofectamine complex, gently mixed, and incubated for 30 minutes. HepG2215 cells seeded at a density of 1 × 105 cells per well were treated to this siRNA:Lipofectamine complex and incubated for 24, 48, 72, and 96 hours at 37°C. The cells also were transfected at these different time points with siGLO Green to assess the transfection efficiency. The cellular RNA was extracted from the cells, reverse-transcribed, and quantitated by real-time PCR as previously described.1 The primers used were commercially available (HsPPIA_1_SG; HsPPIB_1_SG; HsPPIC_1_SG; HsPPIF_1_SG; and β-actin as the house-keeping gene HB-ACTB_1_SG; Qiagen).
Dose-Dependent Inhibition of Secreted HBV DNA by Telbivudine in HepG2215 Cells
Telbivudine stock solutions of 2000× were made in 100% DMSO and diluted to prepare the following working concentrations: 0.04 μmol/L (9.6 × 10-3 μg/mL), 0.16 μmol/L (0.038 μg/mL), 0.65 μmol/L (0.157 μg/mL), 2.5 μmol/L (0.605 μg/mL), 10 μmol/L (2.42 μg/mL), and 20 μmol/L (4.84 μg/mL). HepG2215 cells were treated with these working solutions of telbivudine at BL, and at 24, 48, and 72 hours. The telbivudine-containing media were replaced every 24 hours. The supernatants were collected at every time point and the HBV DNA extracted from the supernatants was quantitated by real-time PCR.
Supplementary Figure 1.
The 3 in vitro HBV models used: stably transfected HepG2215 cells, transiently transfected HuH-7 cells, and HBV-infected HepaRG cells, with the time points of sample collection after treatment with cyclophilin inhibitors: alisporivir (ALV), or NIM (NIM811). During treatment with the compounds used (ALV, NIM, or telbivudine) for all cell lines, the culture medium including fresh compound was replaced every 24 hours. Cells and supernatants were collected at each time point as shown. HepaRG cells were infected with HBV derived from 5-day culture supernatant of AD38 cells, in the presence of 4% PEG8000. After 16 hours, the cells were washed thoroughly and the culture medium replaced with fresh William’s Medium E with HepaRG Maintenance Medium Supplement plus GlutaMAX, containing different concentrations of the test compounds ALV or NIM811. HepaRG cell viability, before adding the compounds and after 7-day culture, was checked with trypan blue exclusion and always exceeded 90%.
Supplementary Figure 2.
Dose-dependent inhibition of secreted HBV DNA by telbivudine (LdT) in 2215 cells. HepG2215 cells were treated at BL, and at 24, 48, and 72 hours with 0.04, 0.16, 0.65, 2.5, 10, and 20 μmol/L of LdT. The levels of secreted-virus (SV) HBV DNA were quantitated by real-time PCR. The mean values and the SD of 3 independent experiments are shown. *P values less than .05, **P values less than .01, and ***P values less than .001 for LdT-treated vs nontreated cells.
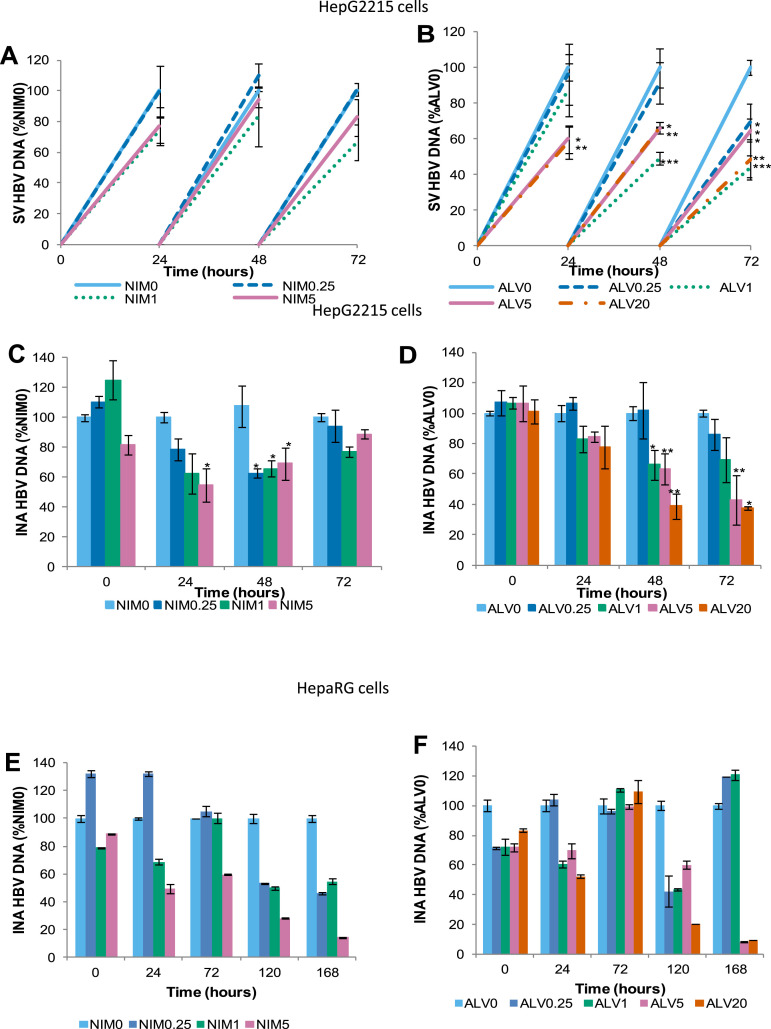
Supplementary Figure 3.
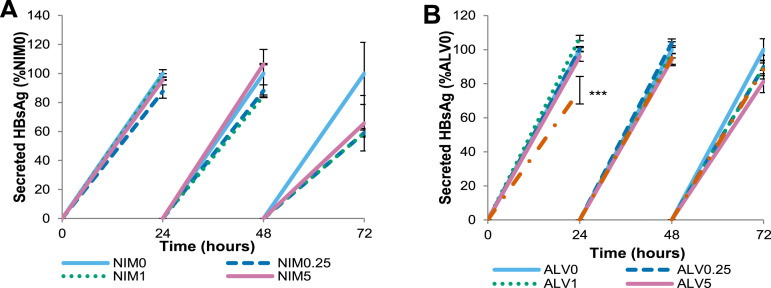
NIM811 also suppresses HBV replication and HBsAg secretion in HepG2215 cells and HepaRG. HepG2215 and HepaRG cells were treated with 0.25, 1, and 5 μg/mL of NIM811 and ALV over 72 hours and HBV DNA was quantitated by real-time PCR. Cells and supernatants were collected every 24 hours. HepG2215 cells: (A and B) secreted-virus (SV) HBV DNA and (C and D) intracellular nucleocapsid-associated (INA) HBV DNA. HepaRG: (E and F) INA HBV DNA. Mean values and the SD of 3 independent experiments are shown. *P values less than .05, **P values less than .01, and ***P values less than .001 for NIM811-treated vs nontreated cells. The ALV-treated figures were added for comparison purposes.
Supplementary Figure 4.
In HepG2215 cells, sHBsAg was measured by ELISA after treatment with (A) NIM811 and (B) ALV. Mean values and the SD of 2 independent experiments are shown. ***P values less than .001 for treated vs nontreated cells.
Supplementary Figure 5.
ALV cytotoxicity. HepG2215 cells were treated with 0.25, 1, 5, and 20 μg/mL of ALV alone, in combination with 10 μmol/L of telbivudine (LdT) or 10 μmol/L of LdT alone. Supernatants were collected every 24 hours. LDH was measured by absorbance in HepG2215 treated with (A) ALV alone, (B) ALV/LdT, and in HBV-transfected HUH-7 treated with (C) ALV alone.
Supplementary Figure 6.
Dose-dependent inhibition of cellular cyclophilin expression by cyclophilin-specific siRNA. Inhibition of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by GAPDH-specific siRNA and transfection efficiency (siGLO). HepG2215 cells were transfected with CypA, B, C, and D siRNA at 30, 60, 100, and 150 nmol/L for 24, 48, 72, and 96 hours. (A) CypA, (B) CypC, and (C) CypD expressions were measured in the cyclophilin siRNA-treated cells by real-time PCR. As controls. HepG2215 cells were transfected with GAPDH siRNA and siGLO transfection indicator for 48 hours. (D) GAPDH expression was measured in GAPDH and CypA siRNA-treated cells at BL (48 h after transfection), and at 24, 48, and 72 hours by real-time PCR. (E) Transfection efficiency was measured by flow cytometry at BL and 72 hours. ND, not done. The mean values and the SD of 2 independent experiments are shown. FITC, fluorescein isothiocyanate; SSC, side scatter.
Supplementary Figure 7.
Cyclophilin inhibition by siRNA. HepG2215 cells were transfected with siRNAs specific for CypA, CypC, or CypD, or scramble siRNA (negative control) at baseline. (A) The cells subsequently were cultured for up to 96 hours and the expression of the respective cyclophilin mRNA was quantitated by real-time PCR in cell extracts collected every 24 hours, relative to the expression in the controls: cells transfected with scramble siRNA. Mean values and the SD of 3 independent experiments are shown. (B) In the same experiment, the knock-down effect of siRNA at a protein level was assessed by quantitation of CYPA in the HepG2215 cells by ELISA. (C) The specificity of siRNA in reducing the CYPA expression in HepG2215 cells was tested by Western blotting to detect CYPA, CYPB, and CYPD. (D) Western blot of HuH-7 cells, transfected with CypA short hairpin RNA (shRNA), confirms the marked reduction of CYPA in these cells. *P values less than .05, and **P values less than .01 for Cyp siRNA-treated vs scramble siRNA-treated cells.
References
- 1.Wang P., Heitman J. The cyclophilins. Genome Biol. 2005;6:226. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin K., Gallay P. Curing a viral infection by targeting the host: the example of cyclophilin inhibitors. Antiviral Res. 2013;99:68–77. doi: 10.1016/j.antiviral.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaither L.A., Borawski J., Anderson L.J. Multiple cyclophilins involved in different cellular pathways mediate HCV replication. Virology. 2010;397:43–55. doi: 10.1016/j.virol.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 4.Dorner M., Horwitz J.A., Donovan B.M. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro A.P., Carvalho T.M., Moussatche N. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J Virol. 2003;77:9052–9068. doi: 10.1128/JVI.77.16.9052-9068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou D., Mei Q., Li J. Cyclophilin A and viral infections. Biochem Biophys Res Commun. 2012;424:647–650. doi: 10.1016/j.bbrc.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y.J., Wu H.H., Weng C.H. Cyclophilin A and nuclear factor of activated T cells are essential in cyclosporine-mediated suppression of polyomavirus BK replication. Am J Transplant. 2012;12:2348–2362. doi: 10.1111/j.1600-6143.2012.04116.x. [DOI] [PubMed] [Google Scholar]
- 8.Pfefferle S., Schopf J., Kogl M. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallay P.A., Lin K. Profile of alisporivir and its potential in the treatment of hepatitis C. Drug Des Devel Ther. 2013;7:105–115. doi: 10.2147/DDDT.S30946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coelmont L., Kaptein S., Paeshuyse J. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrob Agents Chemother. 2009;53:967–976. doi: 10.1128/AAC.00939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterji U., Bobardt M., Selvarajah S. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J Biol Chem. 2009;284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaul A., Stauffer S., Berger C. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson L.J., Lin K., Compton T. Inhibition of cyclophilins alters lipid trafficking and blocks hepatitis C virus secretion. Virol J. 2011;8:329. doi: 10.1186/1743-422X-8-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips S., Chokshi S., Riva A. CD8(+) T cell control of hepatitis B virus replication: direct comparison between cytolytic and noncytolytic functions. J Immunol. 2010;184:287–295. doi: 10.4049/jimmunol.0902761. [DOI] [PubMed] [Google Scholar]
- 15.Rang A., Gunther S., Will H. Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J Hepatol. 1999;31:791–799. doi: 10.1016/s0168-8278(99)80279-7. [DOI] [PubMed] [Google Scholar]
- 16.Hantz O., Parent R., Durantel D. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J Gen Virol. 2009;90:127–135. doi: 10.1099/vir.0.004861-0. [DOI] [PubMed] [Google Scholar]
- 17.Seifer M., Patty A., Serra I. Telbivudine, a nucleoside analog inhibitor of HBV polymerase, has a different in vitro cross-resistance profile than the nucleotide analog inhibitors adefovir and tenofovir. Antiviral Res. 2009;81:147–155. doi: 10.1016/j.antiviral.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Proto S., Taylor J.A., Chokshi S. APOBEC and iNOS are not the main intracellular effectors of IFN-gamma-mediated inactivation of hepatitis B virus replication. Antiviral Res. 2008;78:260–267. doi: 10.1016/j.antiviral.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Southern E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular cloning: a laboratory Manual. [Google Scholar]
- 21.Flisiak R., Horban A., Gallay P. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 22.McHugh M.L. Multiple comparison analysis testing in ANOVA. Biochem Med (Zagreb) 2011;21:203–209. doi: 10.11613/bm.2011.029. [DOI] [PubMed] [Google Scholar]
- 23.Dandri M., Locarnini S. New insight in the pathobiology of hepatitis B virus infection. Gut. 2012;61(Suppl 1):i6–i17. doi: 10.1136/gutjnl-2012-302056. [DOI] [PubMed] [Google Scholar]
- 24.Tian X., Zhao C., Zhu H. Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: possible link to pathogenesis of HBV infection. J Virol. 2010;84:3373–3381. doi: 10.1128/JVI.02555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouchard M.J., Wang L.H., Schneider R.J. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 26.Lucifora J., Arzberger S., Durantel D. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol. 2011;55:996–1003. doi: 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Clippinger A.J., Bouchard M.J. Hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modulates mitochondrial membrane potential. J Virol. 2008;82:6798–6811. doi: 10.1128/JVI.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchard M.J., Puro R.J., Wang L. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J Virol. 2003;77:7713–7719. doi: 10.1128/JVI.77.14.7713-7719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni Y., Lempp F.A., Mehrle S. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Yan H., Zhong G., Xu G. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nkongolo S., Ni Y., Lempp F.A. Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol. 2014;60:723–731. doi: 10.1016/j.jhep.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Watashi K., Sluder A., Daito T. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP) Hepatology. 2014;59:1726–1737. doi: 10.1002/hep.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liaw Y.F., Leung N., Kao J.H. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2008 update. Hepatol Int. 2008;2:263–283. doi: 10.1007/s12072-008-9080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.EASL clinical practice guidelines: management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Lok A.S., McMahon B.J. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 36.Wursthorn K., Jung M., Riva A. Kinetics of hepatitis B surface antigen decline during 3 years of telbivudine treatment in hepatitis B e antigen-positive patients. Hepatology. 2010;52:1611–1620. doi: 10.1002/hep.23905. [DOI] [PubMed] [Google Scholar]
- 37.Scaglione S.J., Lok A.S. Effectiveness of hepatitis B treatment in clinical practice. Gastroenterology. 2012;142:1360–1368. doi: 10.1053/j.gastro.2012.01.044. [DOI] [PubMed] [Google Scholar]
- 38.Griffel L., Bao W., Orsenigo R. Interferon (IFN)-free alisporivir has a better overall safety profile compared to IFN-containing treatment: a pooled analysis of the ALV development program. J Hepatol. 2013;58(Suppl 1):S336–S337. [Google Scholar]
Supplementary Reference
- 1.Evans A., Riva A., Cooksley H. Programmed death 1 expression during antiviral treatment of chronic hepatitis B: impact of hepatitis B e-antigen seroconversion. Hepatology. 2008;48:759–769. doi: 10.1002/hep.22419. [DOI] [PubMed] [Google Scholar]