Abstract
Detection of invading pathogens by pattern recognition receptors (PRRs) is crucial for the activation of the innate immune response. These sensors signal through intertwining signaling cascades which result in the expression of pro-inflammatory cytokines and type I interferons. Conjugation, or binding, of ubiquitin and ubiquitin-like modifiers (UBLs) to a plethora of immune signaling molecules forms a common theme in innate immune regulation. Numerous E3 ligases and deubiquitylating enzymes (DUBs) actively modify signaling components in order to achieve a balanced activation of the innate immune system. This review will discuss how this balance is achieved and which questions remain regarding innate immune regulation by ubiquitin and UBLs.
Keywords: Ubiquitin, E3 ligase, Innate immunity, DUB, Infection
The innate immune response is the first line of defense against invading pathogens and is a crucial part of the immune system. It bridges the time between infection and the delayed activation of the adaptive immune response (Fig. 1A and B). Moreover, cells of the innate immune response, such as monocytes, dendritic cells and NK-cells assist in the proper activation of the adaptive immune response. In order to activate the immune system, recognition of pathogens is required, followed by the secretion of cytokines that induce an antiviral state in the area and recruit innate immune cells. Some of the most critical cytokines that control viral infections are type I and type III interferons, both of which signal through different receptors, yet activate the same downstream signaling partners. Type I interferon can be produced by most cell types, whereas type III interferon production is predominantly restricted to epithelial cells in the intestine [1]. The importance of interferons is underpinned by the numerous studies demonstrating that animals lacking the receptors for these cytokines are hyper-susceptible to infection with numerous different microbes and succumb to infection before mounting an effective adaptive response [2].
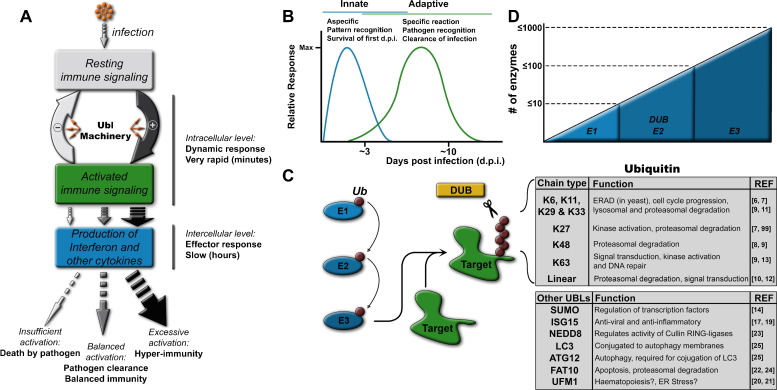
Fig. 1.
Overview of innate immunity and ubiquitylation. (A) Infection triggers the activation of the innate immune signaling. Insufficient activation leads to uncontrolled infection, but hyper-immunity is detrimental to the host. A balanced activation of the innate immune signaling, at least partly regulated by UBLs, is required for pathogen clearance and host survival. (B) The innate immune response is a swift response, which is required for control of the initial infection and activation of the adaptive immune response. (C and D) Conjugation of UBLs involves sequential activity of three classes of enzymes, E1-activating enzymes, E2-conjugases and E3-ligases. DUBs remove conjugated UBLs from substrates. The functions of different ubiquitin chain linkages are listed with corresponding references, as well as the general functions of the other UBLs.
Activation of the innate immune response relies on the detection of incoming pathogens by cellular sensors such as Toll-like- and RIG-I-like receptors (TLRs and RLRs respectively). These sensors, through various signaling cascades, activate the pro-inflammatory transcription factors AP-1, NF-κB and/or one or more members of the interferon-regulatory factor (IRF) family. This ultimately results in the release of pro-inflammatory cytokines and interferons (IFNs). These signaling cascades are divergent and intertwine at several steps. Regulation of innate immune signaling relies on post-translational modifications such as conjugation of ubiquitin and UBLs. This topic has recently been reviewed in [3], [4], [5], however the focus of this review will be on how innate immune signaling is more a dynamic response instead of a sequential signaling pathway and that signaling through the innate immune pathways requires an equilibrium of ubiquitin and UBL conjugation and deconjugation.
1. The ubiquitin system
Ubiquitin is a small 8-kDa protein of 76 amino acids, which is conjugated by covalent attachment of its C-terminal glycine residue to lysine side chains or the N-terminus of substrate proteins. Differently linked ubiquitin chains can be formed, free or on protein substrates, by ubiquitylation of one of the internal lysines in ubiquitin itself (K6, K11, K27, K29, K33, K48, or K63) or on the N-terminal methionine of ubiquitin (M1).
The significance of each of the different ubiquitin chain topologies is not yet fully understood, although several common themes have been established so far [6], [7]. For example, substrates modified with a chain of at least four K48-linked ubiquitin units are selectively degraded by the 26S proteasome, which is an integral part of protein turnover and cellular homeostasis [8], [9]. Although most chain types have since been implicated in proteasomal and/or lysosomal degradation to some extent, K48 linkages are believed to be the predominant proteasomal degradation signal in the cell [10], [11]. In contrast, K63- and linear chains are often implicated in the regulation of signaling pathways and in the activation of signaling kinases (Fig. 1C) [12], [13].
1.1. Ubiquitin-like modifiers
Since the discovery of ubiquitin, more ubiquitin-like modifiers have been identified, among which are ISG15 and SUMO [9], [14], [15], each showing limited sequence homology with ubiquitin, yet sharing conserved structural features, such as the ubiquitin fold and one or two C-terminal glycines for conjugation (Fig. 1C) [16]. SUMOylation requires a consensus sequence in substrates (ψKXE), where ψ is a hydrophobic residue and X is any amino acid. SUMOylation of transcription factors generally results in transcriptional repression, but it can also affect nuclear import and export of proteins [14]. ISG15 is a potent antiviral molecule, which is produced after interferon signaling [17]. It is conjugated co-translationally to newly synthesized proteins to inhibit viral replication, however it also likely has anti-inflammatory effects [18], [19]. General functions of all UBLs are listed in Fig. 1C [20], [21], [22], [23], [24], [25].
1.2. Conjugation
Conjugation of ubiquitin and UBLs is a multi-step process involving a cascade of E1-activating, E2-conjugating and E3 ligase enzymes [9], [26], [27]. There is an astounding degree of diversity in ubiquitylation enzymes, and mammalian genomes have been estimated to contain ≤10 E1 enzymes, ≤100 E2 enzymes and ≤1000 different E3 ligases (Fig. 1D) [28].
Not only conjugation of ubiquitin and UBLs regulates innate immune responses, deconjugation is also a critical component of the system. Approximately 100 ubiquitin and UBL deconjugating enzymes (DUBs) have been identified in the human genome [29]. DUBs are required for generating the mature forms of ubiquitin and UBLs by cleavage of precursor proteins, thereby exposing the C-terminal glycine residue required for conjugation. Additionally, DUBs function in the reversion of regulatory ubiquitin and UBL conjugation (Fig. 1C). They are often specific for a certain UBL, the ubiquitin chain topology and the substrate [29]. DUBs are therefore an integral part of the regulation of the signaling cascades in which ubiquitin conjugation plays a role, which includes several innate immune response pathways. Several classes of DUBs have been recognized [30], and in the case of innate immune regulation, the ovarian tumor domain-containing (OTU) DUBs seem to play a major role. Among them are DUBA, OTUB1, OTUB2 and A20 [31], [32], [33]. Another DUB that is critical in the regulation of the innate immune response is CYLD, which belongs to the USP DUB class [34]. Since so many enzymes are involved in the conjugation and deconjugation, post-translational modification by ubiquitin and UBLs allows for precise and dynamic adjustment of signaling pathways and their ultimate effect in the (infected) organism.
2. PRR signaling pathways
Recognition of incoming pathogens by PRRs is essential for the innate immune response [35]. The best-studied PRRs include TLRs at the plasma membrane and endosomes, as well as Nod-Like Receptors (NLRs), RLRs and DNA sensors in the cytosol (Fig. 2 ). Upon recognition of pathogen associated molecular patterns (PAMPs) by specific PRRs, one or more of several signaling cascades are initiated. PAMPs, such as LPS, 5′-triphosphate ssRNA and dsRNA are microbe-specific, and can thus be distinguished from cellular ‘self’ components by PRRs.
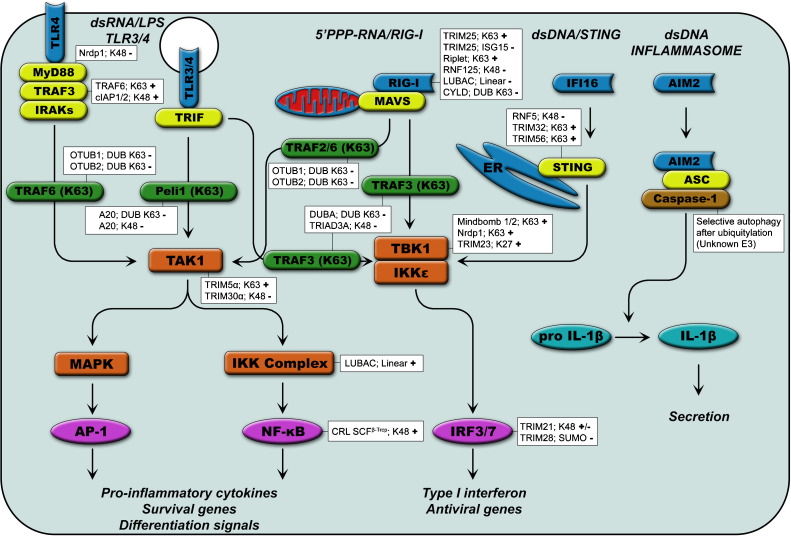
Fig. 2.
PRR signaling pathways and their regulation by ubiquitin. PRR signaling pathways are heavily regulated by UBLs. Basic components of the pathways are shown here, more detailed sub-pathways are depicted in Fig. 3. PRRs are shown in blue, adaptor proteins in yellow, the most important E3 ligases in green, kinases in orange and transcription factors in purple. Other E3 ligases and DUBs that influence PRR signaling are listed along with their chain specificity or UBL and effect (+ or −) on the outcome of the signaling cascade.
NLRs are cytosolic leucine rich repeat (LRR) proteins which mainly recognize bacterial antigens and signal either to activate the AP-1 and NF-κB transcription factors or to activate Caspase-1 and the inflammasome [36]. TLRs are transmembrane PRRs that recognize PAMPs from viruses, bacteria and/or fungi and are expressed mainly by cells of hematopoietic origin [37]. Upon ligand binding, all TLRs signal through the adaptor proteins MyD88 and/or TRIF to activate NF-κB, AP-1 and, in the case of endosomal TLRs, also IRF3/7 [38], [39].
RLRs are ubiquitously expressed cytosolic RNA helicases that recognize viral RNA and signal through the mitochondrial adaptor MAVS [40], [41]. Members of the RLR family include RIG-I, which recognizes 5′-triphosphate RNA and dsRNA [42], and MDA5, which can be activated by (viral) ssRNA cleaved by RNase L [43], as well as (synthetic) dsRNA [44].
DNA sensors, such as IFI16 and AIM2 recognize dsDNA in the cytosol [45], [46]. IFI16 signals through the ER signaling adaptor STING to activate IRFs as well as NF-κB [47]. AIM2 activation does not result in the activation of transcription factors, but instead activates the inflammasome to produce mature IL-1β.
The next step in most of the TLR and RLR pathways that follow the activation of PRRs and their adaptors is the recruitment and activation of the TAK1 kinase complex. This complex acts as a Mitogen Activated Protein Kinase Kinase Kinase (MAPKKK) to activate the AP-1 transcription factor. It also activates the classical IKK complex, which subsequently phosphorylates the inhibitor of the NF-κB transcription factor (IκBα), ultimately resulting in its degradation and release of active NF-κB [38], [48].
In addition to the activation of the TAK1 complex, both the intracellular TLR and RLR signaling pathways also activate the IKK-related kinases TBK1 and IKKɛ. These kinases subsequently phosphorylate members of the IRF transcription factor family, which mediate type I interferon production [49], [50]. Interferon signals neighboring cells through the interferon α/β receptor (IFNAR) to induce expression of interferon stimulated genes (ISGs), many of which have potent antiviral activity [51].
2.1. Innate immunity in diseases
Although the ability to effectively mount an immune response during infection is critical for survival, prolonged or excessive-activation of the innate immune system can have detrimental effects. For example, highly pathogenic influenza virus strains, such as the 1918 H1N1 ‘Spanish flu’ virus and several highly pathogenic H5N1 viruses, are potent activators of pro-inflammatory genes. The resulting ‘cytokine storm’ during infection is thought to contribute to the high mortality associated with these viruses [52]. In addition, regulation of the innate immune system is important to prevent the development of auto-immunity. Several immune disorders, such as Systemic Lupus Erythematosus (SLE), Crohn's Disease, Blau syndrome, type I diabetes, multiple sclerosis and rheumatoid arthritis, have been linked to dysregulation of the innate immune system [53], [54]. Only for a few of these diseases some aspects of the underlying immune dysregulation have been identified. For example, a link between mutations in the cytosolic NOD2 sensor and susceptibility for developing Crohn's disease and Blau syndrome has been found [53], [55]. Also, increased levels of IFN-α have been detected in SLE patients, which correlated with severity of the disease [56]. Onset of type I diabetes could be delayed in non-obese diabetic (NOD) mice, which spontaneously develop type I diabetes, by blocking the IFNAR, suggesting that IFN signaling plays a role in determining type I diabetes onset and progression [57]. In these mice, plasmacytoid dendritic cells in the pancreatic draining lymph nodes produced higher levels of IFN-α compared to control mice, which led to activation of CD4 T Cells and subsequent development of disease [57]. Determining what activates the type I interferon system and why it has adverse effects involved in these diseases will help in the development of new, more specific treatments with less side-effects than current immunosuppressive drugs used to treat some of these diseases.
To facilitate mounting of an effective immune response during infection, but also subsequent restoration to a resting state once the infection is cleared, the type I IFN system should be dynamically regulated. This dynamic nature ensures a balance between the extremes of insufficient activation (resulting in death or persistent infection by the pathogen), on the one hand, and over-activation, resulting in hyper-inflammation and tissue damage on the other hand (Fig. 1A).
Cross-talk between different cells and cell types is one of the main ways to balance pro- and anti-inflammatory signals at the inter-cellular level. The response time resulting from the time it takes to produce and secrete the cytokines is in the range of minutes to hours. However, at the intra-cellular level, post-translational modifications of signaling components ensure even more rapid response times in the range of seconds to minutes. The sum of these signals ultimately determines the cytokine repertoire and functional fate of each particular cell during the immune response.
Phosphorylation and dephosphorylation form a main regulatory mechanism by which innate immune signaling cascades are controlled. In addition, many studies in recent years have established a very significant role for post-translational, covalent modification with ubiquitin and UBL modifiers in the regulation of innate immunity.
2.2. Regulation of RIG-I by ubiquitin-mediated signals
Activation of the cytosolic sensor RIG-I by microbial RNA ultimately results in synthesis of the type I IFN and NF-κB-regulated pro-inflammatory pathways. RIG-I is comprised of two CARD domains at its N-terminus, which are responsible for downstream signaling to the critical mitochondrial adaptor molecule MAVS. Downstream of the CARD domains resides an RNA helicase domain for unwinding of the RNA, followed by a C-terminal regulatory domain (Fig. 3 A).
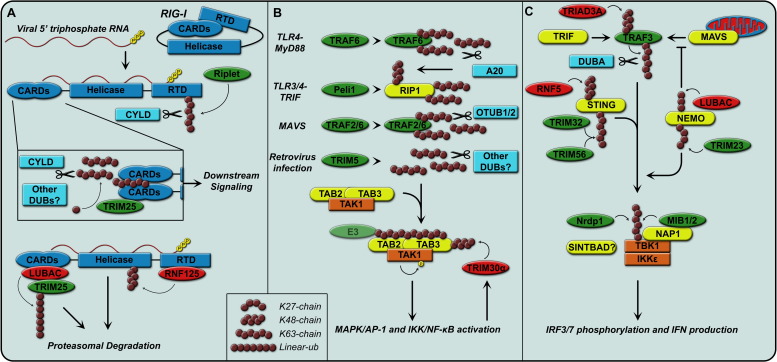
Fig. 3.
Ubiquitin-mediated regulation of RIG-I, TAK1 and TBK1/IKKɛ. (A) Viral 5′-triphosphate RNA is recognized by RIG-I, which leads to a conformational change and then multimerization for downstream signaling (top part). Activation of RIG-I signaling is regulated by several distinct ubiquitylation mechanisms. (B) The TAK1 complex can be activated by K63 chains (bound or free) from a variety of inputs as listed. Once the TAB2 and TAB3 subunits bind K63 chains, TAK1 auto-phosphorylates and initiates downstream signaling. (A–C) Activating E3 ligases are shown in green, deactivating in red and DUBs in light blue. For more detailed descriptions the reader is referred to the text.
Four different E3 ligases have been described to bind RIG-I and regulate its activation: Riplet, TRIM25, RNF125 and LUBAC. Riplet and TRIM25 promote RIG-I signaling, whereas RNF125 and LUBAC negatively affect RIG-I function. The regulatory domain can recognize the 5′-triphosphate on viral RNA, after which this domain is ubiquitylated by Riplet with a K63 chain (Fig. 2, Fig. 3) [41], [58]. This leads to a conformational change in the RIG-I molecule followed by exposure of the N-terminal CARD domains. TRIM25 subsequently binds to the first CARD domain via its SPRY domain and is required for downstream signaling [59]. Direct K63 ubiquitin linkage by TRIM25 onto K172, located in the second CARD domain of RIG-I has been reported to be required for RIG-I activation [60].
However, a recent study using a completely in vitro reconstituted IFN induction pathway, demonstrated that K172 is a critical structural residue, required for the CARD domains to function as a ubiquitin-binding domain (UBD). Specific binding of the CARDs to short unanchored K63 chains was reported to be critical for RIG-I multimerization into a functional hetero-tetrameric complex of four RIG-I molecules and four unanchored K63 chains (Fig. 3A) [61], [62]. Consistently, when cell lysates were treated with the DUB Isopeptidase T, which only cleaves free ubiquitin chains by recognizing the free C-terminus, the ability to activate RIG-I signaling in vitro was completely lost [61], [63].
Thus, K172 of RIG-I was found to be structurally important for multimerization of RIG-I molecules with free ubiquitin-chains in vitro, while the significance of covalent ubiquitylation of K172 for downstream signaling was determined in tissue culture systems. One possibility to explain this apparent discrepancy could be that ubiquitin chains covalently attached to K172 facilitate the binding or synthesis of unanchored ubiquitin chains, and therefore both observations may not be necessarily mutually exclusive to describe the mechanism of RIG-I regulation by ubiquitin. Mutation of K172 decreased RIG-I ubiquitylation [60], which is seemingly more consistent with a model of covalent ubiquitin conjugation to this lysine in RIG-I. Apart from the suggested role of TRIM25 in the ubiquitin conjugation to K172 [60] however, also Riplet was reported to conjugate K63-chains to K154, K164 and K172 in the CARD domains of RIG-I, which was required for downstream signaling [64], and which could therefore also explain the loss of ubiquitylation after K172 mutation.
The exact role of Riplet-mediated modification of the CARD domains remains uncertain, because it was not required for the in vitro RIG-I activation [61]. One possibility is that, due to the nature of in vitro experiments, not all factors are present and Riplet-mediated conjugation might serve to alleviate inhibition by (non ubiquitin-mediated) negative regulators such as NLRX1 and gC1qR [65], [66]. Moreover, knockout mouse models have shown that both TRIM25 and Riplet are both pivotal in the innate immune defense against RNA viruses and loss of either of these E3 ligases abolishes IFN production after viral infection [60], [67].
MDA5 functions similarly to RIG-I, although it does not have a C-terminal regulatory domain. After substrate recognition it also binds to short unanchored K-63 chains to assemble a hetero-tetrameric complex and initiate downstream signaling [62].
Negative regulation of ubiquitin-mediated RLR signaling is accomplished by RNF125, LUBAC, as well as the DUB CYLD. The first two molecules function as E3 ligases, whereas the latter specifically cleaves K63 linked ubiquitin chains from RIG-I and is also able to disassemble free ubiquitin chains (Fig. 3A) [13], [34].
RNF125 expression is induced by RIG-I signaling after which it modifies RIG-I and MDA5 with K48 linked ubiquitin chains, targeting them for proteasomal degradation [68]. LUBAC inhibits RIG-I function by two distinct mechanisms. The HOIL-1 subunit of this E3 ligase complex binds to the CARD domains to limit activation, and the HOIP subunit binds to and modifies TRIM25 with a linear ubiquitin chain, which results in proteasomal degradation of TRIM25 (Fig. 3A) [69].
Another proposed mechanism for negative TRIM25 regulation is mediated by auto-ISGylation of TRIM25 with the ubiquitin like modifier ISG15, potentially though competition with ubiquitin [70]. Taken together, it seems clear RIG-I signaling is regulated by multiple ubiquitin-dependent mechanisms, however how exactly these are related and temporally and spatially determine the activity of RIG-I in vivo remains to be elucidated.
2.3. TAK1 signal transduction
The TAK1 complex is an essential signaling complex in many innate immune pathways and is activated by RIG-I/MAVS and TLRs. It consists of the TAB2 and TAB3 accessory proteins and the TAK1 kinase. The TAK1 complex acts upstream of NF-κB and AP-1 (Fig. 2, Fig. 3) [71], [72]. It is not only involved in PRR signaling, but is a component in many pathways, such as TNFα, T-Cell Receptor, and IL-1 signaling. The TAK1 kinase subunit can be activated by auto-phosphorylation, which requires binding of TAB2 and TAB3 to K63-linked ubiquitin chains (Fig. 3B) [13]. Interestingly, free K63 chains are sufficient for activation of TAK1 in vitro, however, K63 chains conjugated to a substrate are also potent activators of the TAK1 complex [13].
Depending on the signaling pathway involved, the K63 chains can originate from different sources (Fig. 2, Fig. 3). For example in the case of MyD88-mediated TLR4 signaling, the E3-ligase TRAF6 synthesizes the K63 chains either free or on itself. In the case of TRIF-mediated TLR signaling the E3-ligase Peli1 attaches K63 chains to RIP1. Additionally, there is evidence that TRAF2 and/or TRAF6 mediate MAVS signaling to the TAK1 complex through K63-linked Ub chains [73], [74], [75], [76], [77], [78], [79].
TRIM5 is a potent cytosolic restriction factor for HIV. It restricts HIV by binding the HIV capsid, thereby interfering with uncoating of the viral genome and subsequent reverse transcription. Recent work suggests that TRIM5 may act as a PRR for the retrovirus capsid lattice, which in turn results in NF-κB and AP-1 activation. The authors demonstrated that upon HIV-1 infection, TRIM5 promoted unanchored K63 chain formation in cooperation with the Uev1A-Ubc13 E2 enzyme complex in vitro [80]. Subsequently, these synthesized Ub chains bound and activated TAK1 (Fig. 3B).
Even though the TAK1 complex activates two distinct signaling cascades, mediated by either MAPK/AP-1 complex or IKK complex/NF-κB, their activation is spatially and temporally separated (Fig. 2) [48]. In the case of MyD88-mediated TLR4 signaling, activation of the IKK complex by TAK1 occurs at the plasma membrane whereas MAPK signaling is initiated later on in the cytosol after the TAK1 complex has been released from the membrane-bound signaling complex. TRAF6 is anchored to the TLR signaling complex at the plasma membrane by IRAK proteins and by TRAF3, which in that case has a role as a scaffolding protein distinct from its E3 ligase functions (Fig. 2) [81]. TRAF3, and possibly the IRAK proteins as well, are then modified with a K48 chain. This targets them for degradation by the E3-ligases cIAP1 and cIAP2, which are themselves activated by TRAF6-mediated attachment of a K63 chain (Fig. 2).
The many signals that activate the TAK1 complex can also be terminated in several different ways. The signals transduced by TRAF6 and RIP1 can be discontinued by the OTU DUB A20, which is induced by TAK1 signaling and specifically de-conjugates K63 chains (Fig. 2, Fig. 3) [33]. A20 also possesses E3-ligase activity, which specifically modifies RIP1 with a K48 chain targeting it for proteasomal degradation, providing an interesting dual function of both E3-ligase and DUB to serve two complementary roles in negatively regulating TAK1 activation [82]. Furthermore, the DUBs OTUB1 and OTUB2 also negatively regulate TAK1 signaling by removing K63 chains from TRAF6 after viral infection (Fig. 2, Fig. 3) [31]. After TAK1-mediated IKK and NF-κB activation, expression of TRIM30α is induced. This causes TAB2 to be degraded in a ubiquitin-dependent manner and TAK1 signaling to be terminated (Fig. 3B) [83].
Since free K63 chains have been found to regulate at least two distinct signaling components, one possible issue could be the signal specificity. Since free chains do not “encode” any other signal than the linkage specificity, the question arises: what makes chains from TRIM25 activate RIG-I and chains from TRIM5a activate the TAK1 complex? One possibility is that these free chains are very volatile signals that can rapidly activate a signaling complex and are subsequently degraded by the DUBs present in the cytosol. Since in vitro most DUBs can efficiently break down purified free ubiquitin chains (depending on chain topology in some cases) it is likely that free K63 chains are degraded rapidly after synthesis and present a very dynamic mechanism for signaling complex activation [84]. Since DUBs have been reported as integral components of large ubiquitylation complexes, it is also not unconceivable that ubiquitin chains function within the complexes they were synthesized in and do not ‘float’ around free in the cytosol where they could engage unintended targets. Along those lines, one would predict that the number and activity of the conjugating and de-conjugating enzymes within those complexes establish an equilibrium between continuous synthesis and degradation of ubiquitin chains. Such an equilibrium could be very transiently and rapidly shifted ‘left’ or ‘right’ by modifying the number and the activity of the different enzymes in the protein complex or disengaging the intended target of the ubiquitin chains.
2.4. The classical IKK complex
The final signaling component before the activation of NF-κB is the IKK complex, which is activated by TAK1 (Fig. 2). The ubiquitin-binding protein NEMO is part of the classical IKK complex together with IKKα and IKKβ. The latter is required for phosphorylation of IκBα, its K48 ubiquitylation by the CRL (Cullin RING Ligase) complex SCFβ-TrcpI, and its subsequent degradation and release of active NF-κB [85]. NEMO possesses an UBAN (Ubiquitin Binding in ABIN and NEMO) domain that preferentially binds to linear polyubiquitin chains, but can also interact with K63 and K48 chains [86], [87]. Furthermore, NEMO itself can also be modified by LUBAC with a linear ubiquitin chain during TNFα signaling, which is required for NF-κB activation [12].
Recruitment of the IKK complex to the TLR or TNFR signaling complexes is thought to occur by interaction of NEMO with K63 ubiquitin chains [87]. Since IKK is activated by phosphorylation of the IKKα/β subunits and TAK1 is capable of phosphorylating those subunits in vitro (a catalytic mutant was unable to do so), a model was proposed in which TAK1 phosphorylates IKKβ [71]. In contrast, activation and auto-phosphorylation of the IKK complex by binding of free ubiquitin chains to NEMO has been proposed based on in vitro studies [13]. Contrary to the requirement of K63 chains for TAK1 activation, the activation of IKK was independent of K48 or K63 linked ubiquitin. Moreover, the E2-enzyme involved was identified as UbcH5 (instead of Uev1A-Ubc13), which forms chains of various linkages. This suggested that distinct ubiquitin chain types could be involved. Moreover, due to the ability of NEMO to both bind and be modified by linear ubiquitin chains, it is possible that chains bound to one NEMO subunit are recognized by another and then oligomerize due to this interaction. This could then lead to auto-phosphorylation of the bound IKKα/β subunits similarly to how TAK1 can activate itself in vitro [13]. In conclusion, the function of the linear ubiquitin chains in the classical IKK complex activation has not been fully clarified yet.
Most studies involving NEMO and the IKK complex have been carried out in the “canonical” NF-κB signaling pathway downstream of the TNF receptor. One caveat to this approach is that it obscures the role of NEMO and the IKK complex in other parts of the signaling cascades controlling innate immunity. Most knowledge about the activation of the IKK complex is inferred from the canonical pathway, yet the exact functions of NEMO in each different pathway remain unclear. It is not unlikely that as a results of multiple signaling cascades culminating in the activation of the IKK complex (TNFα, TLR signaling, CD40, IL-1, etc.), each pathway differs slightly in its method for activating the IKK complex. Due to the promiscuity of the interaction of NEMO with different ubiquitin chain linkages, as well as the various modifications of the protein itself, different ubiquitin signals could provide an interesting angle for differential and dynamic IKK complex activation.
2.5. TBK1/IKKɛ dependent IRF activation
MAVS and TRIF are two key signaling molecules critical for the activation of IRFs and thus IFN-mediated antiviral immunity (Fig. 3C). TRAF3 is recruited to all MAVS- and TRIF-based signaling complexes to mediate the activation of the IKK-related kinases IKKɛ and TBK1. TRAF3 self-activates by covalent auto-polyubiquitylation with K63 linked ubiquitin chains [81], [88]. TRAF3 activation can in turn be negated by DUBA, a DUB specifically binding TRAF3 and deconjugating its K63 chains [32]. Furthermore, the E3-ligase TRIAD3A can modify TRAF3 with a K48 chain to target it for degradation as another means of down-regulating MAVS-mediated signaling (Fig. 3C) [89].
Signaling through the ER adaptor STING also results in activation of TBK1. STING, which requires modification with K63 chains by the E3 ligases TRIM56 and TRIM32 for signaling [90], [91], is able to signal through TBK1 and IKKɛ to activate IRFs. After activation, STING is targeted for degradation by K48 chain modifications by the E3 ligase RNF5 [92].
The following steps in the pathways downstream of MAVS and STING are not entirely clear, but at least require the E2 enzyme Ubc5 and K63 polyubiquitin [93]. The IRF family members require phosphorylation by the IKK related kinases TBK1 and IKKɛ [49]. TBK1 and IKKɛ were discovered to function as kinases of IRFs with some redundancy. However, functions specific for either TBK1 or IKKɛ have been discovered since [49], [94]. For example, results from IKKɛ knock-out mice have suggested a predominant role in the activation of STAT1 following IFNAR signaling [94]. While TBK1 has been linked to selective autophagy of intracellular pathogens such as Salmonella [95]. How the functions of IKKɛ and TBK1 relate to each other and how their respective functions fit together remains to be elucidated. For downstream signaling, TBK1 also requires K63-ubiquitylation by mind bomb proteins after dsRNA signaling and by Nrdp1 after LPS signaling [96], [97]; yet the major activator for IKKɛ is still unknown (Fig. 3C). How these kinases are activated is also likely dependent on their assembly into a complex [97].
Several NEMO-like adaptor/scaffold proteins have been identified (TANK, NAP1 and SINTBAD) all of which can constitutively bind TBK1 and IKKɛ [98]. These adaptor proteins contain UBDs similar to NEMO and therefore regulation by ubiquitin chains is a feasible possibility. It is worth mentioning that their ability to directly interact with the kinases, distinguishes them from NEMO, which itself does not interact with TBK1 and IKKɛ. It is TANK that mediates interaction of NEMO with the IKK-related kinases. Moreover, TRIM23-mediated K27-linked ubiquitylation of NEMO is important in IRF activation, although the precise molecular mechanism behind this remains to be determined (Fig. 3C) [99], [100].
Recently, linearly ubiquitylated NEMO was reported to inhibit MAVS signaling by interfering with the interactions between MAVS and TRAF3/6 (Fig. 3C) [101]. LUBAC was critical for the NEMO–MAVS interaction and subsequent interference with TRAF3/6 binding. Taken together, LUBAC has so far been found to positively regulate NF-κB signaling (Fig. 2), but negatively regulate antiviral signaling (Fig. 3A and C). These findings indicate that LUBAC is involved in directing the innate immune response to a more pro-inflammatory instead of antiviral response. An E3 ligase that has the opposite effect is Nrdp1, which activates TBK1, but targets TLRs for degradation by K48-linked ubiquitylation [96]. This shifts the balance of the innate immune system more to an interferon instead of a pro-inflammatory response.
The IKK-related kinases and the classical IKK complex also influence each other. For example, TANK inhibits the classical IKK complex and subsequent NF-κB activation by the IKK-related kinases via its interaction with NEMO [102]. Interestingly, K63 chains formed by TRAF3 activate the IKK-related kinases, whereas TRAF2 and/or TRAF6 also form K63 chains but activate the TAK1 complex. These E3-ligases all interact with MAVS, yet how specificity is achieved by their synthesized K63 chains in TAK1 or IKK-related kinase activation remains to be determined. The exact mechanisms regulating TBK1 and IKKɛ activation and the interactions with their adaptor proteins will likely be the subject of extensive studies in the near future.
2.6. IRF3 and IRF7 regulation by ubiquitin and SUMO
The IKK-related kinases phosphorylate two members of the IRF transcription factor family: IRF3 and IRF7, which are required for the production of type I interferons (Fig. 2) [49]. IRF3 is ubiquitously expressed in most cell types whereas IRF7 is an ISG and is only expressed by leukocytes in the absence of infection [50]. Phosphorylated IRF3 and IRF7 can form homo- or heterodimers, each of which has different promoter specificity. IRF3 homodimers are created when there is a low supply of IRF7 (so in the absence of ISG expression) and initiate production of chemokines such as CXCL10 to attract innate immune cells. Conversely, IRF3/IRF7 dimers potently activate type I interferon production, but this can only occur in innate immune cells or cells already stimulated with type I interferon [50]. IRF7 dimers regulate the expression of a subset of ISGs as well as type I IFN by binding to specific ISREs to ensure proper induction of the antiviral state in the absence of IFNAR signaling [103].
TRIM21 has been found to regulate both IRF3 and IRF7. Binding of TRIM21 to IRF3 stabilized IRF3, whereas it targets IRF7 for proteasomal degradation by K48 ubiquitylation [104], [105]. Studies using TRIM21 knockout mice found that loss of TRIM21 increases NF-κB dependent cytokine expression in embryonic fibroblasts, yet no effects were observed in vivo [106]. The net result of TRIM21 activity is therefore still uncertain, and variations in cell types used or different isoforms of TRIM21 might explain its different functions. However, there are only very few examples of transcription factors being regulated by ubiquitylation like this. Another far more common mechanism affecting transcription factors, including the ones involved in innate immune signaling, is SUMOylation.
Attenuation of transcription factors is very important for regulation of the innate immune system in order to prevent harmful auto- and hyper immunity. Both IRF3 and IRF7 contain SUMO consensus sequences, which have been shown to be SUMOylated during viral infection, thereby providing a mechanism for post-activation attenuation by transcriptional repression [107]. TRIM28 was identified as the E3-ligase for IRF7 SUMOylation, yet a SUMO E3 ligase for IRF3 has hitherto not been identified [108]. Protein inhibitor of activated STATγ (PIASγ) was also found to inhibit the transcriptional activity of both NF-κB as well as transcription factors binding the ISRE (IRF7 and ISGF3) [109]. Members of the PIAS protein family are SUMO E3-ligases and are involved in the regulation of a variety of innate immunity transcription factors [110]. The PIAS proteins were identified as inhibitors of STAT proteins, possibly by prohibiting interaction of STAT with DNA or other co-factors through protein–protein interactions. However, the biological relevance of the SUMO E3-ligase function of the PIAS proteins remains unclear. Since the exact mechanism by which SUMO functions as a transcriptional repressor is not yet fully understood, more factors affecting SUMO-dependent regulation of IRF3 and IRF7 are likely to exist.
3. Pathogen evasion strategies targeting ubiquitin and UBL regulation
In order for pathogens to establish a productive infection and to facilitate productive replication, evasion of innate immune responses is pivotal. Different pathogens employ different strategies and many affect the innate immune response by tampering with the ubiquitin machinery regulating this system. For example, several unrelated virus families encode DUBs to modulate innate immune signaling, among which are arteriviruses, coronaviruses, nairoviruses, picornaviruses, adenoviruses and herpesviruses [111], [112], [113], [114], [115].
Some of the DUBs identified in these viruses resemble members of the OTU family of ubiquitin deconjugating enzymes. They are often capable of deconjugating different ubiquitin chain linkages as well as ISG15, whereas mammalian OTU domain-containing DUBs are ubiquitin chain specific [113]. Ectopic expression of these viral DUBs increases global ubiquitin and ISG15 deconjugation, however, it remains to be determined how the promiscuous activity of these enzymes is regulated during infection. Viral proteases of arteriviruses are able to inhibit RIG-I activation by deconjugating the K63 chains required for RIG-I activation [116], and more targets within the innate immune response cascades are likely to exist. Other examples of viral proteins interfering with the function of E3-ligases exist, such as Influenza A Virus NS1, which specifically binds TRIM25 thereby inhibiting RIG-I activation [117]. Some large DNA viruses, such as herpes- and poxviruses even encode their own E3 ligases. Herpesviral E3s target the cellular E3 ligases RNF8 and RNF168 for degradation, which has been proposed to facilitate reactivation from latency [118], [119]. Given that viral enzymes are often hard to recognize purely based on their sequence, more viral E3 ligases and DUBs likely still remain undiscovered.
4. Closing comments
In the case of a severe infection, survival can hinge on the balanced activation of the innate immune response. In order to achieve that balance, numerous E3-ligases and DUBs actively regulate the outcome of the innate immune response. The basic components and signals of the signaling pathways are well characterized, but how the different components interact is less clearly understood. Often signaling molecules are assembled into large complexes. However, the stoichiometry and localization of the different molecules in these complexes in time have remained relatively poorly studied thus far. The sophistication in complex composition is one of the current obstacles that the scientific community will need to overcome in both in vitro and cell based systems.
Because the post-translational modifications of signaling components are transient signals in most cases, monitoring changes in those signals during infection and/or auto-immunity over time will likely yield crucial information in understanding the dynamics of this system. To study these events, advances in mass spectrometry, such as multiplex MS and analysis of ubiquitylation levels and substrates in complex samples from relevant tissue culture or experimental animal models, will be very important [120], [121]. These tools could be used to discern whether patients with autoimmune diseases that are related to type I interferons have a different state of activation of the signaling pathways involved in innate immunity.
Most knowledge on the functions of free ubiquitin chains in the regulation of RIG-I and TAK1 has been deducted from in vitro reconstituted systems [13], [61]. These experiments allowed for precise control of the individual components in the signaling pathways and have been instrumental in unraveling the minimal signals for the pathways to function. Future work should focus to validate the findings of the in vitro systems in the relevant primary cell types and/or disease models. Discrepancies between in vitro experiments and experiments in cell culture, such as the mechanism of K172-mediated RIG-I activation, underline the importance of exploiting the strengths of individual model systems and validating findings in others [60], [61].
A vast array of enzymes is involved in ubiquitin (de)conjugation of proteins. It seems logical that the interpretation of a ubiquitin signal by UBDs, of which over 150 types divided over ∼20 classes have been discovered, would also be very specific [122]. However, the affinity of UBDs for ubiquitin chains is often relatively low, which raises the question how signaling complexes are assembled by ubiquitylated proteins. Either multivalent interactions of multiple UBDs with multiple conjugated ubiquitin molecules or enhancement of a weak existing interaction between two proteins can be considered. Alternatively, the ubiquitin–UBD interactions could be less specific and/or very transient in dynamic molecular structures where a relatively high local ubiquitin (-chain) concentration can be maintained. Under such circumstances, target specificity could be achieved by target availability in the complex, or by e.g. specific interactions between the target and one or more UBLs that would bring it close to the regulating ubiquitin groups.
An interesting possibility is that long ubiquitin chains themselves could form structural ‘meshes’ on/in which UBLs and their interacting targets could ‘dock’ for interaction with other complex components. In depth characterization of the localization and composition of signaling complexes is required to further our knowledge about the plethora of effects ubiquitin and UBLs exert on the innate immune system.
The importance of ubiquitylation in regulating cellular mechanisms is becoming more and more clear and the innate immune system is no exception. PRR signaling relies on the conjugation of K63 chains to activate key signaling components, which results in quick and dynamic activation of signaling cascades. Knowledge gained on the regulation of the innate immune system could very well be used to develop new vaccine adjuvants and new strategies to treat autoimmune diseases.
Biographies

Diede Oudshoorn received his M.Sc. degree from Leiden University after doing research projects in the Molecular Virology Department in the LUMC in Leiden and the Microbiology Department at Mount Sinai School of Medicine in New York. Diede is currently a Ph.D. student at the Department of Medical Microbiology in the LUMC with a focus on the interaction between replication structures of nidoviruses and innate immune signaling.

Gijs Versteeg has had a long-standing interest in investigating how viruses interact with their hosts and negate their innate immune responses. He received his Ph.D. from Leiden University after studying how coronaviruses regulate innate immunity. Currently, Gijs holds a post-doctoral training position in the Laboratory of Adolfo García-Sastre at Mount Sinai School of Medicine in New York. Here he has identified and characterized novel small-molecules that activate antiviral host responses and how tri-partite motif (TRIM) proteins regulate the innate immune system.

Marjolein Kikkert received a M.Sc. degree in Molecular Sciences from Wageningen University and Research Center (The Netherlands) which motivated Marjolein (1969) to pursue a Ph.D. degree, which she achieved in 1999 at the Department of Virology of this University. Her thesis was entitled “Role of the envelope glycoproteins in the infection of tomato spotted wilt virus”. She then switched to mammalian Virology, became a post-doc at the National Institute of Health and the Environment in Bilthoven, The Netherlands, and at Leiden University Medical Center (LUMC), working on immune evasion strategies of the human cytomegalovirus. She subsequently started working with Prof. Eric Snijder at the LUMC as a post-doc, developed into a senior researcher and subsequently became Assistant Professor in the Department of Medical Microbiology. Her research currently focuses on the replication complex and related virus-host interactions of nidoviruses and other +RNA viruses, and the (innate) immune evasion mechanisms which these viruses employ.
References
- 1.Pott J., Mahlakoiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S. IFN-lambda determines the intestinal epithelial antiviral host defense. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 3.Jiang X., Chen Z.J. The role of ubiquitylation in immune defence and pathogen evasion. Nature Reviews Immunology. 2012;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maelfait J., Beyaert R. Emerging role of ubiquitination in antiviral RIG-I signaling. Microbiology and Molecular Biology Reviews. 2012;76:33–45. doi: 10.1128/MMBR.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harhaj E.W., Dixit V.M. Regulation of NF-kappaB by deubiquitinases. Immunological Reviews. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malynn B.A., Ma A. Ubiquitin makes its mark on immune regulation. Immunity. 2010;33:843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershko A., Ciechanover A., Heller H., Haas A.L., Rose I.A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proceedings of the National Academy of Sciences of the United States of America. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annual Review of Cell and Developmental Biology. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S., Ulrich H.D. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7704–7709. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dammer E.B., Na C.H., Xu P., Seyfried N.T., Duong D.M., Cheng D. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. Journal of Biological Chemistry. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nature Cell Biology. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 13.Xia Z.P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay R.T. SUMO: a history of modification. Molecular Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Haas A.L., Ahrens P., Bright P.M., Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. Journal of Biological Chemistry. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 16.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nature Cell Biology. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 17.Lenschow D.J., Lai C., Frias-Staheli N., Giannakopoulos N.V., Lutz A., Wolff T. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee L.A., Lyon N., Seo K., Huibregtse J.M. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Molecular Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werneke S.W., Schilte C., Rohatgi A., Monte K.J., Michault A., Arenzana-Seisdedos F. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathogens. 2011;7:e1002322. doi: 10.1371/journal.ppat.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatsumi K., Yamamoto-Mukai H., Shimizu R., Waguri S., Sou Y.S., Sakamoto A. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nature Communications. 2011;2:181. doi: 10.1038/ncomms1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaire K., Moura R.F., Granvik M., Igoillo-Esteve M., Hohmeier H.E., Hendrickx N. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE. 2011;6:e18517. doi: 10.1371/journal.pone.0018517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidtke G., Kalveram B., Groettrup M. Degradation of FAT10 by the 26S proteasome is independent of ubiquitylation but relies on NUB1L. FEBS Letters. 2009;583:591–594. doi: 10.1016/j.febslet.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Watson I.R., Irwin M.S., Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011;19:168–176. doi: 10.1016/j.ccr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Raasi S., Schmidtke G., Groettrup M. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. Journal of Biological Chemistry. 2001;276:35334–35343. doi: 10.1074/jbc.M105139200. [DOI] [PubMed] [Google Scholar]
- 25.Schmid D., Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickart C.M. Mechanisms underlying ubiquitination. Annual Review of Biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 27.Hershko A., Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 28.Schulman B.A., Harper J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nature Reviews Molecular Cell Biology. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love K.R., Catic A., Schlieker C., Ploegh H.L. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nature Chemical Biology. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 30.Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Li S., Zheng H., Mao A.P., Zhong B., Li Y., Liu Y. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. Journal of Biological Chemistry. 2010;285:4291–4297. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O’Rourke K.M. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 33.Boone D.L., Turer E.E., Lee E.G., Ahmad R.C., Wheeler M.T., Tsui C. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nature Immunology. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 34.Friedman C.S., O’Donnell M.A., Legarda-Addison D., Ng A., Cardenas W.B., Yount J.S. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Reports. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pichlmair A., Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Chen G., Shaw M.H., Kim Y.G., Nunez G. Nod-like receptors: role in innate immunity and inflammatory disease. Annual Review of Pathology. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 37.Gay N.J., Gangloff M. Structure and function of Toll receptors and their ligands. Annual Review of Biochemistry. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 38.Ostuni R., Zanoni I., Granucci F. Deciphering the complexity of Toll-like receptor signaling. Cellular and Molecular Life Sciences. 2010;67:4109–4134. doi: 10.1007/s00018-010-0464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawai T., Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature Immunology. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 41.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hausmann S., Marq J.B., Tapparel C., Kolakofsky D., Garcin D. RIG-I and dsRNA-induced IFNbeta activation. PLoS ONE. 2008;3:e3965. doi: 10.1371/journal.pone.0003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luthra P., Sun D., Silverman R.H., He B. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 45.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S. IFI16 is an innate immune sensor for intracellular DNA. Nature Immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature Immunology. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuzawa A., Tseng P.H., Vallabhapurapu S., Luo J.L., Zhang W., Wang H. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science. 2008;321:663–668. doi: 10.1126/science.1157340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma S., tenOever B.R., Grandvaux N., Zhou G.P., Lin R., Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 50.Honda K., Takaoka A., Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 51.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kash J.C., Tumpey T.M., Proll S.C., Carter V., Perwitasari O., Thomas M.J. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cezard J.P., Belaiche J. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 54.Crow M.K. Type I interferon in organ-targeted autoimmune and inflammatory diseases. Arthritis Research and Therapy. 2010;12(Suppl. 1):S5. doi: 10.1186/ar2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miceli-Richard C., Lesage S., Rybojad M., Prieur A.M., Manouvrier-Hanu S., Hafner R. CARD15 mutations in Blau syndrome. Nature Genetics. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 56.Hooks J.J., Moutsopoulos H.M., Geis S.A., Stahl N.I., Decker J.L., Notkins A.L. Immune interferon in the circulation of patients with autoimmune disease. New England Journal of Medicine. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 57.Li Q., Xu B., Michie S.A., Rubins K.H., Schreriber R.D., McDevitt H.O. Interferon-alpha initiates type 1 diabetes in nonobese diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oshiumi H., Matsumoto M., Hatakeyama S., Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. Journal of Biological Chemistry. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 59.Gack M.U., Kirchhofer A., Shin Y.C., Inn K.S., Liang C., Cui S. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 61.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang X., Kinch L.N., Brautigam C.A., Chen X., Du F., Grishin N.V. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reyes-Turcu F.E., Horton J.R., Mullally J.E., Heroux A., Cheng X., Wilkinson K.D. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 64.Gao D., Yang Y.K., Wang R.P., Zhou X., Diao F.C., Li M.D. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS ONE. 2009;4:e5760. doi: 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu L., Xiao N., Liu F., Ren H., Gu J. Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1530–1535. doi: 10.1073/pnas.0811029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moore C.B., Bergstralh D.T., Duncan J.A., Lei Y., Morrison T.E., Zimmermann A.G. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 67.Oshiumi H., Miyashita M., Inoue N., Okabe M., Matsumoto M., Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host and Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 68.Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T., Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inn K.S., Gack M.U., Tokunaga F., Shi M., Wong L.Y., Iwai K. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Molecular Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou W., Wang J., Zhang D.E. Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochemical and Biophysical Research Communications. 2007;354:321–327. doi: 10.1016/j.bbrc.2006.12.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang C., Deng L., Hong M., Akkaraju G.R., Inoue J., Chen Z.J. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 72.Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto M., Okamoto T., Takeda K., Sato S., Sanjo H., Uematsu S. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nature Immunology. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 74.Deng L., Wang C., Spencer E., Yang L., Braun A., You J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 75.Bhoj V.G., Chen Z.J. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 76.Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Molecular Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 77.Chang M., Jin W., Sun S.C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nature Immunology. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meylan E., Burns K., Hofmann K., Blancheteau V., Martinon F., Kelliher M. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nature Immunology. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 79.Mikkelsen S.S., Jensen S.B., Chiliveru S., Melchjorsen J., Julkunen I., Gaestel M. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: dependence on TRAF2 and TAK1. Journal of Biological Chemistry. 2009;284:10774–10782. doi: 10.1074/jbc.M807272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pertel T., Hausmann S., Morger D., Zuger S., Guerra J., Lascano J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tseng P.H., Matsuzawa A., Zhang W., Mino T., Vignali D.A., Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nature Immunology. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wertz I.E., O’Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 83.Shi M., Deng W., Bi E., Mao K., Ji Y., Lin G., TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nature Immunology. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 84.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual Review of Biochemistry. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maniatis T. A ubiquitin ligase complex essential for the NF-kappaB Wnt/Wingless, and Hedgehog signaling pathways. Genes and Development. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 86.Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 87.Laplantine E., Fontan E., Chiaravalli J., Lopez T., Lakisic G., Veron M. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO Journal. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hacker H., Tseng P.H., Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nature Reviews Immunology. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 89.Nakhaei P., Mesplede T., Solis M., Sun Q., Zhao T., Yang L. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathogens. 2009;5:e1000650. doi: 10.1371/journal.ppat.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 91.Zhang J., Hu M.M., Wang Y.Y., Shu H.B. TRIM32 modulates type I interferon induction and cellular antiviral response by targeting MITA/STING for K63-linked ubiquitination. Journal of Biological Chemistry. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhong B., Zhang L., Lei C., Li Y., Mao A.P., Yang Y. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 93.Zeng W., Xu M., Liu S., Sun L., Chen Z.J. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Molecular Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tenoever B.R., Ng S.L., Chua M.A., McWhirter S.M., Garcia-Sastre A., Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 95.Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nature Immunology. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 97.Li S., Wang L., Berman M., Kong Y.Y., Dorf M.E. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chau T.L., Gioia R., Gatot J.S., Patrascu F., Carpentier I., Chapelle J.P. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends in Biochemical Sciences. 2008;33:171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 99.Arimoto K., Funami K., Saeki Y., Tanaka K., Okawa K., Takeuchi O. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao T., Yang L., Sun Q., Arguello M., Ballard D.W., Hiscott J. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nature Immunology. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- 101.Belgnaoui S., Paz S., Goulet M., Sun Q., Iwai K., Dikic I. Linear ubiquitination of NEMO negatively regulates the IFN antiviral response through the disruption of the MAVS signalosome. Cell Host and Microbe. 2012;12:211–222. doi: 10.1016/j.chom.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 102.Clark K., Takeuchi O., Akira S., Cohen P. The TRAF-associated protein TANK facilitates cross-talk within the IkappaB kinase family during Toll-like receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17093–17098. doi: 10.1073/pnas.1114194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmid S., Mordstein M., Kochs G., Garcia-Sastre A., Tenoever B.R. Transcription factor redundancy ensures induction of the antiviral state. Journal of Biological Chemistry. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang K., Shi H.X., Liu X.Y., Shan Y.F., Wei B., Chen S. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. Journal of Immunology. 2009;182:3782–3792. doi: 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- 105.Higgs R., Lazzari E., Wynne C., Ni Gabhann J., Espinosa A., Wahren-Herlenius M. Self protection from anti-viral responses—Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral Toll-like receptors. PLoS ONE. 2010;5:e11776. doi: 10.1371/journal.pone.0011776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yoshimi R., Chang T.H., Wang H., Atsumi T., Morse H.C., 3rd, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. Journal of Immunology. 2009;182:7527–7538. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kubota T., Matsuoka M., Chang T.H., Tailor P., Sasaki T., Tashiro M. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. Journal of Biological Chemistry. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liang Q., Deng H., Li X., Wu X., Tang Q., Chang T.H. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier e3 ligase and negative regulator of IFN regulatory factor 7. Journal of Immunology. 2011;187:4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J., Xu L.G., Han K.J., Wei X., Shu H.B. PIASy represses TRIF-induced ISRE and NF-kappaB activation but not apoptosis. FEBS Letters. 2004;570:97–101. doi: 10.1016/j.febslet.2004.05.081. [DOI] [PubMed] [Google Scholar]
- 110.Shuai K., Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nature Reviews Immunology. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 111.Balakirev M.Y., Jaquinod M., Haas A.L., Chroboczek J. Deubiquitinating function of adenovirus proteinase. Journal of Virology. 2002;76:6323–6331. doi: 10.1128/JVI.76.12.6323-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gonzalez C.M., Wang L., Damania B. Kaposi's sarcoma-associated herpesvirus encodes a viral deubiquitinase. Journal of Virology. 2009;83:10224–10233. doi: 10.1128/JVI.00589-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host and Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. Journal of Virology. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. Journal of Virology. 2011;85:3758–3766. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Kasteren P.B., Beugeling C., Ninaber D.K., Frias-Staheli N., van Boheemen S., Garcia-Sastre A. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. Journal of Virology. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gack M.U., Albrecht R.A., Urano T., Inn K.S., Huang I.C., Carnero E. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host and Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lilley C.E., Chaurushiya M.S., Boutell C., Landry S., Suh J., Panier S. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO Journal. 2010;29:943–955. doi: 10.1038/emboj.2009.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nerenberg B.T., Taylor J., Bartee E., Gouveia K., Barry M., Fruh K. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. Journal of Virology. 2005;79:597–601. doi: 10.1128/JVI.79.1.597-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ting L., Rad R., Gygi S.P., Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nature Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dikic I., Wakatsuki S., Walters K.J. Ubiquitin-binding domains—from structures to functions. Nature Reviews Molecular Cell Biology. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]