Abstract
A procedure was developed to generate recombinant single chain Fv (scFv) antibody fragments reacting with the extracellular domain of human cell surface antigen CD13 (hCD13; aminopeptidase N) on intact cells. Membrane fractions prepared from a stably transfected hCD13-positive murine NIH/3T3 cell line were used to immunize BALB/c mice, with the intention that hCD13 would be the major immunogenic molecule recognized by the immune system. Spleen RNA from the immunized mice served to generate a combinatorial scFv phage display library. The library was adsorbed against non-transfected NIH/3T3 or Sf21 insect cells to eliminate nonrelevant binders. The supernatant was then used for panning with either hCD13-transfected Sf21 insect cells or a hCD13-expressing human leukemia-derived cell line. Therefore, the key concepts of the procedure were the presentation of hCD13 as the sole human antigen on murine NIH/3T3 cells and a screening strategy where hCD13 was the major common antigen of the material used for immunization and panning. Two different hCD13-reactive phages were isolated and the soluble scFvs were expressed in E. coli and purified. The two scFvs, anti-hCD13-1 and anti-hCD13-3, differed at four amino acid positions in their VH regions and both had high affinities for hCD13 as determined by surface plasmon resonance (KD=7 and 33×10−10 M, respectively). Both efficiently recognized hCD13 on intact cells. Therefore, the procedure allowed the production of high affinity scFvs reacting with a desired antigen in its native conformation without requiring extensive purification of the antigen and should be useful for the preparation of scFvs against other conformation-sensitive cell-surface antigens.
Keywords: Single chain Fv-fragments, Recombinant antibodies, Phage display, CD13, Mixed lineage leukemia, Leukocyte surface antigens, Aminopeptidase N
Abbreviations: BSA, bovine serum albumin; ELISA, enzyme-linked immunosorbent assay; IPTG, isopropyl-β-d-thiogalactopyranoside; PCR, polymerase chain reaction; RT, room temperature; RU, response units; scFv, single chain Fv fragment
1. Introduction
CD13, aminopeptidase N, is a cell surface antigen used in human leukocyte typing (Riemann et al., 1999) and functions as a membrane-bound metalloproteinase that catalyzes the removal of N-terminal amino acids from polypeptides and small peptides (Mizutani et al., 1993, Hooper, 1994). CD13 is expressed in epithelial cells in liver, kidney and intestine, in placenta, brain and spleen, and in a variety of hematopoietic cells. Recently, roles for CD13 in metastasis and in the degradation of extracellular matrix components by tumor cells were reported (Saiki et al., 1993, Fujii et al., 1995). In addition, CD13 was identified as a receptor for tumor-homing peptides in the vasculature of tumors and appears to play a role in the aberrant angiogenesis of tumor tissues. Therefore, CD13 was proposed as a useful target for the delivery of drugs to tumors that aim at inhibiting angiogenesis (Pasqualini et al., 2000). Furthermore, CD13 is a receptor for cytomegalovirus (CMV; Soderberg et al., 1993, Giugni et al., 1996) and certain coronaviruses (Delmas et al., 1992, Yeager et al., 1992) and future antiviral strategies directed against these viruses may also benefit from including CD13 as a target (Larsson et al., 1998).
In the hematopoietic system, CD13 is expressed predominantly in myelo-monocytic cells (Shapiro et al., 1991, Shipp and Look, 1993, Olsen et al., 1997). The antigen is present on early hematopoietic progenitors including progenitors of the B- and T-lymphoid lineages, and expression is progressively reduced with increasing maturation of these cells. The antigen is not detectable on mature lymphocytes from peripheral blood, spleen and tonsils (Spits et al., 1995). CD13 is often upregulated on malignant cells, particularly on blasts from acute and chronic B-cell leukemias (ALLs and CLLs), including infant pro-B cell leukemias (Makrynikola et al., 1995). The pro-B ALLs frequently express CD19, CD22 and CDw75 as lymphoid markers and CD13, CD15, CD33, CDw65 as myeloid markers (Stong et al., 1985, Greil et al., 1994). The expression of myeloid markers on a pro-B cell background is unusual and highly characteristic for these particular leukemias. Therefore, special terms including ‘biphenotypic expression’ and ‘mixed-lineage phenotype’ have been coined to designate this property.
The mixed lineage phenotype provides a unique opportunity to direct novel immuno-therapeutic approaches against the pro-B ALLs that are often refractory to other forms of treatment. In particular, infants with leukemias associated with translocations t(4;11) and t(11;19) affecting the MLL-gene on chromosome 11q23 have poor prognosis and new therapeutic approaches for these leukemias are needed (Behm et al., 1996, Heerema et al., 1999). Immunotherapeutic approaches targeting individual antigens on tumor cells have suffered in the past from the fact that the same antigens were also present on normal cells. By contrast, the mixed-lineage antigen spectrum is preferentially found on the tumor cells and therefore offers novel possibilities to specifically target the tumor cells. The long-range objective of our research is to contribute to the development of immunotherapeutic approaches against leukemic blasts with chromosomal translocations t(4;11) or t(11;19) to the MLL gene. For this purpose cDNAs coding for antibodies or antibody fragments reactive with several of the component antigens of the mixed lineage spectrum are needed.
The immediate purpose of the present study was to generate recombinant single chain Fv fragments (scFv) that bind human CD13 in its native conformation on intact human leukemic blasts. To our knowledge, no reports on scFvs reactive against hCD13 on intact cells have been published, although a number of anti-human CD13 hybridoma antibodies have been reported (Goyert, 1997). A standard procedure for the generation of scFvs consists in the bacterial expression of the antigen followed by biochemical purification and the use of the purified antigen for the immunization of mice. However, hCD13 is heavily glycosylated (Look et al., 1989) and an authentic glycosylation is not achieved by bacterial expression. Therefore, mice immunized with bacterially expressed antigen may produce antibodies reacting only with the recombinant protein, but not with the antigen in its native conformation on human cells (Chowdhury et al., 1998, Huls et al., 1999). An alternative strategy that avoids the use of bacterially expressed hCD13 would involve the preparation of cell surface membranes from CD13 positive human cells, such as CD13-expressing leukemia cells (Stong et al., 1985, Greil et al., 1994). The disadvantage of this approach is that the immunized mice are likely to produce an excess of antibodies against other human cell surface antigens and antibodies against hCD13 may be of low abundance. Anti-hCD13 scFvs would, therefore, be difficult to isolate from combinatorial phage libraries prepared from immunized mice without further specific modifications of the procedure added to maximize the probability of isolating scFvs against the desired antigen. To avoid these problems, a novel procedure was designed here with the purpose of maximizing the yield of anti-hCD13 scFv antibodies while avoiding the production of antibodies against other human cell surface antigens. Indeed, this procedure produced recombinant scFvs against human CD13 at high frequency. An advantage of the new procedure is that it does not require the extensive biochemical purification of the antigen, which can result in the partial or total loss of its native conformation. We anticipate that this procedure will be applicable to a number of other conformation-sensitive cell surface antigens.
2. Materials and methods
2.1. Bacterial strains and plasmids
E. coli strain XL1-Blue (Stratagene, Amsterdam, Netherlands) was used for the amplification of plasmids, and E. coli strain TG1 (from Dr. G. Winter, MRC Cambridge, UK) served for the screening of antibody libraries. Libraries were generated in phagemid vector pAK100 and pAK400 was employed for the expression of soluble scFvs (Krebber et al., 1997). Plasmid sk3.4-CD13 (from Dr. L. Shapiro, St. Jude Children’s Hospital, Memphis, TN, USA) harboring human CD13 cDNA (Look et al., 1989) was digested with SpeI/EcoRV and the insert was ligated to SpeI/EcoRV digested pcDNA3.1(+) (Invitrogen, Groningen, Netherlands), resulting in construct pcDNA-hCD13. For stable transfection of Sf21 cells, the EcoRV/NotI fragment of hCD13 cDNA was ligated into pIZT-V5-His (Invitrogen), resulting in pIZT-hCD13. To express soluble hCD13, a cDNA fragment lacking the transmembrane and intracellular domains was amplified by PCR from pIZT-hCD13. The primers used for this amplification were: CD13-Fc-EcoRV (5′-tac gat atc aga aga aca aga acg cca ac-3′) and CD13-Fc-NotI (5′-ctg ggc ggc cgc tcg tgt ttt ctg tga acc ac-3′). The PCR amplimer was ligated into the vector pSecTag-C-Fc containing the coding sequences for the Fc portion of a human IgG1 heavy chain (M. Peipp, unpublished data). The resulting chimeric protein with an N-terminal portion consisting of the extracellular domain of hCD13 and the Fc domain of human IgG1 at its C-terminus was designated s-hCD13.
2.2. Culture of eukaryotic cells
NIH/3T3 murine fibroblasts and human 293T kidney cells were cultured in DMEM-Glutamax-I medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal calf serum (FCS) and penicillin and streptomycin (Life Technologies) at concentrations of 100 U/ml and 100 μg/ml, respectively. Leukemia-derived SEM cells (Greil et al., 1994) were cultured in RPMI 1640-Glutamax-I medium containing 10% FCS and penicillin and streptomycin at concentrations of 100 U/ml and 100 μg/ml, respectively. Sf21 insect cells were cultured in TC100 medium (Life Technologies) supplemented with 10% FCS, penicillin, streptomycin and gentamycin at concentrations of 50 U/ml, 50 μg/ml, and 50 μg/ml, respectively, at 27°C in a humidified incubator.
2.3. Establishment of NIH/3T3 and Sf21 cell lines stably expressing hCD13
One day prior to transfection, a total of 4×105 NIH/3T3 cells were seeded in 60-mm dishes. Five μg of linearized pcDNA3-hCD13 plasmid were combined with Superfect reagent (Qiagen, Hilden, Germany) and transfection was performed following the manufacturer’s protocol. Selection medium containing 600 μg/ml of G418 was exchanged every 3–4 days. After 2 weeks of selection, single cells were subcloned. Isolated clones were analyzed for surface expression of hCD13 by fluorescent-activated cell sorting (FACS). Sf21 cells were seeded in 60-mm plates at 4×105 cells/well. NdeI-linearized pIZT-hCD13 vector (3.6 μg) and BstEII linearized pIZT-V5-His vector carrying a Zeocin resistance gene (0.4 μg) were cotransfected using the TransFast reagent (Promega, Mannheim, Germany). After 3 weeks of continuous selection with 400 μg/ml Zeocin, the mixed cell population was examined by FACS for surface expression of hCD13. The population expressing hCD13 was designated Sf21-hCD13. No attempt was made to isolate single-cell subcloned lines, because for the intended application this was not required.
2.4. Preparation and gradient purification of plasma membrane fractions
For the preparation of plasma membranes, 5–10×108 cells from the hCD13-expressing NIH/3T3 line were suspended in lysis buffer consisting of 1 mM NaHCO3, pH 8, 1 mM MgCl2 and 100 μM Pefablock proteinase inhibitor (Roth, Karlsruhe, Germany). After incubation on ice for 15–20 min, the cells were disrupted in a dounce homogenizer. The lysate was cleared by centrifugation and sucrose was added to a final concentration of 1.59 M. Ten-ml aliquots were overlayed in SW28 rotor tubes (Beckman, Munich, Germany) with 9 ml of 1.175 M sucrose in carbonate buffer (1 mM NaHCO3; pH 8). A second layer of 7 ml of 0.98 M sucrose and a third layer of 2 ml of 0.8 M sucrose were added. Gradient centrifugation was performed for 16 h at 70,000×g in an SW28 rotor. Membrane fractions were collected, diluted in 5 volumes of phosphate buffered saline (PBS), and sedimented for 1 h at 70,000×g as previously described (Eylar and Hagopian, 1971). Membranes were resuspended in 100 μl of PBS, and fractions containing hCD13 were identified by ELISA.
2.5. Immunization of mice
BALB/c mice (Charles River, Sulzfeld, Germany) were maintained according to the European guidelines for the protection of laboratory animals. Approximately 500 μg of gradient-purified membrane protein were combined with TiterMax Gold™ adjuvant (Sigma, Deisenhofen, Germany) and injected subcutaneously on day 0. On days 25 and 49, mice were boosted with 300 μg of membrane protein. A final injection of 150 μg of membrane protein followed on day 60. Four days later, the mice were sacrificed and spleens were recovered under sterile conditions. The serum was tested at different timepoints for the presence of anti-hCD13 antibodies by FACS analysis using 293T cells that were transiently transfected with hCD13 in the expression plasmid pcDNA3-hCD13.
2.6. Preparation of combinatorial scFv libraries and conversion to phage display libraries
RNA was prepared from spleens of the immunized mice using Trizol reagent (Life Technologies) and 10–15 μg of total RNA were used to prepare first strand cDNA following standard procedures (Krebber et al., 1997). PCR amplification of immunoglobulin variable region cDNAs and cloning into the phagemid vector pAK100 was performed as described (Krebber et al., 1997). The initial combinatorial scFv library was propagated in SB-medium (1% glucose; 30 μg/ml chloramphenicol) at 37°C under vigorous shaking. After the density of the culture reached an OD600 of 0.5, 100 ml of the culture were infected with helper phage M13KO7 (Stratagene). The culture was incubated at 37°C for another 30 min without shaking. Then, 400 ml of SB-medium were added and isopropyl β-d-thiogalactopyranoside (IPTG) was supplied at a final concentration of 0.5 mM. The culture was incubated at 37°C for another 2 h under vigorous shaking (250 rev./min) and then kanamycin was added (25 μg/ml). Finally, the culture was incubated at 30°C overnight. Phages were precipitated with polyethylene glycol (20% PEG 6000, 2.5 M NaCl) and resuspended in 2 ml PBS.
2.7. Panning of phage display libraries with intact cells
Panning of phage display libraries with intact cells was carried out using two different approaches. In both cases, the phage display library was first adsorbed against non-transfected cells that were blocked in 2% nonfat dry milk in PBS (NM-PBS) for 30 min. In the first approach, 0.5 ml of the phage display library was resuspended in NM-PBS and incubated for 1 h at RT with 3×106 blocked non-transfected NIH/3T3 cells. The pre-adsorbed library was then used for positive selection on SEM leukemic cells. For this purpose, 106 SEM cells were blocked with NM-PBS and then incubated with the pre-adsorbed scFv library for 1 h at RT under slow agitation. Cells were washed 10 times with 2% NM-PBS and twice with PBS. Bound phages were eluted with 1.5 ml of 50 mM HCl for 10 min. After neutralization with 0.5 ml of 1 M Tris–HCl at pH 8, cells were pelleted and the supernatant was used to infect 10 ml of exponentially growing E. coli TG1 cells. To the culture, 20 ml 2×YT medium containing 1% glucose and 30 μg/ml chloramphenicol were added and the cells were incubated for 2 h at 37°C under vigorous shaking (250 rev./min). Cells were superinfected with helper phage. Seventy ml of 2×YT medium were then added and the IPTG concentration was adjusted to 0.5 mM. Two hours after infection, kanamycin was added at a final concentration of 25 μg/ml and the culture was allowed to grow overnight at 30°C. On the following day, phages were prepared as described above. In the second approach, non-transfected Sf21 insect cells were used to pre-adsorb the phage display library and positive selection was performed on Sf21-hCD13 cells by the panning procedure described above. Individual phages were clonally purified and sequence analysis of the inserts was performed using dideoxynucleotide sequencing (Sambrook et al., 1989) on an Applied Biosystems automated DNA sequencer (ABI Prism 310 Genetic Analyzer; Perkin Elmer, Ueberlingen, Germany).
2.8. Bacterial expression and purification of soluble scFv fragments
For the soluble expression of antibody fragments, plasmids were propagated in E. coli HB2151 (from Dr. G. Winter, MRC Cambridge, UK). Overnight cultures were diluted in 2×YT medium supplemented with 1% glucose and 30 μg/ml chloramphenicol until the extinction at 600 nm (OD600) was ≤0.1. Cultures were grown to an OD600 of 0.6–0.8 at 37°C and expression was induced by the addition of IPTG at a final concentration of 1 mM and by lowering the temperature to 30°C. After 4–6 h, the bacteria were collected by centrifugation. Periplasmic extracts were prepared using established protocols (Kipriyanov, 1997) and dialyzed extensively against buffer containing 50 mM NaH2PO4, pH 8, 300 mM NaCl, and 10 mM imidazole at 4°C. Purification of 6×His tagged scFv fragments was achieved by affinity chromatography with nickel-nitrilotriacetic acid (Ni-NTA) magnetic agarose beads (Qiagen).
2.9. Flow cytometric analysis
For monitoring of scFv binding to intact cells, 5×105 cells were washed with PBS containing 0.1% BSA and 7 mM Na-azide (PBA). The cells were then incubated with either 50 μl of bacterial periplasmic extracts or 50 μl of purified scFv (10 μg/ml) for 1 h on ice. Periplasmic extracts from cells producing nonrelevant scFv fragments served as isotype controls. The cells were then washed with PBA and incubated with 20 μl of 0.2 μg/ml Penta-His antibody (Qiagen) for 30 min on ice. Unbound antibodies were removed by washing with PBA and 10 μl of PE conjugated rat-anti-mouse-IgG1 antibody (Becton-Dickinson, Heidelberg, Germany) were added to the cell pellets. The cells were incubated for 30 min on ice. After a final wash, FACS analysis was performed on a FACSCalibur using CellQuest software (Becton-Dickinson). Ten thousand events were collected for each sample and all analyses of whole cells were performed using appropriate scatter gates to exclude cellular debris and aggregates. For monitoring surface expression of hCD13 on transfected cell lines, 5×105 cells were incubated for 30 min on ice with 20 μl of anti-hCD13 antibody (Clone WM-47; DAKO Diagnostica GmbH, Hamburg, Germany) at a concentration of 5 μg/ml. Mouse IgG1 served as the isotype control. The cells were washed in PBA and 10 μl of PE conjugated rat-anti-mouse-IgG1 antibody were added. After a final wash, cells were analyzed as described above.
2.10. Expression of soluble hCD13 (s-hCD13) in mammalian cells
293T cells were transiently transfected with a plasmid containing the cDNA coding for s-hCD13 using Fugene6 reagent (Roche, Mannheim, Germany). After 24 h, medium containing 10% FCS with reduced IgG content (Life Technologies) was added. Beginning at 48 h after transfection, the medium was exchanged every 24 h and culture supernatant containing secreted s-hCD13 was collected each day for 7 days. The purification was performed on protein A agarose columns (Biorad, Munich, Germany) as described elsewhere (Harlow and Lane, 1988). A commercial anti-hCD13 antibody (Clone WM-47) was used to identify s-hCD13 by ELISA.
2.11. SDS–PAGE and Western blot analysis
SDS–PAGE was carried out according to Laemmli (1970). Gels were stained with Coomassie brilliant blue R250. Western blots (Towbin et al., 1979) were performed with secondary antibodies coupled to horseradish peroxidase (HRP; Dianova, Hamburg, Germany) and detected using ECL reagents (Amersham, Freiburg, Germany). ScFvs were detected with the Penta-His antibody (Qiagen). S-hCD13 was detected using a HRP conjugated anti-human-Fc antibody (Sigma).
2.12. Surface plasmon resonance and determination of binding constants
S-hCD13 was immobilized on a CM5 sensor chip by standard amine coupling (Biacore AB, Freiburg, Germany). The amount of deposited s-hCD13 was monitored by surface plasmon resonance. Coupling was continued until the amount deposited corresponded to ∼800 resonance units (RU). For binding analyses, the purified scFv fragments CD13-1 and CD13-3 were dialyzed against running buffer (10 mM Hepes pH 7.4, 3.4 mM EDTA, 150 mM NaCl, and 0.005% surfactant P20). The scFvs in the fluid phase were allowed to interact with the immobilized s-hCD13 at RT and a flow rate of 10 μl/min. Varying concentrations of the scFvs were analyzed with the same chip after regeneration. Binding constants were calculated using the BIAevaluation 3.0 software (Kazemier et al., 1996; Langmuir 1:1 model, Biacore AB).
3. Results
3.1. Generation of NIH/3T3-derived cell lines expressing hCD13
Two NIH/3T3-derived cell lines stably expressing human CD13 (hCD13) were established by transfection and single cell cloning. The lines, NIH/3T3-hCD13 clones 4 and 6, expressed elevated levels of hCD13 antigen on their surface, as judged by FACS staining with a commercial anti-hCD13 antibody (Fig. 1 ). The mean fluorescence intensity of NIH/3T3 clones 4 and 6 stained with anti-hCD13 antibody was ∼20–30 times greater than non-transfected NIH/3T3 cells and cells stained with an isotype control antibody (Fig. 1). Although the absolute surface density of hCD13 expressed in number of molecules per cell was not determined, the distribution of the cells with respect to their fluorescence intensity was comparable to the distribution obtained for the leukemia-derived SEM cell line (data not shown). Therefore, clones 4 and 6 expressed sufficient surface densities of hCD13 to be useful for the preparation of membrane fractions for the immunization of mice.
Fig. 1.

Expression of hCD13 on stably transfected cell lines. NIH/3T3 cells (A, B, C) or Sf21 cells (D, E) were transfected with cDNA clones carrying the coding sequences for the extracellular and transmembrane domains of hCD13 and two stably transfected NIH/3T3-derived lines, clones 4 and 6, were obtained. Cells were stained with a commercial anti-hCD13 antibody (WM-47) (dark shading) or an isotype-matched control (light shading) and analyzed by FACS. (A) Non-transfected NIH/3T3 cells; (B) NIH/3T3-hCD13 clone 4; (C) NIH/3T3-hCD13 clone 6. (D) Non-transfected Sf21 insect cells; (E) mixture of Sf21 cell clones, stably transfected with hCD13 cDNA.
3.2. Preparation and enrichment of plasma membranes from hCD13-expressing NIH/3T3 cells and immunization of BALB/c mice
The procedure is outlined in Fig. 2 . After generating an NIH/3T3 murine line expressing hCD13, the membrane fraction was prepared under gentle conditions to avoid denaturation of the antigen. The enriched plasma membrane fraction was used for the immunization of mice. With this procedure, we intended to avoid the production of antibodies against human plasma membrane proteins other than hCD13. The immunized BALB/c mice were expected to be tolerant against most membrane antigens from NIH/3T3 cells, with the possible exception of alloantigens. Membranes were prepared from NIH/3T3 clones 4 and 6 by gentle hypotonic disruption and subsequent density centrifugation in a sucrose gradient. Several membrane fractions were collected from the gradient and the hCD13-positive fractions were identified by ELISA (data not shown). The peak fraction (at 0.98 M) was used to immunize BALB/c mice. In an average preparation, 400 μg of hCD13-positive plasma membrane protein were recovered from 5×108 cells and the estimated enrichment of plasma membrane proteins was 20- to 50-fold. After 64 days and three boosts, the immunized mice developed a high titer of circulating anti-hCD13 antibodies, as judged by sampling aliquots of their peripheral blood at various times and performing flow-cytometric assays with hCD13-positive cells (transiently transfected 293T cells).
Fig. 2.

Flow diagram of the improved procedure for the isolation of anti hCD13 scFvs. Key steps of the procedure are outlined. Three sequential rounds of panning were employed for the isolation of scFvs anti hCD13-1 and -3.
3.3. Preparation of combinatorial phage libraries displaying scFv antibody fragments on their pIII tail fiber proteins
Combinatorial scFv cDNA libraries were produced from total spleen RNA of the immunized mice according to published procedures (Krebber et al., 1997). From 10 μg of total spleen RNA, combinatorial libraries with ∼1–3×105 independent chloramphenicol-resistant clones were obtained before amplification. Only mouse κ light chain cDNAs were amplified and included in the library. The λ light chains were omitted because they represented only a small proportion of mouse antibodies. Six separate libraries were produced, characterized, and combined for panning. The combined library contained ∼8–10×105 independent clones.
3.4. Isolation of CD13-reactive phages by differential panning
Panning was performed by employing two different variants of the protocol. In variant 1, the libraries were pre-adsorbed with non-transfected NIH/3T3 cells and the supernatant was panned with intact hCD13-expressing SEM leukemia cells. In variant 2, the libraries were pre-adsorbed with non-transfected Sf21 insect cells and the supernatant was panned with hCD13-expressing Sf21 cells. Therefore, Sf21 insect cells were transfected with a cDNA construct coding for hCD13 and a mixture of stably transfected clones was used for the panning procedure. This mixture expressed hCD13 with sufficient stability as judged by flow cytometry and obviated the need for single cell subcloning. The mean fluorescence intensity of Sf21 cells stained with anti-CD13 antibody was ∼50–100 fold greater than the intensity of the same cells stained with an isotype-control antibody or that of non-transfected Sf21 cells (Fig. 1).
The phages that specifically bound to the cells were eluted with 50 mM hydrochloric acid (HCl) and immediately neutralized. The titer of eluted phages was determined and an enrichment factor was calculated relative to the first round of panning. After three rounds, reactive phages were enriched 16-fold after panning with SEM cells and 66-fold after panning with hCD13 positive Sf21 cells (Table 1 ). Individually isolated phages were clonally purified and their specificity of binding was evaluated by cellular ELISA. As a result, 12 of 14 individually isolated phages from the third round of panning specifically reacted with NIH/3T3 cells stably expressing hCD13 and showed no binding to non-transfected NIH/3T3 cells (data not shown). Characterization of the cDNA inserts of these phages revealed that two different scFv clones, anti-hCD13-1 and anti-hCD13-3, were isolated multiple times independently, using both variants of the panning protocol. Both had identical light chain variable regions (VL), but differed in four amino acid positions of their heavy chain variable regions (VH). Two of these changes were located in the complementarity-determining regions 1 and 2 and the other two were found in framework 3 (Fig. 3 ).
Table 1.
Enrichment of phages by three consecutive rounds of panning
| Cells | Rounda | Phage input | Phage eluted | Input/elution | Factor of |
|---|---|---|---|---|---|
| (cfu)×1012 | (cfu)×105 | ×106 | enrichmentb | ||
| SEM | 1 | 16 | 7 | 23 | 1 |
| 2 | 1.5 | 6.5 | 2.3 | 10 | |
| 3 | 6.5 | 47 | 1.4 | 16 | |
| Sf21-hCD13 | 1 | 16 | 1.6 | 10 | 1 |
| 2 | 9.5 | 27 | 3.5 | 2.8 | |
| 3 | 9.5 | 620 | 0.15 | 66 |
Round of panning.
The factor of enrichment is calculated by setting the input/elution ratio of the first round to 1 and computing the ratios measured in subsequent rounds as multiples of the first round ratio (normalization to the first round).
Fig. 3.

Amino acid sequence of scFvs anti-hCD13-1 and -3. Amino acid sequences deduced from cDNA sequence. Row A, anti-hCD13-1. Row B, anti hCD13-3. Dashes, identical residues; deviating residues are boxed. Complementarity determining regions (CDR) of the VL (L-CDR) and VH (H-CDR) chains are delineated. Numbers in the left margin, continuous numbering of amino acids in the scFv polypeptides; positions 117–136, synthetic linker consisting of serine (S) and glycine (G) residues; positions 261–266, hexa-histidine tag.
3.5. Expression and purification of soluble anti-hCD13 scFvs
For the purpose of large-scale expression of the recombinant antibody fragments, the cDNAs coding for the scFvs were subcloned into the expression vector pAK400. E. coli strain HB2151 was chosen for high level expression of the scFvs. Intact recombinant scFvs were secreted to the periplasmic compartment, as visualized by Western blot with an antibody reactive against the hexa-histidine tag (Fig. 4A , lane 2). No antigenic material was observed in the supernatant (Fig. 4, lane 1) or in periplasmic extracts from non-transformed E. coli HB2151 cells grown under the same conditions (Fig. 4, lane 3). A periplasmic extract from E. coli HB2151 harboring a nonrelevant scFv served as a positive control. From previous work (M. Peipp; unpublished observations), it was known that this latter scFv was efficiently secreted into the periplasmic space. Typically, 5–6 g of packed bacterial cells were obtained from a culture volume of 1 l, resulting in ∼100–150 μg of purified soluble scFv. The scFvs contained a hexa-histidine tag as a C-terminal extension, which allowed affinity purification with magnetic Ni-NTA beads. The protocol for the preparation of the periplasmic compartment and the purification with NTA beads was reduced to a one-step procedure resulting in the typical yield described above. The one-step purification procedure resulted in scFvs essentially free of contaminants detectable by staining with Coomassie blue (Fig. 4B, lanes 1 and 2). Both purified soluble scFvs reacted with the hCD13 antigen on intact cells, as shown by FACS analysis (Fig. 5 ; data shown for scFv anti-hCD13-1), indicating that the two scFvs reacted with hCD13 in the native state.
Fig. 4.

Bacterial expression and purification of soluble scFvs. Recombinant scFvs were expressed in E. coli HB2151, purified from the periplasmic space and analyzed by SDS–PAGE by Western blot or staining with Coomassie blue. (A) Western blot using an antibody against the hexa-histidine tag as the first antibody. Lane 1, culture supernatant; lane 2, periplasmic extract from E.coli culture harboring anti-hCD13-1 scFv in vector pAK400; lane 3, periplasmic extract from untransfected E. coli host; lane 4, a control scFv, also carrying a C-terminal hexa-histidine tag, was loaded as a positive control for effective secretion into the periplasmic space. Lane 5, a recombinant transcription factor (rat STAT5b) that carries a hexa-His tag was loaded as a positive control for reaction with the anti-His-tag antibody. Lane 6, BSA was loaded as a negative control. (B) Evaluation of the purification of scFvs anti hCD13-1 and -3. Coomassie blue stained SDS–PAGE gel. Left track, size marker; 10 kDa ladder; lanes 1 and 2, purified scFvs anti-hCD13-1 and -3, respectively. Lanes 3–6, calibration with 0.5, 1, 2 and 4 μg of BSA.
Fig. 5.
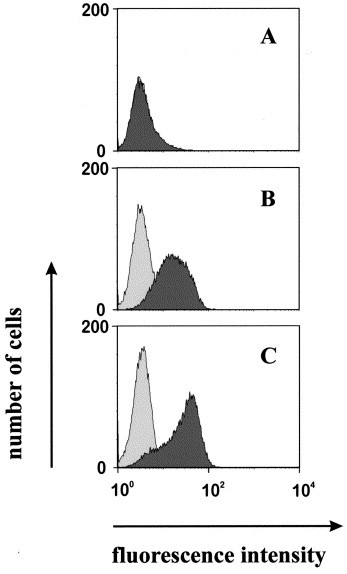
Specific binding of soluble anti-hCD13 scFvs to hCD13-expressing Sf21 insect cells and human SEM leukemia cells. Soluble anti-hCD13-1 scFv (dark shade) prepared and purified from the periplasmic space of E. coli was allowed to react with Sf21 insect cells (A and B) or human leukemia derived SEM cells (C). A control scFv, purified from periplasmic extracts of the same E. coli strain using the same procedure was used as a negative control (light shade). Flow-cytometric analysis. Panel A, non-transfected Sf21 cells. Panel B, Sf21 cells transfected with hCD13. Panel C, SEM leukemia cells.
3.6. Expression of soluble CD13 in its native conformation by fusion with an Fc-portion from a human IgG1 antibody
The two anti-hCD13 scFvs isolated here reacted with hCD13 in its native conformation and failed to react with extracts from hCD13-positive cells on Western blots (data not shown). Therefore, to determine molecular binding constants for scFvs anti-hCD13-1 and anti-hCD13-3, it was necessary to generate the antigen in its native conformation and in a pure state. To achieve these objectives, the antigen was fused to the Fc-domain of a human IgG1 antibody, since this procedure was known to generate soluble antigens in a native conformation (M. Peipp, unpublished data). In this procedure, chimeric molecules are secreted from mammalian cells and receive the post-translational modifications (e.g. glycosylation) characteristic of mammalian cells. For the purpose of secretion, sequences coding for a leader peptide from a murine immunoglobulin κ light chain were fused to the coding sequences for the extracellular domain of hCD13 (Fig. 6A ). For the purpose of high level expression, 293T cells were transiently transfected with the construct and soluble human CD13 (s-hCD13) was then produced in high quantities for several days after transfection. The chimeric protein was collected from the culture supernatant and purified by affinity chromatography on a protein A agarose column. Using this procedure, typically 50-100 μg of the recombinant protein were recovered from 1 l of culture supernatant. The chimeric molecule reacted on a Western blot with an antibody against the Fc-portion (Fig. 6B), and no contaminants or breakdown products were visible by staining with Coomassie blue (Fig. 6C). The material reacted in an ELISA with the commercial anti-hCD13 (WM-47) antibody, as well as with the two isolated scFvs (data not shown). Thus, the integrity of the antigenic epitope in the chimeric s-hCD13 molecule resembled that on intact cells and this material was considered suitable as a ligand for the determination of molecular binding constants of the scFv antibodies.
Fig. 6.

Expression of soluble hCD13 chimeric protein (s-hCD13) in 293T cells. (A) Block-structure of the chimeric protein. A leader peptide from murine immunoglobulin κ light chains was fused to the extracellular domain of hCD13 (∼100 kDa), and this in turn was fused to the Fc-portion of a human IgG1 (∼28 kDa). The combined molecular mass of the CD13 and Fc-domains amounts to 130 kDa, approximately. The difference to the electrophoretic mobility of ∼148 kDa is due to glycosylation. (B) Verification of effective secretion of intact s-hCD13 into the culture supernatant by Western blot analysis with an antibody against the Fc portion of human IgG1. Lane 1, supernatant from non-transfected 293T cells; lane 2, supernatant from 293T cells transfected with the expression construct for s-hCD13. (C) Evaluation of the purity and integrity of the purified s-hCD13 by SDS–PAGE and staining with Coomassie blue. Left track, size marker; 10 kDa ladder; lanes 1 and 2, eluted fractions 1 and 2 from the protein A agarose column.
3.7. The two anti-hCD13 scFvs have high affinities as judged by surface plasmon resonance
The soluble chimeric ligand s-hCD13 was deposited on a Biacore CM5 sensor chip and chemically coupled by standard procedures. Sequential rounds of loading were performed until the total amount coupled corresponded to ∼800 relative units (RU). Then the scFvs were allowed to flow over the chip in the fluid phase, and the association and dissociation reactions were monitored by surface plasmon resonance. This procedure allowed a reliable measurement of the association (k a) and dissociation rate constants (k d). The affinity K D was determined for each scFv antibody from the quotient k d/k a. The procedure was performed with several different concentrations of the same scFv in the fluid phase and with separate chips for anti-hCD13-1 and anti-hCD13-3 (Fig. 7 ). Each scFv was also injected over a blank inactivated surface and this signal was subtracted from the signal obtained on the s-hCD13 surface to remove contribution from potential unspecific binding. The resulting affinities were 7.27×10−10 M for anti-hCD13-1 and 32.9×10−10 M for anti-hCD13-3, suggesting that anti-hCD13-3 had a lower affinity (Table 2 ). Therefore, the sequence differences in the VH regions of both scFvs may have resulted in measurably different affinities. The numerical values of the affinities were in the nanomolar range and below, which is typical for high affinity antibodies resulting from a secondary immunization (Griffiths et al., 1994). Thus, the novel procedure reported here was efficient in capturing scFvs with high affinities from immunized mice.
Fig. 7.

Determination of association, dissociation rate constants, and affinities for recombinant anti-hCD13 scFvs by surface plasmon resonance. The soluble chimeric protein s-hCD13 was deposited as a ligand on a Biacore CM5 sensor chip and purified soluble scFvs anti-hCD13-1 and -3 were then applied to the chip in increasing concentrations and allowed to bind. Binding was monitored by surface plasmon resonance using the BIACORE X™ system. (A, B) Association and dissociation profiles for scFvs hCD13-1 and -3, respectively. Numbers in the right margins are concentrations of purified soluble scFv used for each curve. Data are representative of two separate experiments.
Table 2.
Measurement of association and dissociation rate constants and affinities of scFvs anti-hCD13-1 and -3 by surface plasmon resonance
| scFv | ka (M−1 s−1) | kd (s−1) | KD (M) |
|---|---|---|---|
| ×105 | ×10−4 | ×10−10 | |
| CD13-1 | 4.42 | 3.13 | 7.27±1.75 |
| CD13-3 | 1.71 | 5.51 | 32.9±6.6 |
4. Discussion
The main result of this study was the establishment of a novel procedure for the generation of recombinant scFvs reactive with pre-selected antigens on the surface of human leukocytes. The procedure delivered scFvs reacting with hCD13 in its native state on the surface of intact cells with high affinity and without requiring an extensive purification of the antigen. The principle components and distinctive features of the procedure were the following: (a) the establishment of stably transfected NIH/3T3 mouse and Sf21 insect cell lines expressing high levels of hCD13 on their cell surface; (b) the purification of membranes from NIH/3T3 mouse cells expressing hCD13 by gentle extraction that maintained the antigen in a native state; (c) the immunization of mice with membranes containing hCD13 as the sole human antigen; (d) the pre-adsorption of the phage display library with non-transfected NIH/3T3 or Sf21 cells that allowed the removal of antibodies against alloantigens, as well as non-specific proteins; and (e) panning the pre-adsorbed library with hCD13-expressing human leukemia cells or insect cells.
Other related methods producing scFvs against cell surface antigens have been previously reported (Portolano et al., 1993, Cai and Garen, 1995, Cai and Garen, 1996, Cai and Garen, 1997, Siegel et al., 1997, Huls et al., 1999, Winthrop et al., 1999, Ridgway et al., 1999, Topping et al., 2000). In the majority of these cases, the libraries were panned subtractively with tumor- and non-tumor cell lines and the resulting scFvs were analyzed without prior knowledge of their identity and without specific features of the procedure designed to augment the efficiency of isolating antibodies against predetermined specific antigens. Only a few cases have been reported where scFvs against pre-selected specific cell surface antigens have been isolated by panning with intact cells (Portolano et al., 1993; Hoogenboom et al., 1999). The key feature of the procedure described here is the presentation of a defined single human antigen in its native state on membrane fragments purified from a transfected murine cell line. When membrane preparations from the hCD13-transfected NIH/3T3 cells were injected into recipient mice, we reasoned that the human CD13 antigen would be the major immunogenic molecule recognized by the immune system. A second important point was the screening strategy where hCD13 was the major common antigen between the material used for the immunization and the cells used for panning, allowing a more stringent positive selection on intact cells. To the best of our knowledge, a method employing a similar combination of steps has not been previously reported.
It is of interest to contrast the advantages and disadvantages of our procedure with those previously reported. One frequently employed method is the panning of complex non-immune (naive) or synthetic libraries (Griffiths et al., 1994, Vaughan et al., 1996, Knappik et al., 2000). The advantage of this procedure is that one universal library can be used to screen for scFvs reacting with many different antigens (Nissim et al., 1994). Synthetic libraries do not necessarily represent the naive repertoire, but often also contain high affinity scFvs because generation of these libraries includes hypermutation of the CDR3 region. Such libraries have been expanded to considerable complexity and libraries containing up to 109–1011 different scFvs have been reported (Vaughan et al., 1996, Knappik et al., 2000). One disadvantage of this approach is that the libraries, in spite of hypermutated CDR3 regions, have not been isolated from animals undergoing a secondary or tertiary immune response against a specific antigen. Although high affinity scFvs have been isolated using this approach, the probability of finding scFvs against a specific antigen is reduced, entailing the need to screen a large library of 109–1011 different clones (Perelson, 1989). Another disadvantage is that the isolation of specific binders to a defined antigen on the cell surface may be compromised using non-immune or synthetic libraries, because it has been reported that non-immune antibody libraries contain binders to dominant epitopes (Noronha et al., 1998, Hoogenboom et al., 1999). The binders to dominant epitopes may be preferentially amplified during screening and decrease the probability of isolating the desired binders to specific, predetermined antigens, especially when screening is performed using complex antigens, such as whole cells. In our procedure, mice were immunized with a specific antigen (hCD13) and then given multiple boosts until a secondary and possibly even a tertiary immune response was well underway. This strategy increased the likelihood of affinity maturation of antibodies in vivo and subsequent isolation of high affinity antibodies against hCD13. Furthermore, two different scFvs reacting specifically with hCD13 were obtained after screening a library of only 106 clones. Thus, the strategy described here allowed us to isolate high affinity scFvs against our antigen of choice from comparatively small libraries. The procedure should be useful for the preparation of scFvs against other conformation-sensitive cell surface antigens, for which cloned cDNA is available.
The procedure was not designed as a general procedure to isolate novel scFvs against a large variety of previously unrecognized cell surface antigens. Existing procedures involving subtractive whole cell-panning without the labor-intensive refinements added here may be sufficient or superior for this purpose. In addition, the scFvs isolated here are murine scFvs, with known disadvantages when used in human clinical applications. Although some of these disadvantages can be relieved by chimerizing or humanizing the murine scFvs, it may still be preferable for a majority of clinical applications to generate human scFvs from the beginning. Finally, we do not wish to detract from the major advantage of large combinatorial or synthetic naive human scFv-libraries. Clearly, their major advantage is, that from one library human scFvs against many different antigens can be isolated without the need for prior immunization. For a majority of general applications these existing procedures may be equal or superior to the procedure reported here. The strength of the novel procedure specifically lies in the generation of recombinant scFvs against conformation sensitive antigens, such as CD13, for which cDNAs are available, but for which no recombinant scFvs have previously been obtained, that recognize the antigen in its native state on intact cells. In this case, the advantage of our procedure is, that it avoids the need for an extensive biochemical purification of the antigen, which may destroy the conformation-sensitive epitope. This epitope may only exist when the antigen is present in its native conformation on the cell surface and may disappear when it is purified away from its nearest neighbor membrane components. For this narrow segment of applications, the procedure reported here presents significant progress, as demonstrated for the example of CD13, for which no scFvs had previously been isolated, that recognized the antigen in its native state.
The two scFvs reported here reacted with the antigen in its native state on living leukemia cells. Their affinities were high in comparison with published values (Griffiths et al., 1994). Antibodies with affinities in the nanomolar range or higher are high affinity antibodies (Vaughan et al., 1996). However, caution must be exerted when comparing the numbers obtained here with previously published figures, since different measurement procedures were utilized. In our case, soluble scFvs were used that were monovalent and association and dissociation rate constants were measured by surface plasmon resonance. In this method, one of the two binding partners was immobilized (in the solid phase); the other was freely mobile in the fluid phase. Reactions with immobilized partners have different kinetic parameters compared with reactions where both interacting partners remain in the fluid phase (Nieba et al., 1996). Furthermore, many published affinity constants were obtained by measurement procedures in which both the antibody and ligand were in fluid phase. Therefore, the numerical affinity values reported here are not strictly comparable to previously published values and are not intended to represent absolute values. They must be considered approximate values. With this proviso they indicate that scFvs of high affinity have been isolated by our procedure.
The antibodies against hCD13 isolated here may become useful for purposes similar to other monoclonal anti-hCD13 antibodies that have been previously described by others. These purposes include human cell- and tissue-typing (Goyert, 1997) and the intervention against cytomegalovirus including the removal of hCD13-positive cells from transplants in order to reduce the likelihood of transmission of hCMV with the transplant (Larsson et al., 1998). These scFvs may also become useful in the design of novel therapeutic approaches against human cancers aimed at interfering with angiogenesis (Pasqualini et al., 2000). Anti-hCD13 antibodies have been used successfully by other authors in a mouse model as active agents against HL60 myeloid leukemia cells (Xu and Scheinberg, 1995). While it is clear that scFvs of murine origin represent problems in human clinical applications, they may still be clinically useful, as demonstrated by antibodies against human CD20, that have recently been approved as human therapeutical agents and that were derived from murine monoclonal antibodies. In our perspective, these scFvs and the corresponding cDNAs may become valuable for the design of novel therapeutic strategies against t(4;11) and t(11;19) leukemia cells.
Acknowledgements
This work was supported by research grants from the Deutsche Forschungsgemeinschaft (DFG; SFB 473-B6) and grants 96.047.1 from the Wilhelm Sander Foundation to GHF and SJZ. We thank Prof. M. Gramatzki for providing the CEM cell line; Prof. A. Plückthun for the scFv vectors; Dr. L. Shapiro for the human CD13 cDNA clone; and Dr. G. Winter for E. coli TG1. J. Dörrie, A. Birkmann and Dr. R. Slany are acknowledged for valuable discussions and Th. Lange for administrative assistance.
References
- Behm F.G., Raimondi S.C., Frestedt J.L., Liu Q., Crist W.M., Downing J.R., Rivera G.K., Kersey J.H., Pui C.H. Rearrangement of the MLL gene confers a poor prognosis in childhood acute lymphoblastic leukemia, regardless of presenting age. Blood. 1996;87:2870–2877. [PubMed] [Google Scholar]
- Cai X., Garen A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: selection of specific antibodies from single-chain Fv fusion phage libraries. Proc. Natl. Acad. Sci. USA. 1995;92:6537–6541. doi: 10.1073/pnas.92.14.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Garen A. A melanoma-specific VH antibody cloned from a fusion phage library of a vaccinated melanoma patient. Proc. Natl. Acad. Sci. USA. 1996;93:6280–6285. doi: 10.1073/pnas.93.13.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Garen A. Comparison of fusion phage libraries displaying VH or single-chain Fv antibody fragments derived from the antibody repertoire of a vaccinated melanoma patient as a source of melanoma-specific targetting molecules. Proc. Natl. Acad. Sci. USA. 1997;94:9261–9266. doi: 10.1073/pnas.94.17.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury P.S., Viner J.L., Beers R., Pastan I. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with anti-tumor activity. Proc. Natl. Acad. Sci. USA. 1998;95:669–674. doi: 10.1073/pnas.95.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L’Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eylar E., Hagopian A. Isolation of plasma membranes from mammalian cells. In: Jakoby W.B., editor. Vol. XXII. Academic Press; London: 1971. (Methods in Enzymology: Enzyme Purification and Related Techniques). [Google Scholar]
- Fujii H., Nakajima M., Saiki I., Yoneda J., Azuma I., Tsuruo T. Human melanoma invasion and metastasis enhancement by high expression of aminopeptidase N/CD13. Clin. Exp. Metastasis. 1995;13:337–344. doi: 10.1007/BF00121910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giugni T.D., Soderberg C., Ham D.J., Bautista R.M., Hedlund K.O., Moller E., Zaia J.A. Neutralization of human cytomegalovirus by human CD13-specific antibodies. J. Infect. Dis. 1996;173:1062–1071. doi: 10.1093/infdis/173.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyert S.M. CD13 workshop panel report. In: Kishimoto T., Kikutani H., von dem Borne A.E.G.K., Goyert S.M., Mason D.Y., Miyasaka M., Moretta L., Okumura K., Shaw S., Springer T.A., Sugamura K., Zola H., editors. Vol. VI. Garland Publishing; New York: 1997. pp. 962–963. (Leukocyte Typing). [Google Scholar]
- Greil J., Gramatzki M., Burger R., Marschalek R., Peltner M., Trautmann U., Hansen-Hagge T.E., Bartram C.R., Fey G.H., Stehr K. The acute lymphoblastic leukaemia cell line SEM with t(4;11) chromosomal rearrangement is biphenotypic and responsive to interleukin-7. Br. J. Haematol. 1994;86:275–283. doi: 10.1111/j.1365-2141.1994.tb04726.x. [DOI] [PubMed] [Google Scholar]
- Griffiths A.D., Williams S.C., Hartley O., Tomlinson I.M., Waterhouse P., Crosby W.L., Kontermann R.E., Jones P.T., Low N.M., Allison T.J. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. Antibodies — A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Heerema N.A., Sather H.N., Ge J., Arthur D.C., Hilden J.M., Trigg M.E., Reaman G.H. Cytogenetic studies of infant acute lymphoblastic leukemia: poor prognosis of infants with t(4;11) — a report of the Children’s Cancer Group. Leukemia. 1999;13:679–686. doi: 10.1038/sj.leu.2401413. [DOI] [PubMed] [Google Scholar]
- Hoogenboom H.R., Lutgerink J.T., Pelsers M.M., Rousch M.J., Coote J., Van Neer N., De Bruine A., Van Nieuwenhoven F.A., Glatz J.F., Arends J.W. Selection-dominant and nonaccessible epitopes on cell-surface receptors revealed by cell-panning with a large phage antibody library. Eur. J. Biochem. 1999;260:774–784. doi: 10.1046/j.1432-1327.1999.00214.x. [DOI] [PubMed] [Google Scholar]
- Hooper N.M. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Huls G.A., Heijnen I.A., Cuomo M.E., Koningsberger J.C., Wiegman L., Boel E., van der Vuurst de Vries A.R., Loyson S.A., Helfrich W., van Berge Henegouwen G.P., van Meijer M., de Kruif J., Logtenberg T. A recombinant, fully human monoclonal antibody with antitumor activity constructed from phage-displayed antibody fragments. Nat. Biotechnol. 1999;17:276–281. doi: 10.1038/7023. [DOI] [PubMed] [Google Scholar]
- Kazemier B., de Haard H., Boender P., van Gemen B., Hoogenboom H. Determination of active single chain antibody concentrations in crude periplasmic fractions. J. Immunol. Methods. 1996;194:201–209. doi: 10.1016/0022-1759(96)00086-5. [DOI] [PubMed] [Google Scholar]
- Kipriyanov S. Purification, characterization and biotinylation of single chain antibodies. In: Reischl U., editor. Vol. 13. Humana Press; Totowa, NY: 1997. (Methods in Molecular Medicine: Molecular Diagnosis of Infectious Diseases). [DOI] [PubMed] [Google Scholar]
- Knappik A., Ge L., Honegger A., Pack P., Fischer M., Wellnhofer G., Hoess A., Wolle J., Pluckthun A., Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- Krebber A., Bornhauser S., Burmester J., Honegger A., Willuda J., Bosshard H.R., Pluckthun A. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods. 1997;201:35–55. doi: 10.1016/s0022-1759(96)00208-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson S., Soderberg-Naucler C., Moller E. Productive cytomegalovirus (CMV) infection exclusively in CD13-positive peripheral blood mononuclear cells from CMV-infected individuals: implications for prevention of CMV transmission. Transplantation. 1998;65:411–415. doi: 10.1097/00007890-199802150-00021. [DOI] [PubMed] [Google Scholar]
- Look A.T., Ashmun R.A., Shapiro L.H., Peiper S.C. Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J. Clin. Invest. 1989;83:1299–1307. doi: 10.1172/JCI114015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrynikola V., Favaloro E.J., Browning T., Bianchi A., Bradstock K.F. Functional and phenotypic upregulation of CD13/aminopeptidase-N on precursor-B acute lymphoblastic leukemia after in vitro stimulation. Exp. Hematol. 1995;23:1173–1179. [PubMed] [Google Scholar]
- Mizutani S., Goto K., Nomura S., Ino K., Goto S., Kikkawa F., Kurauchi O., Goldstein G., Tomoda Y. Possible action of human placental aminopeptidase N in feto-placental unit. Res. Commun. Chem. Pathol. Pharmacol. 1993;82:65–80. [PubMed] [Google Scholar]
- Nieba L., Krebber A., Pluckthun A. Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal. Biochem. 1996;234:155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- Nissim A., Hoogenboom H.R., Tomlinson I.M., Flynn G., Midgley C., Lane D., Winter G. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 1994;13:692–698. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha E.J., Wang X., Desai S.A., Kageshita T., Ferrone S. Limited diversity of human scFv fragments isolated by panning a synthetic phage-display scFv library with cultured human melanoma cells. J. Immunol. 1998;161:2968–2976. [PubMed] [Google Scholar]
- Olsen J., Kokholm K., Noren O., Sjostrom H. Structure and expression of aminopeptidase N. Adv. Exp. Med. Biol. 1997;421:47–57. doi: 10.1007/978-1-4757-9613-1_7. [DOI] [PubMed] [Google Scholar]
- Pasqualini R., Koivunen E., Kain R., Lahdenranta J., Sakamoto M., Stryhn A., Ashmun R.A., Shapiro L.H., Arap W., Ruoslahti E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- Perelson A.S. Immune network theory. Immunol. Rev. 1989;110:5–36. doi: 10.1111/j.1600-065x.1989.tb00025.x. [DOI] [PubMed] [Google Scholar]
- Portolano S., McLachlan S.M., Rapoport B. High affinity, thyroid-specific human autoantibodies displayed on the surface of filamentous phage use V genes similar to other autoantibodies. J. Immunol. 1993;151:2839–2851. [PubMed] [Google Scholar]
- Ridgway J.B., Ng E., Kern J.A., Lee J., Brush J., Goddard A., Carter P. Identification of a human anti-CD55 single-chain Fv by subtractive panning of a phage library using tumor and nontumor cell lines. Cancer Res. 1999;59:2718–2723. [PubMed] [Google Scholar]
- Riemann D., Kehlen A., Langner J. CD13 — not just a marker in leukemia typing. Immunol. Today. 1999;20:83–88. doi: 10.1016/S0167-5699(98)01398-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki I., Fujii H., Yoneda J., Abe F., Nakajima M., Tsuruo T., Azuma I. Role of aminopeptidase N (CD13) in tumor-cell invasion and extracellular matrix degradation. Int. J. Cancer. 1993;54:137–143. doi: 10.1002/ijc.2910540122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbour Laboratory Press; Cold Spring Harbour, NY: 1989. [Google Scholar]
- Shapiro L.H., Ashmun R.A., Roberts W.M., Look A.T. Separate promoters control transcription of the human aminopeptidase N gene in myeloid and intestinal epithelial cells. J. Biol. Chem. 1991;266:11999–12007. [PubMed] [Google Scholar]
- Shipp M.A., Look A.T. Hematopoietic differentiation antigens that are membrane-associated enzymes: cutting is the key! Blood. 1993;82:1052–1070. [PubMed] [Google Scholar]
- Siegel D.L., Chang T.Y., Russell S.L., Bunya V.Y. Isolation of cell surface-specific human monoclonal antibodies using phage display and magnetically-activated cell sorting: applications in immunohematology. J. Immunol. Methods. 1997;206:73–85. doi: 10.1016/s0022-1759(97)00087-2. [DOI] [PubMed] [Google Scholar]
- Soderberg C., Giugni T.D., Zaia J.A., Larsson S., Wahlberg J.M., Moller E. CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J. Virol. 1993;67:6576–6585. doi: 10.1128/jvi.67.11.6576-6585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H., Lanier L.L., Phillips J.H. Development of human T and natural killer cells. Blood. 1995;85:2654–2670. [PubMed] [Google Scholar]
- Stong R.C., Korsmeyer S.J., Parkin J.L., Arthur D.C., Kersey J.H. Human acute leukemia cell line with the t(4;11) chromosomal rearrangement exhibits B lineage and monocytic characteristics. Blood. 1985;65:21–31. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping K.P., Hough V.C., Monson J.R., Greenman J. Isolation of human colorectal tumor reactive antibodies using phage display technology. Int. J. Oncol. 2000;16:187–195. doi: 10.3892/ijo.16.1.187. [DOI] [PubMed] [Google Scholar]
- Vaughan T.J., Williams A.J., Pritchard K., Osbourn J.K., Pope A.R., Earnshaw J.C., McCafferty J., Hodits R.A., Wilton J., Johnson K.S. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- Winthrop M.D., DeNardo S.J., DeNardo G.L. Development of a hyperimmune anti-MUC-1 single chain antibody fragments phage display library for targeting breast cancer. Clin. Cancer Res. 1999;5:3088s–3094s. [PubMed] [Google Scholar]
- Xu Y., Scheinberg D.A. Elimination of human leukemia by monoclonal antibodies in an athymic nude mouse leukemia model. Clin. Cancer Res. 1995;1:1179–1187. [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]


