Abstract
Advancements in molecular technologies have provided new platforms that are being increasingly adopted for use in the clinical microbiology laboratory. Among these, microarray methods are particularly well suited for diagnostics as they allow multiplexing, or the ability to test for multiple targets simultaneously from the same specimen. Microarray technologies commonly used for the detection and identification of microbial targets include solid-state microarrays, electronic microarrays and bead suspension microarrays. Microarray methods have been applied to microbial detection, genotyping and antimicrobial resistance gene detection. Microarrays can offer a panel approach to diagnose specific patient presentations, such as respiratory or gastrointestinal infections, and can discriminate isolates by genotype for tracking epidemiology and outbreak investigations. And, as more information has become available on specific genes and pathways involved in antimicrobial resistance, we are beginning to be able to predict susceptibility patterns based on sequence detection for particular organisms. With further advances in automated microarray processing methods and genotype–phenotype prediction algorithms, these tests will become even more useful as an adjunct or replacement for conventional antimicrobial susceptibility testing, allowing for more rapid selection of targeted therapy for infectious diseases.
Keywords: Solid-state microarrays, Suspension microarrays, Pathogen detection, Genotyping, Antimicrobial resistance, Microbial genomics
1. Introduction
Microarrays can be most simply defined as a collection of microscopic spots (or features) of biological capture molecules (such as DNA or proteins) spatially arranged in a predefined order on a solid substrate. In DNA microarrays (also known as DNA chips or biochips), each spot contains predefined short (25- to 70-nucleotide) single-stranded DNA oligonucleotides or larger (200- to 800-basepair) double-stranded DNA, which are known as probes. A microarray experiment consists of the hybridisation of a sample of target DNA in solution, whose identity or abundance is being sought, with a typically large number of probes fixed to the substrate. Target DNA molecules are prepared with fluorescent tags, the microarray is exposed to the resulting mixture under conditions that favour hybridisation, and any unbound DNA fragments are washed away. Microarray positions with DNA–DNA hybrids are then detected optically, indicating the components that were present in the starting sample. Each microarray experiment is therefore a large number of hybridisation experiments done in parallel and as such, microarrays can be thought of as ‘highly parallel’ processors of biological queries. Various microarray methods are available and differ in the type of the probe, the solid support and the methods used for addressing the probe and detecting the target (Miller & Tang, 2009). Microarray technologies commonly used for the detection and identification of microbial targets include solid-state microarrays, electronic microarrays and suspension bead microarrays. Details of these technologies, including probe length, number of possible features (or density) and surface chemistry employed, are described below and compared in Table 1 . In this chapter, we review the concepts behind each of these microarray technologies and describe clinical applications of microarray analysis for pathogen detection, genotyping and antimicrobial resistance testing in the microbiology laboratory.
Table 1.
Comparison of Microarray Technologies
| Technology | Principle | Probe Length | Features | Solid Support | References |
|---|---|---|---|---|---|
| Printed | Pre-synthesised probes are deposited onto solid surface and attached by covalent or electrostatic chemistry | 25–80 base | ≤ 30,000 | Glass slide | Goldmann and Gonzalez (2000) and Lausted et al. (2004) |
| In situ synthesised with photolithographic mask | Probes are synthesised in parallel directly on solid surface with phosphoramidite protection/deprotection chemistry using mask-directed photolithography | 20–25 base | > 1,000,000 | Silicon chip | Beier and Hoheisel (2000) and Pease et al. (1994) |
| In situ synthesised with digital micromirror | Probes are synthesised in parallel directly on solid surface with phosphoramidite protection/deprotection chemistry using maskless photodeprotection with digital micromirror devices | 60–100 base | 15,000–>1,000,000 | Silicon chip | Singh-Gasson et al. (1999) |
| In situ synthesised with inkjet | Probes are synthesised in parallel directly on solid surface with phosphoramidite protection/deprotection chemistry using acid-mediated chemical deprotection with inkjet printing | 60 base | 15,000–>1,000,000 | Glass slide | Hughes et al. (2001) |
| Electronic | Pre-synthesised probes are transported to specific locations on the solid surface by electric field and immobilised by streptavidin-biotin bonding | 20–30 base | 400 | Silicon chip | Sosnowski, Tu, Butler, O'Connell, and Heller (1997) and Edman et al. (1997) |
| Suspension bead array | Pre-synthesised probes are covalently coupled to solid surface | 15–100 base | ≤ 500 | Polystyrene microspheres | Dunbar (2013) and Dunbar and Jacobson (2007) |
2. Solid-State Microarrays
Solid-state (also called solid-phase) microarrays, in particular, use rigid, impermeable substrates (such as glass or silicon wafers) as the solid support (Foglieni et al., 2010, Palmieri et al., 2008). The microscopic features, each with thousands of identical probes immobilised to the solid surface, are arranged in an orderly fashion at high density. Solid-state microarrays are the technological descendants of ‘dot-blotting’ techniques (Kafatos, Jones, & Efstratiadis, 1979), which involved the application of macroscopic spots over porous membranes (i.e. nitrocellulose or gel pads). The introduction of solid substrates was critical to the success of DNA microarrays for high-throughput large-scale biological applications (Khrapko et al., 1989). With probes tethered to a solid surface rather than being buried in pores, the rate of hybridisation is significantly increased and the subsequent washing steps are unhampered by diffusion effect. The rigidity of the solid support improves image acquisition and processing by producing sharper images, and high image definition is indispensable to the micron-scale feature sizes achieved on these microarrays.
2.1. Printed Microarrays
Printed (spotted) microarrays are built by depositing pre-synthesised probes onto glass slides using fine-pointed pins (Figure 1 ; Goldmann and Gonzalez, 2000, Lausted et al., 2004). The probes can be short oligonucleotides, cDNA or PCR products, and the surface attachment chemistry can be either covalent or non-covalent. Covalent attachment requires the addition of an aliphatic amine (NH2) group to the 5′-end of the probe. The probes are then attached to the glass from their 5′-end. Non-covalent attachment involves electrostatic attraction between the amine groups on the glass slide and the phosphate group of the probe's backbone. Non-covalent interactions are not strong enough to tether short oligonucleotide probes to glass, thus non-covalent attachment is more appropriate for cDNA microarrays.
Figure 1.

Printed microarrays. (A) Pre-synthesised oligonucleotide or DNA probes are spotted onto glass slides using fine-pointed pins. After printing, the probes are chemically fixed to the slide. (B) Samples containing target DNA are processed through extraction, labelling and hybridisation to the microarray. (C) The hybridised microarray is scanned, and the raw images are processed and analysed.
The delivery of probes onto the glass slide is achieved with spotting robots using either contact or non-contact microarray printing. Contact printing technologies have been used since the early days of microarray technology. They are based on pin tools, which are dipped into the sample solution to take up a small volume of sample and then brought in contact with the slide surface to deliver the sample as a small spot. The currently available pins deliver spot volumes in 0.5–12 nl range, resulting in spot diameters from 62.5 to 600 μm. Non-contact printing enables microarray printing without direct contact to the surface. It uses piezoelectric, bubble-generated and micro-solenoid-driven dispensers that work with the same principle as inkjet printers. These systems are capable of dispensing single drops down to a volume of several hundred picolitres.
After the probes are printed, the surface of the array is usually chemically modified (fixed) making the attachment of additional DNA unfeasible. Fixing is desired to avoid non-specific binding of the DNA target from the sample to the glass slide during hybridisation. Post-production modification of the surface to make it more hydrophilic is also common so as to facilitate the mixing of the target DNA during hybridisation.
2.2. In Situ-Synthesised Microarrays
Probes for very high-density microarrays are synthesised base-by-base and in situ on the solid surface of the microarray. The most straightforward method for synthesising oligonucleotides involves adding nucleotides one at a time to the growing end of an oligonucleotide chain. In this method, the sequence of the oligonucleotide chain is dictated by the order in which dNTPs are added to the reaction. For solid-phase in situ chemical synthesis of DNA, an alternative chemical oligonucleotide synthesis based on phosphoramidite (protection/deprotection) chemistry is coupled with a solid support containing covalent linker molecules (Beier & Hoheisel, 2004). Phosphoramidite building blocks (phosphoramidite nucleosides) derived from chemically protected 2′-deoxynucleosides (dA, dC, dG and dT) are used in phosphoramidite protection/deprotection chemistry. The protection of the amine, phosphate and hydroxyl reactive groups ensures the prevention of unwanted side reactions. For the protection of the exocyclic amino group, standard schemes use a benzoyl group for adenosine and cytidine and an isobutyryl group for guanosine. Thymidine does not have a reactive exocyclic amino group and hence does not need any protection.
The in situ synthesis of the oligonucleotide chain using protection/deprotection chemistry consists of reaction cycles, where each cycle of reactions results in the addition of a single base. The base added has a protective group on its 5′-end to prevent the addition of more than one base during each round of synthesis. At the initial stage, the terminal 3′-base is covalently attached to the solid support. Oligonucleotides are synthesised in the 3′- to 5′-direction (as opposed to the 5′- to 3′-direction in DNA replication) by repeating the synthesis cycle, as many times as required until the desired length of DNA is achieved. A protection/deprotection synthesis cycle has the following four conceptual steps:
-
1.
Deprotection: the protecting group of the 5′-terminal nucleotide is removed, yielding a reactive 5′-hydroxyl (OH) ready to bond with the as yet to be added phosphoramidite.
-
2.
Coupling: an activated nucleoside phosphoramidite is brought in contact with the support-bound oligonucleotide precursor. The 5′-OH group reacts with the activated phosphoramidite moiety forming a phosphite triester bond. This is an unstable linkage for the oligonucleotide at the end of the coupling reaction and a small fraction of the 5′-OH groups remains unreacted. The fraction of the newly added bases that are actually incorporated into the growing DNA chain is called coupling efficiency. One desired property of a high-yield nucleotide synthesis scheme is a high coupling efficiency.
-
3.
Capping: the permanent blocking of the unreacted 5′-OH groups by treating with a chemical catalyst mixture.
-
4.
Oxidation: the phosphite triester bond formed after coupling is unstable under the oligonucleotide synthesis conditions. An oxidation step involves the oxidation of the trivalent phosphite to a more stable pentavalent phosphate.
Different methods used in the deprotection step result in three technologies for in situ-synthesised microarrays. These three technologies differ in whether they use photo- or acid-labile protecting groups for 5′-OH group of the nucleoside phosphoramidite and in how they achieve the combinatorial chemistry involved in parallel synthesis of thousands of probes.
2.2.1. Photodeprotection using photolithographic mask
In this method, which forms the basis of Affymetrix® GeneChip® technology, light is used to convert the protective group on the 5′-terminal nucleotide into an OH group (Figure 2 ). During each round of synthesis, the selective exposure of specific array locations (features) to light is achieved with a physical chromium mask that allows light to reach only certain areas of the microarray. This technique, known as photolithography, was adapted from the manufacturing of silicon microprocessor chips (Beier and Hoheisel, 2000, Pease et al., 1994). Those features exposed to light become active for further synthesis. The microarray surface is then flushed with a solution containing a single type of nucleotide, but the chemical coupling occurs only in those features that have been deprotected. The newly coupled nucleotides also have a protecting group so the process can be repeated with the application of another mask and subsequent flushing of another nucleotide, until all probes have been fully synthesised. Each probe on the microarray requires four masks per round of synthesis. The probes on an Affymetrix GeneChip microarray are 25 nucleotides long, requiring nearly 100 masks per microarray. The photolithographic masks are expensive to produce but once designed and manufactured, they can be reused to make a large number of identical microarrays with high fidelity. Thus, Affymetrix GeneChip technology is well positioned for making large numbers of standardised microarrays.
Figure 2.
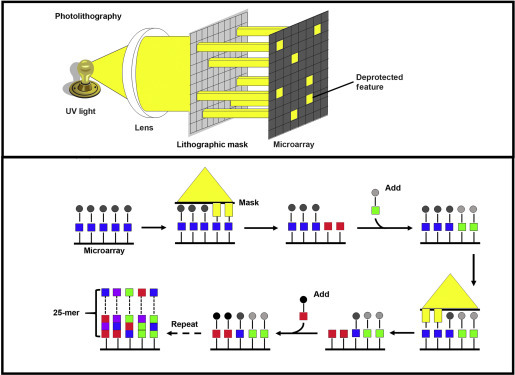
In situ-synthesised microarray using mask-assisted photolithography. (Top) A lithographic mask is used to transmit or block UV light from chemically protected nucleotides on the microarray surface. (Bottom) Protecting groups (circles) are removed by UV exposure, allowing addition of a single protected nucleotide (square). The cycle is repeated using different masks sequentially to dictate the order of nucleotide sequence in the oligonucleotide probes.
2.2.2. Maskless photodeprotection using digital micromirror devices
Maskless photodeprotection is also based on light-directed synthesis but uses a digital micromirror device for selective light exposure of array features (Singh-Gasson et al., 1999). This is the method behind the Roche NimbleGen Maskless Array Synthesiser (MAS) technology. A digital micromirror device is a solid-state silicon device with an array of aluminium mirrors that can be computer controlled to direct light to desired locations of the solid microarray surface (Figure 3 ). It effectively creates virtual masks replacing standard photolithographic masks. NimbleGen's digital micromirror device is able to pattern up to 786,000 individual pixels of light which enables minimum feature sizes of 17 μm. Since there are no physical masks to produce, the primary expense of the maskless approach is the chemistry. The flexibility of the virtual masks enables production of small batches of high-density microarrays with different array layouts in a matter of days.
Figure 3.
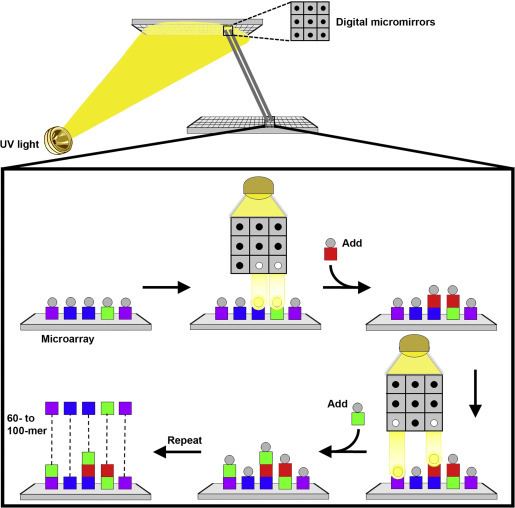
Maskless in situ-synthesised microarray using digital micromirrors. Digital micromirror technology is used to create virtual masks for photodeprotection. UV light is directed through the mask image onto the microarray surface to remove the protecting group (circles) and allow addition of a single nucleotide (square). The steps are repeated with different virtual masks to direct the synthesis of the desired oligonucleotides in a selected pattern.
2.2.3. Chemical deprotection with inkjet-mediated synthesis
This method underlying Agilent Technologies’ microarrays couples chemical acid-mediated deprotection with inkjet printing to deposit 60 basepair oligonucleotides one base at a time on standard glass slides (Hughes et al., 2001). The deprotection of the DMT-protected nucleoside phosphoramidite is achieved with the removal of the DMT with trichloroacetic acid. In the coupling step of the synthesis cycle, the inkjet printer uses tiny nozzles for each monomer and chemical activators to place picolitre drops of reagents in specified locations on the microarray (Figure 4 ).
Figure 4.

In situ-synthesised microarray using chemical deprotection with inkjet-mediated synthesis. This method uses acid-mediated deprotection with inkjet printing for oligonucleotide probe synthesis. The printer uses tiny nozzles to deposit picolitre amounts of nucleotides and reagents in specified locations of the microarray. Repeated rounds of printing extend the probe length to 60 nucleotides.
2.2.4. Comparison of in situ synthesis technologies
Micromirror and inkjet technologies allow computer controlled synthesis of each feature at the time of array production, while photolithography requires physical pre-made masks. This makes the former technologies highly flexible but these technologies are less efficient for making large batches of identical arrays compared to photolithographic masking. Variants of phosphoramidite-based synthesis have different coupling efficiencies. The yield of the desired probes is dependent on the length of the oligonucleotide being synthesised. With low coupling efficiencies and longer probes, eventually the yield of full-length oligos becomes unacceptably low. Photodeprotection and acid-mediated deprotection of dimethoxytrityl protecting group have coupling efficiencies of approximately 95% and 98%, respectively (McGall et al., 1997). The chemical deprotection used in inkjet-mediated synthesis allows the inkjet technology to be more suitable for high-fidelity synthesis of longer oligonucleotides.
2.3. Electronic Microarrays
The printed and in situ-synthesised microarrays rely on passive transport for the hybridisation of nucleic acids. In contrast, electronic microarrays utilise active hybridisation via electric fields to control nucleic acid transport (Figure 5 ). Commercially available as NanoChip® XL by Savyon Diagnostics (formerly NanoChip® 400 from Nanogen, Inc.), the core technology of the microelectronic cartridge uses complementary metal oxide semiconductor technology for the electronic addressing of nucleic acids (Sosnowski et al., 1997). The cartridge has 12 connectors that control 400 individual test sites. The microarray is generated by transport of negatively charged nucleic acids to specific sites when an electric field is applied to one or more test sites on the microarray (Edman et al., 1997). The surface of the microarray contains streptavidin, which allows the formation of streptavidin–biotin bonds with biotinylated probes reaching each location through the application of a specific electrical field, thus immobilising the probe onto the surface. The electric field is then removed from the active features, and new test sites can be activated.
Figure 5.
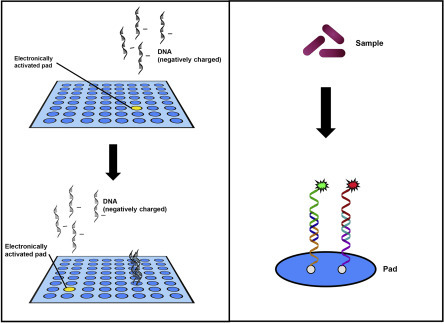
Electronic microarray. (Left) Positive current is applied to specific test sites (pads) to assist movement of negatively charged DNA probes to the activated pads. Probes are attached by streptavidin–biotin binding. The site is deactivated and the current applied to a different pad to localise a new probe. The process is repeated until all of the probes are arrayed. (Right) Samples are extracted, amplified and hybridised to the microarray surface. Target-specific secondary probes and fluorescent detector oligonucleotides are used to measure positive hybridisation reactions.
Once the probes have been hybridised at discrete test sites, the microarray is ready for the application of fluorescently labelled target DNA. Target DNA passively hybridises with the immobilised probes on the microarray or is concentrated electronically (Barlaan et al., 2005, Kumar et al., 2008). Electronically addressing the capture probe first is the most commonly used assay format, but amplicon first and sandwich assays have also been described (Feng & Nerenberg, 1999). If hybridisation occurs between the probe and the target DNA, fluorescent reporters will be present at the positive test site(s) and will be detected when the electronic microarray is scanned and analysed by an imaging instrument using a charge-coupled device (CCD) camera with data collection and analysis software.
3. Suspension Bead Arrays
Suspension bead microarray technologies are three-dimensional arrays that use microparticles (microspheres or beads) as the solid support for the binding reaction and flow cytometry or CCD imaging for detection of the bead and associated target (Figure 6 ). The particles can be distinguished into subsets by different classification features, such as internally absorbed fluorophores, size or diameter and surface or composition characteristics, and multiplexing is achieved through the unique features of the particle subsets. The xMAP® Technology platform by Luminex® is a bead-based suspension microarray technology commonly used for detection of nucleic acids and utilises polystyrene microspheres that are internally dyed with two or three spectrally distinct fluorochromes (Dunbar, 2006). Using precise amounts of each of these fluorochromes, an array is created consisting of different bead sets with specific spectral addresses. A specific capture oligonucleotide probe is covalently coupled to the surface of the specific bead sets or bead sets precoupled with unique capture oligonucleotides are commercially available for nucleic acid microarray assay development. The target nucleic acid is captured by hybridisation to the complementary probe sequence and labelled with a reporter molecule (i.e. fluorescent dye) to allow detection and quantitation of the specific binding that has occurred at the particle surface. Multiple readings are made per bead set or target, providing a fluorescent output signal of the hybridisation results.
Figure 6.
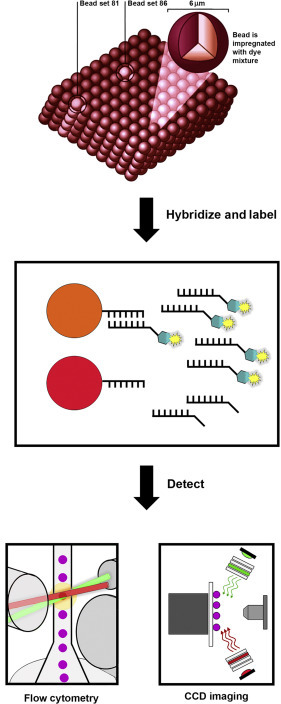
Suspension bead array. Microspheres (beads) are internally dyed with different intensities of red and infrared dyes to create 500 beads sets with unique spectral identities. Bead sets coupled with specific oligonucleotide probes are hybridised to amplified targets generated from the starting sample. Hybridised targets are labelled using a fluorescent reporter dye, and the microsphere suspension is analysed by flow cytometry or CCD imaging to identity each bead and quantify the reporter probe-target reaction on the microsphere surface.
A variety of assay chemistries can be used for generating the DNA targets for suspension bead hybridisation, including direct capture, competitive hybridisation and solution-based enzymatic chemistries in combination with universal capture probes (Figure 7 ).
Figure 7.
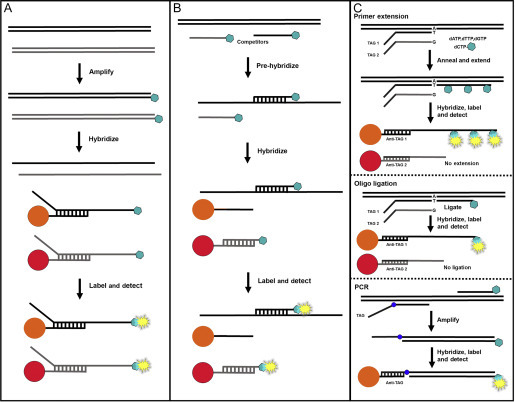
Suspension bead assay formats. (A) In direct hybridisation, target DNA is amplified and biotin labelled. The amplified products are denatured, hybridised to specific probe-coupled bead sets and labelled with streptavidin-linked fluorescent reporter. (B) In competitive hybridisation, labelled oligonucleotides containing the target sequences compete with target DNA in the test sample for hybridisation to specific probe-coupled bead sets. The sample target DNA concentration is inversely proportional to the bead-associated reporter signal, which decreases as sample concentration increases. (C) Enzymatic assay chemistries (primer extension, oligonucleotide ligation and PCR) are used to identify the target sequences in the sample and incorporate specific capture tag sequences. The targets are hybridised to bead sets bearing complementary anti-TAG probes, and labelled for detection.
3.1. Direct Hybridisation
Direct hybridisation of a labelled PCR-amplified target DNA to bead sets bearing specific oligonucleotide capture probes is a simple and commonly used assay chemistry and typically employs probes of approximately 20 nucleotides in length (Dunbar, 2006, Dunbar and Jacobson, 2007). The probes are complementary in sequence to the labelled strand of the PCR product and the polymorphic nucleotide (for single-nucleotide discrimination) is located at or near the centre of the probe. The oligo probes are modified with a 5′-terminal amine for covalent attachment to the functional carboxyl group on the microsphere surface and contain a spacer (usually C-12) between the amine and probe sequence to minimise steric hindrance of hybridisation at the microsphere surface. PCR primers are typically designed to amplify 50- to 300-basepair regions of target sequence with one primer of each pair biotinylated at the 5′ end for labelling the target strand of the amplicon. Using a small target DNA minimises the potential for steric hindrance to affect hybridisation efficiency adversely at the bead surface (Dunbar, 2006).
3.2. Competitive Hybridisation
Competitive hybridisation is similar to direct hybridisation in probe and target design. However, in the competitive assay format, unlabelled double-stranded PCR-amplified targets compete with labelled single-stranded oligonucleotide targets for annealing to the sequence-specific capture probes on the microspheres (Fulton, McDade, Smith, Kienker, & Kettman, 1997).
3.3. Enzymatic Chemistries with Universal Capture Probes
Another approach is to use a sequence-specific enzymatic reaction in solution to identify the target DNA, followed by capture onto a universal probe sequence on the bead surface for detection. This format involves the incorporation of a capture sequence during the enzymatic step that allows hybridisation to a complementary sequence attached to the bead surface. Commonly used enzymatic methods for sequence determination rely on the discriminating ability of DNA polymerases and DNA ligases, and include allele-specific or target-specific primer extension, oligonucleotide ligation, single-base chain extension and target-specific PCR (Dunbar, 2013, Taylor et al., 2001, Ye et al., 2001). These chemistries are used in combination with bead reagents precoupled with universal oligonucleotide capture sequences that are chosen to have identical hybridisation parameters with minimal cross-reactivity (Mahony et al., 2007). This approach takes advantage of the speed of solution-phase hybridisation and permits the universal bead sets to be used in many different assays where new sequences can be targeted by adding the appropriate capture sequence to the target-specific oligonucleotide used in the enzymatic step.
4. Clinical Applications of Microarray Testing
The detection and genetic characterisation of microbial pathogens by microarray methods have been developed for several applications in the clinical microbiology laboratory. The large number of microarray probes available in several commercial platforms allows for highly multiplexed assays, enabling detection of a broad range of organisms and/or discrimination of multiple genetic elements within the targeted species (Miller & Tang, 2009). At the current time, the clinical utility of microarray methods is primarily limited by our lack of detailed understanding about how genetic changes affect organism phenotype and outcomes of infections. The rapid expansion of knowledge about multigenic virulence and resistance traits will allow for clinically actionable interpretation of complex microarray data and additional uses for this testing in patient care. While other molecular methods such as next-generation sequencing can yield the same or additional genetic data for interpretation, targeted microarray methods will likely continue to be used in conjunction with high-throughput sequencing, as they can provide more rapid or cheaper tests for particular indications (Sibley, Peirano, & Church, 2012).
The combination of microbial detection and genotypic characterisation in a single assay is one of the attractions of microarray testing in the clinical laboratory. Probes specific for individual pathogens can be utilised, along with genetic markers such as resistance elements, to provide information enabling early optimisation of therapy. For some pathogens such as Mycobacterium tuberculosis, phenotypic susceptibility testing can take days or weeks to complete, and empiric treatment must be initiated in the interim (Neonakis et al., 2008, Schanne et al., 2008). Often in bacterial infections, phenotypic susceptibility test results are available within 24–48 h, and the utilisation of microarray testing will depend on factors such as turnaround time, cost and the failure rate of empiric antibiotic coverage. It should be noted that the sensitivity may vary for species detection versus resistance markers, so that resistance prediction could be less reliable for organisms present in low titers (Thissen et al., 2014). Guidelines have been developed for validation and implementation of microarrays for diagnosis of infectious disease in clinical laboratories (Clinical and Laboratory Standards Institute, 2014).
4.1. Pathogen Detection
Most microarrays detect target organisms based on conserved gene targets that are amplified and labelled prior to hybridisation and detection. Therefore, the spectrum of detection depends on both the number of available probes covering the desired species and the ability to amplify the regions of interest using conserved primers. No single target gene or genetic region will allow for detection of all pathogens of interest, so microarrays will typically target one or more classes of organisms. The most commonly used gene targets are listed in Table 2 , and include 16S ribosomal DNA (rDNA) (bacteria), internal transcribed spacers for 18S or 28S rDNA (fungi) and rpoB (mycobacteria) (Baker et al., 2003, Gingeras et al., 1998, McCabe et al., 1999). Because it is not possible to design a primer pair that is completely conserved among all prokaryotic species for any gene target, PCR may be performed using degenerate primers or under conditions that allow for primer mismatches to still amplify the expected product. Therefore, species with lower sequence identity to primer regions can be identified, but require higher numbers of organisms for detection. More distantly related organisms may not be detectable, and the diversity of detectable species can be predicted based on known sequences or determined experimentally (Baker et al., 2003, McCabe et al., 1999).
Table 2.
Conserved Gene Targets Used for Microarray Detection
| Gene | Function | Organism Class | References |
|---|---|---|---|
| 16S rDNA | Protein synthesis | Bacteria, Mycobacteria | McCabe, Zhang, Huang, Wagar, and McCabe (1999), Baker, Smith, and Cowan, (2003) and Gingeras et al. (1998) |
| 23S rDNA | Protein synthesis | Bacteria, Mycobacteria | Yoo et al. (2010) and Lee, Jelfs, Sintchenko, and Gilbert, (2009) |
| rpoB | RNA polymerase | Bacteria, Mycobacteria | Adekambi, Colson, and Drancourt, (2003) and Gingeras et al. (1998) |
| gyrB | Topoisomerase | Bacteria, Mycobacteria | Jarvinen et al. (2009) and Fukushima et al. (2003) |
| 16S–23S rRNA internal transcribed spacer | Protein synthesis | Bacteria, Mycobacteria | Kirschner et al. (1996) and Wang et al. (2009) |
| cpn60 | Chaperonin | Bacteria | Maynard et al. (2005) |
| parE | Topoisomerase | Bacteria | Jarvinen et al. (2009) |
| recA | DNA repair | Bacteria | Champagne et al. (2011) |
| wecE | Lipid synthesis | Enterobacteriaceae | Maynard et al. (2005) |
| hsp65 | Chaperonin | Mycobacteria | Ringuet et al. (1999) |
| dnaJ | Chaperonin | Mycobacteria | Nasr Esfahani, Rezaei Yazdi, Moghim, Ghasemian Safaei, and Zarkesh Esfahani (2012) |
| IS6110 | Promoter | Mycobacteria | Thierry et al. (1990) |
| sodA | Superoxide dismutase | Mycobacteria | Kooken et al. (2014) |
| 32 kDa protein | Secreted protein | Mycobacteria | Soini, Skurnik, Liippo, Tala, and Viljanen (1992) |
| 18S and 28S rRNA internal transcribed spacers | Protein synthesis | Fungi | Leinberger, Schumacher, Autenrieth, and Bachmann (2005), Campa et al. (2008) and Landlinger et al. (2009) |
| Genus-specific viral family targets | Various | Viruses | Xiao-Ping et al. (2009) |
4.1.1. Identification of organisms isolated by culture-based methods
Microarrays can be useful for the identification of unusual or difficult to identify organisms isolated by culture-based methods. Arrays are designed for a particular pathogen type, with species diversity dependent on the microarray targets and amplification strategy used. Microarrays can be applied to common bacterial pathogens, but are more often used when alternative techniques such as biochemical tests, MALDI-TOF mass spectrometry or 16S gene sequencing are not available (Yoo et al., 2009).
Bacterial identification by microarray has been demonstrated for potential biothreat organisms, such as Bacillus anthracis, where reliable differentiation from environmental Bacillus species is necessary (Burton, Oshota, & Silman, 2006) and for food or waterborne pathogenic bacteria (Huang et al., 2014, Maynard et al., 2005, Zhou et al., 2011). Substantial genotype information can be derived from these isolates at a single-nucleotide polymorphism (SNP) level (Thierry et al., 2013). Comparative genomic arrays can allow for sensitive discrimination of related isolates and detect virulence-associated genes in pathogens such as Staphylococcus aureus (Spence et al., 2008, Witney et al., 2005).
Slow growing and fastidious organisms can be efficiently identified by microarrays, including mycobacteria (Fukushima et al., 2003, Tobler et al., 2006) and fungi (Campa et al., 2008, Leinberger et al., 2005). A commercially available line probe system uses the 16S–23S mycobacterial spacer for identification of 17 mycobacterial species with high sensitivity and specificity (Mijs et al., 2002, Tortoli et al., 2003). Microarrays have also been applied directly on patient samples to aid diagnosis, where assay sensitivity is a critical factor (Lazzeri, Santoro, Oggioni, Iannelli, & Pozzi, 2012). Microarrays can also be applied to the identification of viruses isolated in culture, using conserved targets within viral families, which can be applied to atypical or highly pathogenic viruses (Fitzgibbon and Sagripanti, 2006, Xiao-Ping et al., 2009).
4.1.2. Identification of organisms in positive blood cultures
The identification of organisms growing in blood cultures and the detection of resistance mechanisms may facilitate earlier and more targeted antimicrobial therapy. This has the potential to improve patient outcomes while reducing the need for the use of broad-spectrum antibiotic therapy. Microarray technology has been successfully applied to positive blood cultures, where the organism concentration is high enough that successful detection can be demonstrated. Molecular diagnostic techniques and MALDI-TOF mass spectrometry can yield similar information 1–3 days sooner than standard biochemical tests, and there are now several commercial assays available for clinical use (Buchan et al., 2013, Laakso et al., 2011, Liesenfeld et al., 2014).
Microarray targets can include many clinically relevant organisms, or target-specific pathogen types as determined by the initial Gram stain. The differentiation of Gram-positive cocci into virulent S. aureus versus the much less pathogenic Staphylococcus epidermidis can lead to significant treatment changes, and these panels typically contain resistance elements such as mecA for staphylococcal methicillin resistance and vanA for vancomycin-resistant enterococci (Palka-Santini et al., 2007, Pasko et al., 2012, Samuel et al., 2013). Gram-negative bacterial microarray panels typically identify Enterobacteriaceae species and Pseudomonas aeruginosa, and detect resistance mechanisms such as beta-lactamases and carbapenemases (Sullivan et al., 2014). The accuracy of these assays in predicting cephalosporin and carbapenemase susceptibility will depend on the extent of the panel targeting various classes of beta-lactamase genes and the local circulating strain epidemiology. Panels can also target fungal pathogens, typically identifying Candida species to enable optimised antifungal therapy (Aittakorpi et al., 2012, Yoo et al., 2010).
Data are still emerging about the optimal approach to translate blood culture microarray results into improved clinical care. While the presence of resistance elements such as mecA and bla beta-lactamase gene influence outcomes, host factors such as age and comorbidities are major factors as well (Rieg et al., 2013). It appears that intervention by an infectious disease or pharmacy practitioner is needed to realise maximum benefit from earlier identification and resistance marker results (Sango et al., 2013). As clinicians become more familiar with these tests and decision algorithms are developed, specialist consultation may become less necessary to achieve their potential improvements in outcomes.
4.1.3. Application of microarrays directly to patient samples
4.1.3.1. Respiratory infections
The use of multiplexed respiratory pathogen detection panels has become routine in many clinical laboratories. Several commercial multiplex panels, including microarray assays, are available and have been approved by regulatory agencies. These typically contain a variety of respiratory virus targets and differ in the scope of detection and ability to subtype (i.e. inclusion of human coronaviruses and subtyping of parainfluenza or adenovirus), as well as the workflow and hands-on time required (Krunic et al., 2011, Mahony et al., 2011). Comparative studies have shown some differences in assay sensitivities for particular targets, particularly adenovirus and influenza B (Balada-Llasat et al., 2011, Popowitch et al., 2013). Some assays may in addition contain bacterial targets such as Bordetella pertussis, Chlamydophila pneumoniae and Mycoplasma pneumoniae, and they all can be adapted to diagnose lower tract as well as upper tract respiratory infections (Lodes et al., 2007, Ruggiero et al., 2014).
Larger microarray panels allow for the diagnosis of more cases of respiratory infection (Bierbaum et al., 2012, Lin et al., 2006, Simoes et al., 2013), but can be difficult to develop and validate for use in the clinical laboratory. Resequencing microarrays allow for detection of co-infections, strain discrimination and epidemiologic tracking of respiratory pathogens (Lin, Blaney, et al., 2007, Lin, Malanoski, et al., 2007). Microarrays using pan-pathogen detection methods such as random amplification followed by broad-spectrum target hybridisation demonstrate a large amount of diversity in viral flora and have diagnosed atypical infections such as human parainfluenzavirus 4, rubella, measles and influenza C (Chiu et al., 2006, Kistler et al., 2007, Shen et al., 2013).
Fungal respiratory infections are an attractive target for microarray testing, since these pathogens can be difficult to grow and some require relatively long incubation periods. Species-specific identification of yeasts, moulds and dimorphic organisms including Histoplasma capsulatum and Coccidioides immitis can be detected in a single test and correlate with clinical and microbiological disease diagnosis (Landlinger et al., 2009). Microarrays can be particularly useful for diagnosis of immunocompromised patients with suspected invasive fungal infection, where detection is difficult by conventional methods (Spiess et al., 2007).
Rapid detection of mycobacterial pathogens can also improve treatment decisions, and commercially available assays have been evaluated for direct patient sample testing, yielding high specificity but lower sensitivity than when compared to conventional culture-based methods (Tortoli & Marcelli, 2007).
The clinical interpretation of respiratory microarray testing is relatively straightforward in the case of viral detection in a symptomatic patient, although the significance of co-infections is less certain. Rapid molecular methods can be useful to guide therapy for pneumonia based on the type of organism detected and resistance genes present, but clinical trial data have not yet been published (Endimiani et al., 2011).
4.1.3.2. Gastrointestinal infections
Conventional methods of identification for gastrointestinal pathogens are typically time-consuming and limited in scope to relatively few organisms. Several different tests are employed (i.e. bacterial culture, microscopy for parasite identification, and viral antigen detection or PCR testing), but additional tests may still be needed to detect less common pathogens. By combining multiple organism types onto a single panel, microarray methods have the potential to simplify testing protocols, while providing high sensitivity and the test panels can include species not commonly tested for by conventional methods (Jin, Wen, Chen, Lin, & Wang, 2006). Mixed infections may also be detected and information regarding the infecting parasite species or bacterial subspecies can be generated and used for epidemiologic studies (Marotta et al., 2013, Wang et al., 2005). Commercially available microarray tests have been shown to increase the number of pathogens detected compared to conventional methods and may provide a faster turnaround time for bacterial and parasitic pathogens (Halligan et al., 2014, Mengelle et al., 2013, Navidad et al., 2013). Future uses may include identification of the intestinal microbiota associated with certain disease states and manipulation of this through probiotic therapy (Carey, Kirk, Ojha, & Kostrzynska, 2007).
4.1.3.3. Bloodstream infections
The full benefit of rapid molecular diagnosis of sepsis requires use of direct patient blood samples to allow for the shortest time to actionable results. These panels can include fastidious organisms that are difficult to diagnose using conventional methods (Fenollar & Raoult, 2007). However, many septic patients have less than one bacteria cell per millilitre of blood, and the extraction of large amounts of blood is technically difficult (Klouche & Schroder, 2008). Therefore, the sensitivity of direct bloodstream detection is a limiting factor, but these results can still be useful to guide more rapid treatment decisions (Huttunen et al., 2013, Schrenzel, 2007). Some molecular panels have been shown to increase the number of detected bacterial infections over culture, assisting the diagnosis in immunocompromised and in patients where antimicrobial therapy has already been initiated (Burdino et al., 2014, Negoro et al., 2013).
Several microarray-based assays for the detection of bacteria, yeast and viruses have been described, some with sensitivities of 100% compared to blood culture (Debaugnies et al., 2014, Shang et al., 2005, Wiesinger-Mayr et al., 2007). Optimised DNA extraction methods, primer sets and amplification conditions are needed to achieve sensitivities down to 101 bacterial cells per millilitre of whole blood (Palka-Santini et al., 2009, Wiesinger-Mayr et al., 2011). Microarrays can also be used to interrogate the host transcriptomic profile and generate gene expression patterns that correlate with bacterial sepsis or other inflammatory states (Sutherland et al., 2011). However, validated clinical algorithms using direct bloodstream pathogen detection and confirmatory data from large prospective studies demonstrating improvements in clinical outcomes are not yet available.
4.1.3.4. Urinary tract infections
Microarrays have been applied to the direct detection of uropathogens with high sensitivity, but since they are not quantitative, the ability to differentiate a true pathogen in urine containing mixed flora is limited (Liao et al., 2006). Therefore, most studies have focused on the detection of resistant organisms such as carbapenem-resistant Enterobacteriaceae and fluoroquinolone-resistant Escherichia coli or nosocomial bacteria such as P. aeruginosa and Acinetobacter baumannii (Keum et al., 2006, Peter et al., 2012, Yu et al., 2007). Novel approaches including electronic biosensor arrays are in development for rapid detection of uropathogens with minimal sample processing, with the eventual goal of providing point-of-care testing capability (Lam et al., 2013).
4.1.3.5. Genital tract infections
Detection of sexually transmitted disease pathogens through microarray panels has been described for Neisseria gonorrhoeae, Chlamydia trachomatis and Ureaplasma urealyticum using a multiplex primer-amplification approach (Cao et al., 2006, Lee et al., 2013, Shi et al., 2005). However, commercial development has focused on human papillomavirus (HPV) detection and genotyping as part of cervical cancer diagnosis and screening. Genotyping is needed to distinguish high-risk HPV types (16 and 18) from other intermediate and low-risk types. Microarrays in a variety of formats have been developed to identify up to 120 known genotypes of HPV infection in cervical cytology and swab specimens (Albrecht et al., 2006, Feng et al., 2009, Shen-Gunther and Rebeles, 2013). For this application, detection sensitivity is less important than the ability to discriminate between low-risk patients and those with carcinomas that are likely to progress clinically, which is typically assessed using abnormal histology at cervical intraepithelial neoplasia (CIN) grade 2 + or higher. Most tests have been compared to the Hybrid Capture II assay and show similar or improved analytical sensitivity (Cho et al., 2011, Park et al., 2013). Currently, only the real-time PCR-based Cobas HPV assay has been evaluated in a large trial and specifically approved by the U.S. Food and Drug Administration for primary cervical cancer screening, which improved sensitivity from 52% to 88% for CIN2 or greater and from 53% to 92% for CIN3 or greater compared to liquid-based cytology (Castle et al., 2011, Wright et al., 2012).
4.1.3.6. Skin, soft tissue and joint infections
Detection microarrays are best suited to identify non-commensal pathogens in non-sterile site infections or a wide range of organisms in sterile sites. They have been reported to detect cutaneous mycobacterial infection, and superficial and invasive mycoses (Campa et al., 2008, Carlos et al., 2012, Sato et al., 2010). Profiling of the microbiota of healing wounds shows an association of Acinetobacter with wound failure, and enteric-associated flora with successful healing (Be et al., 2014). The identification of prosthetic joint infection can be enhanced by molecular techniques such as 16S PCR sequencing, and a microarray-based assay enhances detection in patients previously treated with antibiotics (Metso et al., 2014).
4.1.3.7. Central nervous system infections
Microarrays have been applied to diagnosis of bacterial meningitis, where they have been demonstrated to detect more cases than conventional methods alone, although methods have not been standardised, and there is potential for false-positives through sample contamination (Ben et al., 2008, Liu et al., 2005, Rafi et al., 2010). Identification and differentiation of the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii are possible in cerebrospinal fluid, but methods need to be optimised for sensitive detection (Bovers et al., 2007). Organisms which cause viral meningitis and encephalitis can be targeted through viral microarrays with probes for human herpesviruses, measles, mumps and enteroviruses (Boriskin et al., 2004, Leveque et al., 2011). Specialised microarrays have also been developed for highly pathogenic viruses such as chikungunya, yellow fever, dengue, hantavirus and emerging zoonotic agents (Xiao-Ping et al., 2009).
4.2. Pathogen Genotyping
4.2.1. Bacterial genotyping
A large number of research studies have used microarray methods to determine the presence of virulence factors and other genetic elements in bacteria. These techniques are important in investigating pathogens public health significance such as Salmonella and Legionella, and for outbreak investigations, for example, a multistate Listeria outbreak associated with cantaloupe (Gomgnimbou et al., 2014, Gronlund et al., 2011, Laksanalamai et al., 2012). From a clinical diagnostic standpoint, bacterial genotyping is rarely needed. In special circumstances, genotyping can assist investigation of infections in immunodeficient patients, for instance to determine whether an individual with Streptococcus pneumoniae infection who has been previously vaccinated has a strain included by the vaccine (Wang et al., 2007). As we acquire more knowledge about the clinical relevance of certain bacterial genotypes and virulence factors in predicting disease severity, genotyping could become useful to track disease epidemiology and indicate more aggressive treatment for certain strains, such as Clostridium difficile ribotype 027 (Knetsch et al., 2013).
4.2.2. Mycobacterial genotyping
M. tuberculosis strain typing is important for public health control efforts. PCR-based testing for the presence of variable direct repeat spacer oligonucleotides (spoligotyping) is a widely accepted technique, and has been adopted on microarray platforms as a rapid affordable method (Cowan et al., 2004, Ruettger et al., 2012). Genome deletions and SNPs are stable in M. tuberculosis clones, making these potential surveillance methods and approaches to study the pathogenesis of strains with known deletions (Kato-Maeda et al., 2001, Mahasirimongkol et al., 2009). Clinical microarrays often will contain markers for antibiotic resistance as well (see Section 4.3.2).
4.2.3. Fungal genotyping
Like bacteria, microarrays have been used to determine strain epidemiology and investigate virulence factors in fungal pathogens (Balajee et al., 2007, Garaizar et al., 2006). For example, gene expression analysis yielded candidate virulence factors in C. gattii from outbreak strains (Ngamskulrungroj, Price, Sorrell, Perfect, & Meyer, 2011).
4.2.4. Viral genotyping
With the exception of HPV genotyping (described in Section 4.1.3.5), clinical viral genotyping yields prognostic information that can be used to guide treatment decisions or can be used for epidemiological and research purposes. Microarrays can be targeted to specific viral families or pan-viral using unbiased amplification with hybridisation to conserved sequence regions (Wang et al., 2002). Hepatitis C virus (HCV) genotyping is important to determine the optimal treatment regimen, and microarray-based methods are commercially available (Duarte et al., 2010, Nadarajah et al., 2007, Park et al., 2010). Hepatitis B virus (HBV) genotype can influence treatment response, and microarray methods can differentiate the various genotypes (Gauthier et al., 2010). For other viruses, microarray assays can generate epidemiological information about circulating strains, including segment origin of influenza A or rotavirus genomes, or typing of emerging viruses (Gardner and Jaing, 2013, Honma et al., 2007, Paulin et al., 2014).
4.3. Antimicrobial Resistance Gene Detection
4.3.1. Bacterial resistance testing
The ability to accurately predict phenotypic resistance from bacteria genetic makeup depends on having a large panel of resistance markers, along with prediction algorithms for each antibiotic drug or class of drugs. Microarrays are well suited to detect resistance genes and cassettes, notably for beta-lactamases and carbapenemases, and can also detect SNPs associated with drug resistance. The tests have reasonably high sensitivity and specificity for detection of known resistance elements, but often will not differentiate functional from non-functional genes and fail to identify other mutation types, such as porin mutants, that can affect resistance profiles (Cuzon et al., 2012, Frye et al., 2006, Naas et al., 2010).
Currently available panels for Gram-negative organisms yield a correlation of approximately 90% for positive microarray results with antimicrobial resistance, but microarray-negative results correlate only about 65% with antimicrobial sensitivity (Card et al., 2013). These predictive rates vary per drug class and are generally better for plasmid-mediated resistance elements than for chromosomal mutations. They are well suited for identification of bacterial producing specific extended-spectrum beta-lactamases or carbapenemases for infection control and resistance monitoring (Naas, Cuzon, Bogaerts, Glupczynski, & Nordmann, 2011). Quinolone resistance in E. coli can be identified with known SNPs in gyrA, though correlation with phenotypic resistance has been shown for a limited number of isolates (Barl et al., 2008, Yu et al., 2004). Microarray targets for genotyping antibiotic resistance from nosocomial Gram-negative pathogens include efflux regulators, DNA gyrase, beta-lactamases, aminoglycoside-modifying enzymes and carbapenemases, with genotype–phenotype correlations of 88% for P. aeruginosa and > 95% for selected strains of multidrug-resistant A. baumannii (Dally et al., 2013, Weile et al., 2007). Mutations in Helicobacter pylori 23S rRNA gene associated with clarithromycin resistance can also be detected by microarray testing (Chen, Li, & Yu, 2008).
For Gram-positive pathogens such as S. aureus, microarrays can be used to detect clinically relevant resistance mutations for several antibiotics (Strommenger et al., 2007). Correlations between array detection and phenotypic resistance are approximately 90% for penicillin, methicillin, aminoglycoside, macrolide and lincosamide resistance by phenotypic testing (Zhu et al., 2007). Other techniques, such as next-generation sequencing, have a high correlation with microarray methods for detection of bacterial resistance mutations. In general, the current data support the ability of these molecular techniques to detect known resistance mutations, but they have not been prospectively tested with large numbers of clinical isolates from a variety of locations to determine the performance characteristics relative to conventional phenotypic susceptibility testing (Veenemans et al., 2014). Assay turnaround time and manual manipulation steps generally have to be reduced for these tests results to be available in a clinically relevant time frame.
4.3.2. Mycobacterial resistance testing
Mycobacterial resistance microarray platforms have focused on M. tuberculosis, the most clinically significant pathogen in this group. Phenotypic testing is delayed due to the organism's slow growth, so that molecular techniques can yield rapid detection of resistance mutations, allowing earlier transition to second-line treatment. About 95% of rifampin-resistant strains have certain known point mutations and rearrangements in rpoB gene (Caoili et al., 2006, Yue et al., 2004). Approximately 80% of isoniazid-resistant strains have a katG mutation and about 20% have inhA mutations (Kim et al., 2006, Yao et al., 2010). Pyrazinamide resistance is associated with mutations in the pncA gene, though about 40% of resistant strains have no mutation in this gene (Denkin, Volokhov, Chizhikov, & Zhang, 2005). Microarray-detected embB mutations are found in about 75% of ethambutol-resistant strains (Moure et al., 2014).
For second-line M. tuberculosis therapies, mutations in rpsL were found associated with streptomycin resistance, though sensitivity was low due to limited microarray gene coverage. The same study found sensitivities for detection of resistance at 100% for rifampin, 90% for isoniazid and 70% for ethambutol (Linger et al., 2014). Other second-line drugs for which microarrays can yield detection of resistance mutations include fluoroquinolones, kanamycin and capreomycin (Zimenkov et al., 2013). Other than tuberculosis, a microarray has been developed to detect resistance polymorphisms in Mycobacterium leprae (Matsuoka et al., 2008). Overall, microarray methods for detecting resistance in mycobacterial species are promising, but still require testing of a large number of geographically diverse isolates to determine their ability to guide therapeutic choices.
4.3.3. Fungal resistance testing
Several mechanisms of antifungal resistance in yeast have been identified, and many of these involve overexpression of genes encoding for drug targets or efflux pump transporters. Gene expression microarrays can identify genes associated with azole resistance in Candida albicans and are useful in identifying specific mutations affecting promoter expression (Yan et al., 2008). Increased expression of ergosterol biosynthesis genes is important for amphotericin B resistance in C. albicans, while azole resistance in Candida glabrata is associated with upregulation of the CgPDR1 pleiotropic drug resistance network locus (Barker et al., 2004, Tsai et al., 2010). As more knowledge is gained about the specific mutations causing these upregulations, microarrays can be used to identify strains carrying known resistance mutations.
4.3.4. Viral resistance testing
Detection of known resistance mutations in viruses is amenable to microarray testing and can be implemented as a more economical method than full gene sequencing in resource-limited settings. Testing for HIV resistance in protease and reverse transcriptase genes yields > 95% concordance with sequencing, with a few discrepant cases due to increased sensitivity of microarray over sequencing methods (Zhang, Cai, et al., 2013). For HBV, microarray targets can accurately identify mutations associated with lamivudine resistance (Chen et al., 2005). Microarrays designed to detect mutations in the neuraminidase and matrix genes of influenza A can identify these strains with high accuracy (Zhang, Liu, Wang, Chen, & Wang, 2013). As more viral resistance mutations conferring resistance to therapy are discovered, microarrays will offer a cost-effective method to detect them in clinical and epidemiological settings.
Conclusions
Microarrays offer a convenient method for interrogating specific pathogen sequences on scales of tens to hundreds of thousands of oligonucleotide probes. Several technologies and platforms are available, including solid-state, electronic and suspension bead microarrays, each with multiple variations in probe synthesis chemistries, linkage and data collection and analysis formats. Samples are typically amplified using multiplex primer sets, followed by hybridisation, washing and signal detection. Microarrays have been adopted for microbial detection, genotyping and antimicrobial resistance gene detection directly from clinical samples or from isolated organisms. Limits of detection are comparable to other molecular techniques, but still may not be equivalent to large-volume culture techniques such as blood cultures.
Microarray assays are limited by the set of available probe sequences, so they may not detect unusual sequence variants or novel genetic elements of clinical significance. Careful consideration must be taken during assay design and validation to distinguish between closely related species differing in virulence (i.e. S. pneumoniae vs. viridans Streptococcus), and to ensure that relevant circulating strains containing sequence variations are adequately detected and identified. Ideally, probe sets should be updated to reflect changing genotypes and strain epidemiology. The performance of microarray tests needs to be determined for different specimen types or matrices that may be encountered during clinical testing.
Microarray tests are well suited to offer a panel approach to diagnose specific patient presentations, such as respiratory viral infection or infectious diarrhoea. They can also discriminate between clinical isolates by genotype, which can be useful for tracking epidemiology and for outbreak investigations. As more information has become available regarding specific genes and pathways involved in antimicrobial resistance, we are beginning to be able to predict susceptibility patterns based on sequence interrogation for particular organisms. With further advances in automated microarray processing methods and genotype–phenotype prediction algorithms, these tests will become useful as an adjunct or replacement for conventional susceptibility testing, allowing for more rapid selection of targeted therapy for infectious disease.
References
- Adekambi T., Colson P., Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. Journal of Clinical Microbiology. 2003;41(12):5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aittakorpi A., Kuusela P., Koukila-Kahkola P., Vaara M., Petrou M., Gant V., et al. Accurate and rapid identification of Candida spp. frequently associated with fungemia by using PCR and the microarray-based Prove-it Sepsis assay. Journal of Clinical Microbiology. 2012;50(11):3635–3640. doi: 10.1128/JCM.01461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V., Chevallier A., Magnone V., Barbry P., Vandenbos F., Bongain A., et al. Easy and fast detection and genotyping of high-risk human papillomavirus by dedicated DNA microarrays. Journal of Virological Methods. 2006;137(2):236–244. doi: 10.1016/j.jviromet.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Baker G.C., Smith J.J., Cowan D.A. Review and re-analysis of domain-specific 16S primers. Journal of Microbiological Methods. 2003;55(3):541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Balada-Llasat J.M., LaRue H., Kelly C., Rigali L., Pancholi P. Evaluation of commercial ResPlex II v2.0, MultiCode-PLx, and xTAG respiratory viral panels for the diagnosis of respiratory viral infections in adults. Journal of Clinical Virology. 2011;50(1):42–45. doi: 10.1016/j.jcv.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee S.A., Sigler L., Brandt M.E. DNA and the classical way: Identification of medically important molds in the 21st century. Medical Mycology. 2007;45(6):475–490. doi: 10.1080/13693780701449425. [DOI] [PubMed] [Google Scholar]
- Barker K.S., Crisp S., Wiederhold N., Lewis R.E., Bareither B., Eckstein J., et al. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. The Journal of Antimicrobial Chemotherapy. 2004;54(2):376–385. doi: 10.1093/jac/dkh336. [DOI] [PubMed] [Google Scholar]
- Barl T., Dobrindt U., Yu X., Katcoff D.J., Sompolinsky D., Bonacorsi S., et al. Genotyping DNA chip for the simultaneous assessment of antibiotic resistance and pathogenic potential of extraintestinal pathogenic Escherichia coli. International Journal of Antimicrobial Agents. 2008;32(3):272–277. doi: 10.1016/j.ijantimicag.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Barlaan E.A., Sugimori M., Furukawa S., Takeuchi K. Electronic microarray analysis of 16S rDNA amplicons for bacterial detection. Journal of Biotechnology. 2005;115(1):11–21. doi: 10.1016/j.jbiotec.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Be N.A., Allen J.E., Brown T.S., Gardner S.N., McLoughlin K.S., Forsberg J.A., et al. Microbial profiling of combat wound infection through detection microarray and next-generation sequencing. Journal of Clinical Microbiology. 2014;52(7):2583–2594. doi: 10.1128/JCM.00556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier M., Hoheisel J.D. Production by quantitative photolithographic synthesis of individually quality checked DNA microarrays. Nucleic Acids Research. 2000;28(4):E11. doi: 10.1093/nar/28.4.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier M., Hoheisel J.D. Synthesis of 5′-O-phosphoramidites with a photolabile 3′-O-protecting group. Current Protocols in Nucleic Acid Chemistry. 2004 doi: 10.1002/0471142700.nc1203s17. 17:12.3:12.3.1–12.3.10. [DOI] [PubMed] [Google Scholar]
- Ben R.J., Kung S., Chang F.Y., Lu J.J., Feng N.H., Hsieh Y.D. Rapid diagnosis of bacterial meningitis using a microarray. Journal of the Formosan Medical Association. 2008;107(6):448–453. doi: 10.1016/S0929-6646(08)60152-7. [DOI] [PubMed] [Google Scholar]
- Bierbaum S., Konigsfeld N., Besazza N., Blessing K., Rucker G., Kontny U., et al. Performance of a novel microarray multiplex PCR for the detection of 23 respiratory pathogens (SYMP-ARI study) European Journal of Clinical Microbiology & Infectious Diseases. 2012;31(10):2851–2861. doi: 10.1007/s10096-012-1639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriskin Y.S., Rice P.S., Stabler R.A., Hinds J., Al-Ghusein H., Vass K., et al. DNA microarrays for virus detection in cases of central nervous system infection. Journal of Clinical Microbiology. 2004;42(12):5811–5818. doi: 10.1128/JCM.42.12.5811-5818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovers M., Diaz M.R., Hagen F., Spanjaard L., Duim B., Visser C.E., et al. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. Journal of Clinical Microbiology. 2007;45(6):1874–1883. doi: 10.1128/JCM.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan B.W., Ginocchio C.C., Manii R., Cavagnolo R., Pancholi P., Swyers L., et al. Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: Evaluation of an automated microarray-based nucleic acid test. PLoS Medicine. 2013;10(7):e1001478. doi: 10.1371/journal.pmed.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdino E., Ruggiero T., Allice T., Milia M.G., Gregori G., Milano R., et al. Combination of conventional blood cultures and the SeptiFast molecular test in patients with suspected sepsis for the identification of bloodstream pathogens. Diagnostic Microbiology and Infectious Disease. 2014;79(3):287–292. doi: 10.1016/j.diagmicrobio.2014.03.018. [DOI] [PubMed] [Google Scholar]
- Burton J.E., Oshota O.J., Silman N.J. Differential identification of Bacillus anthracis from environmental Bacillus species using microarray analysis. Journal of Applied Microbiology. 2006;101(4):754–763. doi: 10.1111/j.1365-2672.2006.02991.x. [DOI] [PubMed] [Google Scholar]
- Campa D., Tavanti A., Gemignani F., Mogavero C.S., Bellini I., Bottari F., et al. DNA microarray based on arrayed-primer extension technique for identification of pathogenic fungi responsible for invasive and superficial mycoses. Journal of Clinical Microbiology. 2008;46(3):909–915. doi: 10.1128/JCM.01406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Wang Y.F., Zhang C.F., Gao W.J. Visual DNA microarrays for simultaneous detection of Ureaplasma urealyticum and Chlamydia trachomatis coupled with multiplex asymmetrical PCR. Biosensors & Bioelectronics. 2006;22(3):393–398. doi: 10.1016/j.bios.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Caoili J.C., Mayorova A., Sikes D., Hickman L., Plikaytis B.B., Shinnick T.M. Evaluation of the TB-Biochip oligonucleotide microarray system for rapid detection of rifampin resistance in Mycobacterium tuberculosis. Journal of Clinical Microbiology. 2006;44(7):2378–2381. doi: 10.1128/JCM.00439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card R., Zhang J., Das P., Cook C., Woodford N., Anjum M.F. Evaluation of an expanded microarray for detecting antibiotic resistance genes in a broad range of gram-negative bacterial pathogens. Antimicrobial Agents and Chemotherapy. 2013;57(1):458–465. doi: 10.1128/AAC.01223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey C.M., Kirk J.L., Ojha S., Kostrzynska M. Current and future uses of real-time polymerase chain reaction and microarrays in the study of intestinal microbiota, and probiotic use and effectiveness. Canadian Journal of Microbiology. 2007;53(5):537–550. doi: 10.1139/W07-039. [DOI] [PubMed] [Google Scholar]
- Carlos C.A., Tang Y.W., Adler D.J., Kovarik C.L. Mycobacterial infection identified with broad-range PCR amplification and suspension array identification. Journal of Cutaneous Pathology. 2012;39(8):795–797. doi: 10.1111/j.1600-0560.2012.01934.x. [DOI] [PubMed] [Google Scholar]
- Castle P.E., Stoler M.H., Wright T.C., Jr., Sharma A., Wright T.L., Behrens C.M. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: A subanalysis of the ATHENA study. The Lancet. Oncology. 2011;12(9):880–890. doi: 10.1016/S1470-2045(11)70188-7. [DOI] [PubMed] [Google Scholar]
- Champagne J., Diarra M.S., Rempel H., Topp E., Greer C.W., Harel J., et al. Development of a DNA microarray for enterococcal species, virulence, and antibiotic resistance gene determinations among isolates from poultry. Applied and Environmental Microbiology. 2011;77(8):2625–2633. doi: 10.1128/AEM.00263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Y., Huang J., Zhang X.P., Qiao P., Zhang W., Yang N.M., et al. Clinical evaluation of oligonucleotide microarrays for the detection of HBV mutants associated with lamivudine resistance. Pharmacogenomics. 2005;6(7):721–730. doi: 10.2217/14622416.6.7.721. [DOI] [PubMed] [Google Scholar]
- Chen S., Li Y., Yu C. Oligonucleotide microarray: A new rapid method for screening the 23S rRNA gene of Helicobacter pylori for single nucleotide polymorphisms associated with clarithromycin resistance. Journal of Gastroenterology and Hepatology. 2008;23(1):126–131. doi: 10.1111/j.1440-1746.2007.04900.x. [DOI] [PubMed] [Google Scholar]
- Chiu C.Y., Rouskin S., Koshy A., Urisman A., Fischer K., Yagi S., et al. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clinical Infectious Diseases. 2006;43(8):e71–e76. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Do J.H., Kim Y.S., Bae S., Ahn W.S. Evaluation of a liquid bead array system for high-risk human papillomavirus detection and genotyping in comparison with Hybrid Capture II, DNA chip and sequencing methods. Journal of Medical Microbiology. 2011;60(Pt. 2):162–171. doi: 10.1099/jmm.0.021642-0. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . Clinical and Laboratory Standards Institute; Wayne, PA: 2014. Microarrays for diagnosis and monitoring of infectious diseases; approved guideline: CLSI document MM22-A; p. 84. [Google Scholar]
- Cowan L.S., Diem L., Brake M.C., Crawford J.T. Transfer of a Mycobacterium tuberculosis genotyping method, Spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. Journal of Clinical Microbiology. 2004;42(1):474–477. doi: 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon G., Naas T., Bogaerts P., Glupczynski Y., Nordmann P. Evaluation of a DNA microarray for the rapid detection of extended-spectrum beta-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM) The Journal of Antimicrobial Chemotherapy. 2012;67(8):1865–1869. doi: 10.1093/jac/dks156. [DOI] [PubMed] [Google Scholar]
- Dally S., Lemuth K., Kaase M., Rupp S., Knabbe C., Weile J. DNA microarray for genotyping antibiotic resistance determinants in Acinetobacter baumannii clinical isolates. Antimicrobial Agents and Chemotherapy. 2013;57(10):4761–4768. doi: 10.1128/AAC.00863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaugnies F., Busson L., Ferster A., Lewalle P., Azzi N., Aoun M., et al. Detection of Herpesviridae in whole blood by multiplex PCR DNA-based microarray analysis after hematopoietic stem cell transplantation. Journal of Clinical Microbiology. 2014;52(7):2552–2556. doi: 10.1128/JCM.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkin S., Volokhov D., Chizhikov V., Zhang Y. Microarray-based pncA genotyping of pyrazinamide-resistant strains of Mycobacterium tuberculosis. Journal of Medical Microbiology. 2005;54(Pt. 12):1127–1131. doi: 10.1099/jmm.0.46129-0. [DOI] [PubMed] [Google Scholar]
- Duarte C.A., Foti L., Nakatani S.M., Riediger I.N., Poersch C.O., Pavoni D.P., et al. A novel hepatitis C virus genotyping method based on liquid microarray. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012822. e12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar S.A. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clinica Chimica Acta. 2006;363(1–2):71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar S.A. In: Advanced techniques in diagnostic microbiology. 2nd ed. Tang Y.W., Stratton C.W., editors. Springer; New York: 2013. Bead-based suspension arrays for the detection and identification of respiratory viruses; pp. 813–834. [Google Scholar]
- Dunbar S.A., Jacobson J.W. Quantitative, multiplexed detection of Salmonella and other pathogens by Luminex xMAP suspension array. Methods in Molecular Biology. 2007;394:1–19. doi: 10.1007/978-1-59745-512-1_1. [DOI] [PubMed] [Google Scholar]
- Edman C.F., Raymond D.E., Wu D.J., Tu E., Sosnowski R.G., Butler W.F., et al. Electric field directed nucleic acid hybridization on microchips. Nucleic Acids Research. 1997;25(24):4907–4914. doi: 10.1093/nar/25.24.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endimiani A., Hujer K.M., Hujer A.M., Kurz S., Jacobs M.R., Perlin D.S., et al. Are we ready for novel detection methods to treat respiratory pathogens in hospital-acquired pneumonia? Clinical Infectious Diseases. 2011;52(Suppl. 4):S373–S383. doi: 10.1093/cid/cir054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Cherne S., Winer R.L., Balasubramanian A., Lee S.K., Hawes S.E., et al. Development and evaluation of a liquid bead microarray assay for genotyping genital human papillomaviruses. Journal of Clinical Microbiology. 2009;47(3):547–553. doi: 10.1128/JCM.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Nerenberg M. Electronic microarray for DNA analysis. Gene Therapy and Molecular Biology. 1999;4:183–191. [Google Scholar]
- Fenollar F., Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. International Journal of Antimicrobial Agents. 2007;30(Suppl. 1):S7–S15. doi: 10.1016/j.ijantimicag.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J.E., Sagripanti J.L. Simultaneous identification of orthopoxviruses and alphaviruses by oligonucleotide macroarray with special emphasis on detection of variola and Venezuelan equine encephalitis viruses. Journal of Virological Methods. 2006;131(2):160–167. doi: 10.1016/j.jviromet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Foglieni B., Brisci A., San Biagio F., Di Pietro P., Petralia S., Conoci S., et al. Integrated PCR amplification and detection processes on a Lab-on-Chip platform: A new advanced solution for molecular diagnostics. Clinical Chemistry and Laboratory Medicine. 2010;48(3):329–336. doi: 10.1515/CCLM.2010.063. [DOI] [PubMed] [Google Scholar]
- Frye J.G., Jesse T., Long F., Rondeau G., Porwollik S., McClelland M., et al. DNA microarray detection of antimicrobial resistance genes in diverse bacteria. International Journal of Antimicrobial Agents. 2006;27(2):138–151. doi: 10.1016/j.ijantimicag.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Fukushima M., Kakinuma K., Hayashi H., Nagai H., Ito K., Kawaguchi R. Detection and identification of Mycobacterium species isolates by DNA microarray. Journal of Clinical Microbiology. 2003;41(6):2605–2615. doi: 10.1128/JCM.41.6.2605-2615.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton R.J., McDade R.L., Smith P.L., Kienker L.J., Kettman J.R., Jr. Advanced multiplexed analysis with the FlowMetrix system. Clinical Chemistry. 1997;43(9):1749–1756. [PubMed] [Google Scholar]
- Garaizar J., Brena S., Bikandi J., Rementeria A., Ponton J. Use of DNA microarray technology and gene expression profiles to investigate the pathogenesis, cell biology, antifungal susceptibility and diagnosis of Candida albicans. FEMS Yeast Research. 2006;6(7):987–998. doi: 10.1111/j.1567-1364.2006.00108.x. [DOI] [PubMed] [Google Scholar]
- Gardner S.N., Jaing C.J. Bioinformatics for microbial genotyping of equine encephalitis viruses, orthopoxviruses, and hantaviruses. Journal of Virological Methods. 2013;193(1):112–120. doi: 10.1016/j.jviromet.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Gauthier M., Bonnaud B., Arsac M., Lavocat F., Maisetti J., Kay A., et al. Microarray for hepatitis B virus genotyping and detection of 994 mutations along the genome. Journal of Clinical Microbiology. 2010;48(11):4207–4215. doi: 10.1128/JCM.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T.R., Ghandour G., Wang E., Berno A., Small P.M., Drobniewski F., et al. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Research. 1998;8(5):435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- Goldmann T., Gonzalez J.S. DNA-printing: Utilization of a standard inkjet printer for the transfer of nucleic acids to solid supports. Journal of Biochemical and Biophysical Methods. 2000;42(3):105–110. doi: 10.1016/s0165-022x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- Gomgnimbou M.K., Ginevra C., Peron-Cane C., Versapuech M., Refregier G., Jacotin N., et al. Validation of a microbead-based format for spoligotyping of Legionella pneumophila. Journal of Clinical Microbiology. 2014;52(7):2410–2415. doi: 10.1128/JCM.00219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronlund H., Riber L., Vigre H., Lofstrom C., Folling L., Huehn S., et al. Microarray-based genotyping of Salmonella: Inter-laboratory evaluation of reproducibility and standardization potential. International Journal of Food Microbiology. 2011;145(Suppl. 1):S79–S85. doi: 10.1016/j.ijfoodmicro.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Halligan E., Edgeworth J., Bisnauthsing K., Bible J., Cliff P., Aarons E., et al. Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: A comparative diagnostic and clinical utility study. Clinical Microbiology and Infection. 2014;20(8):O460–O467. doi: 10.1111/1469-0691.12476. [DOI] [PubMed] [Google Scholar]
- Honma S., Chizhikov V., Santos N., Tatsumi M., Timenetsky Mdo C., Linhares A.C., et al. Development and validation of DNA microarray for genotyping group A rotavirus VP4 (P[4], P[6], P[8], P[9], and P[14]) and VP7 (G1 to G6, G8 to G10, and G12) genes. Journal of Clinical Microbiology. 2007;45(8):2641–2648. doi: 10.1128/JCM.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A., Qiu Z., Jin M., Shen Z., Chen Z., Wang X., et al. High-throughput detection of food-borne pathogenic bacteria using oligonucleotide microarray with quantum dots as fluorescent labels. International Journal of Food Microbiology. 2014;185:27–32. doi: 10.1016/j.ijfoodmicro.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nature Biotechnology. 2001;19(4):342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Huttunen R., Syrjanen J., Vuento R., Aittoniemi J. Current concepts in the diagnosis of blood stream infections. Are novel molecular methods useful in clinical practice? International Journal of Infectious Diseases. 2013;17(11):e934–e938. doi: 10.1016/j.ijid.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Jarvinen A.K., Laakso S., Piiparinen P., Aittakorpi A., Lindfors M., Huopaniemi L., et al. Rapid identification of bacterial pathogens using a PCR- and microarray-based assay. BMC Microbiology. 2009;9:161. doi: 10.1186/1471-2180-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D.Z., Wen S.Y., Chen S.H., Lin F., Wang S.Q. Detection and identification of intestinal pathogens in clinical specimens using DNA microarrays. Molecular and Cellular Probes. 2006;20(6):337–347. doi: 10.1016/j.mcp.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Kafatos F.C., Jones C.W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Research. 1979;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Maeda M., Rhee J.T., Gingeras T.R., Salamon H., Drenkow J., Smittipat N., et al. Comparing genomes within the species Mycobacterium tuberculosis. Genome Research. 2001;11(4):547–554. doi: 10.1101/gr166401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum K.C., Yoo S.M., Lee S.Y., Chang K.H., Yoo N.C., Yoo W.M., et al. DNA microarray-based detection of nosocomial pathogenic Pseudomonas aeruginosa and Acinetobacter baumannii. Molecular and Cellular Probes. 2006;20(1):42–50. doi: 10.1016/j.mcp.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Khrapko K.R., Lysov Yu P., Khorlyn A.A., Shick V.V., Florentiev V.L., Mirzabekov A.D. An oligonucleotide hybridization approach to DNA sequencing. FEBS Letters. 1989;256(1–2):118–122. doi: 10.1016/0014-5793(89)81730-2. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Park Y.J., Song E., Jang H., Kim C., Yoo J., et al. Evaluation of the CombiChip Mycobacteria drug-resistance detection DNA chip for identifying mutations associated with resistance to isoniazid and rifampin in Mycobacterium tuberculosis. Diagnostic Microbiology and Infectious Disease. 2006;54(3):203–210. doi: 10.1016/j.diagmicrobio.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Kirschner P., Rosenau J., Springer B., Teschner K., Feldmann K., Bottger E.C. Diagnosis of mycobacterial infections by nucleic acid amplification: 18-month prospective study. Journal of Clinical Microbiology. 1996;34(2):304–312. doi: 10.1128/jcm.34.2.304-312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S., et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. The Journal of Infectious Diseases. 2007;196(6):817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klouche M., Schroder U. Rapid methods for diagnosis of bloodstream infections. Clinical Chemistry and Laboratory Medicine. 2008;46(7):888–908. doi: 10.1515/CCLM.2008.157. [DOI] [PubMed] [Google Scholar]
- Knetsch C.W., Lawley T.D., Hensgens M.P., Corver J., Wilcox M.W., Kuijper E.J. Current application and future perspectives of molecular typing methods to study Clostridium difficile infections. Euro Surveillance. 2013;18(4):20381. doi: 10.2807/ese.18.04.20381-en. [DOI] [PubMed] [Google Scholar]
- Kooken J., Fox K., Fox A., Altomare D., Creek K., Wunschel D., et al. Identification of staphylococcal species based on variations in protein sequences (mass spectrometry) and DNA sequence (sodA microarray) Molecular and Cellular Probes. 2014;28(1):41–50. doi: 10.1016/j.mcp.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Krunic N., Merante F., Yaghoubian S., Himsworth D., Janeczko R. Advances in the diagnosis of respiratory tract infections: Role of the Luminex xTAG respiratory viral panel. Annals of the New York Academy of Sciences. 2011;1222:6–13. doi: 10.1111/j.1749-6632.2011.05964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Wang L., Fan J., Kraft A., Bose M.E., Tiwari S., et al. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. Journal of Clinical Microbiology. 2008;46(9):3063–3072. doi: 10.1128/JCM.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso S., Kirveskari J., Tissari P., Maki M. Evaluation of high-throughput PCR and microarray-based assay in conjunction with automated DNA extraction instruments for diagnosis of sepsis. PLoS One. 2011;6(11):e26655. doi: 10.1371/journal.pone.0026655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laksanalamai P., Joseph L.A., Silk B.J., Burall L.S., Tarr C.L., Gerner-Smidt P., et al. Genomic characterization of Listeria monocytogenes strains involved in a multistate listeriosis outbreak associated with cantaloupe in US. PLoS One. 2012;7(7):e42448. doi: 10.1371/journal.pone.0042448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam B., Das J., Holmes R.D., Live L., Sage A., Sargent E.H., et al. Solution-based circuits enable rapid and multiplexed pathogen detection. Nature Communications. 2013;4:2001. doi: 10.1038/ncomms3001. [DOI] [PubMed] [Google Scholar]
- Landlinger C., Preuner S., Willinger B., Haberpursch B., Racil Z., Mayer J., et al. Species-specific identification of a wide range of clinically relevant fungal pathogens by use of Luminex xMAP technology. Journal of Clinical Microbiology. 2009;47(4):1063–1073. doi: 10.1128/JCM.01558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lausted C., Dahl T., Warren C., King K., Smith K., Johnson M., et al. POSaM: A fast, flexible, open-source, inkjet oligonucleotide synthesizer and microarrayer. Genome Biology. 2004;5(8):R58. doi: 10.1186/gb-2004-5-8-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzeri E., Santoro F., Oggioni M.R., Iannelli F., Pozzi G. Novel primer-probe sets for detection and identification of mycobacteria by PCR-microarray assay. Journal of Clinical Microbiology. 2012;50(11):3777–3779. doi: 10.1128/JCM.02300-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.S., Jelfs P., Sintchenko V., Gilbert G.L. Identification of non-tuberculous mycobacteria: Utility of the GenoType Mycobacterium CM/AS assay compared with HPLC and 16S rRNA gene sequencing. Journal of Medical Microbiology. 2009;58(Pt. 7):900–904. doi: 10.1099/jmm.0.007484-0. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Jeong J.S., Ahn J.J., Lee S.W., Seo H., Ahn Y., et al. Development of electrochemical microbiochip for the biological diagnosis of Neisseria gonorrhoeae. Analytical Sciences. 2013;29(12):1203–1208. doi: 10.2116/analsci.29.1203. [DOI] [PubMed] [Google Scholar]
- Leinberger D.M., Schumacher U., Autenrieth I.B., Bachmann T.T. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. Journal of Clinical Microbiology. 2005;43(10):4943–4953. doi: 10.1128/JCM.43.10.4943-4953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveque N., Van Haecke A., Renois F., Boutolleau D., Talmud D., Andreoletti L. Rapid virological diagnosis of central nervous system infections by use of a multiplex reverse transcription-PCR DNA microarray. Journal of Clinical Microbiology. 2011;49(11):3874–3879. doi: 10.1128/JCM.01214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J.C., Mastali M., Gau V., Suchard M.A., Moller A.K., Bruckner D.A., et al. Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. Journal of Clinical Microbiology. 2006;44(2):561–570. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesenfeld O., Lehman L., Hunfeld K.P., Kost G. Molecular diagnosis of sepsis: New aspects and recent developments. European Journal of Microbiology & Immunology. 2014;4(1):1–25. doi: 10.1556/EuJMI.4.2014.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Blaney K.M., Malanoski A.P., Ligler A.G., Schnur J.M., Metzgar D., et al. Using a resequencing microarray as a multiple respiratory pathogen detection assay. Journal of Clinical Microbiology. 2007;45(2):443–452. doi: 10.1128/JCM.01870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Malanoski A.P., Wang Z., Blaney K.M., Ligler A.G., Rowley R.K., et al. Application of broad-spectrum, sequence-based pathogen identification in an urban population. PLoS One. 2007;2(5):e419. doi: 10.1371/journal.pone.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Wang Z., Vora G.J., Thornton J.A., Schnur J.M., Thach D.C., et al. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Research. 2006;16(4):527–535. doi: 10.1101/gr.4337206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger Y., Kukhtin A., Golova J., Perov A., Lambarqui A., Bryant L., et al. Simplified microarray system for simultaneously detecting rifampin, isoniazid, ethambutol, and streptomycin resistance markers in Mycobacterium tuberculosis. Journal of Clinical Microbiology. 2014;52(6):2100–2107. doi: 10.1128/JCM.00238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Han J.X., Huang H.Y., Zhu B. Development and evaluation of 16S rDNA microarray for detecting bacterial pathogens in cerebrospinal fluid. Experimental Biology and Medicine (Maywood, N.J.) 2005;230(8):587–591. doi: 10.1177/153537020523000810. [DOI] [PubMed] [Google Scholar]
- Lodes M.J., Suciu D., Wilmoth J.L., Ross M., Munro S., Dix K., et al. Identification of upper respiratory tract pathogens using electrochemical detection on an oligonucleotide microarray. PLoS One. 2007;2(9):e924. doi: 10.1371/journal.pone.0000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahasirimongkol S., Yanai H., Nishida N., Ridruechai C., Matsushita I., Ohashi J., et al. Genome-wide SNP-based linkage analysis of tuberculosis in Thais. Genes and Immunity. 2009;10(1):77–83. doi: 10.1038/gene.2008.81. [DOI] [PubMed] [Google Scholar]
- Mahony J., Chong S., Merante F., Yaghoubian S., Sinha T., Lisle C., et al. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. Journal of Clinical Microbiology. 2007;45(9):2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J.B., Petrich A., Smieja M. Molecular diagnosis of respiratory virus infections. Critical Reviews in Clinical Laboratory Sciences. 2011;48(5–6):217–249. doi: 10.3109/10408363.2011.640976. [DOI] [PubMed] [Google Scholar]
- Marotta F., Zilli K., Tonelli A., Sacchini L., Alessiani A., Migliorati G., et al. Detection and genotyping of Campylobacter jejuni and Campylobacter coli by use of DNA oligonucleotide arrays. Molecular Biotechnology. 2013;53(2):182–188. doi: 10.1007/s12033-012-9512-0. [DOI] [PubMed] [Google Scholar]
- Matsuoka M., Aye K.S., Kyaw K., Tan E.V., Balagon M.V., Saunderson P., et al. A novel method for simple detection of mutations conferring drug resistance in Mycobacterium leprae, based on a DNA microarray, and its applicability in developing countries. Journal of Medical Microbiology. 2008;57(Pt. 10):1213–1219. doi: 10.1099/jmm.0.2008/002600-0. [DOI] [PubMed] [Google Scholar]
- Maynard C., Berthiaume F., Lemarchand K., Harel J., Payment P., Bayardelle P., et al. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Applied and Environmental Microbiology. 2005;71(12):8548–8557. doi: 10.1128/AEM.71.12.8548-8557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe K.M., Zhang Y.H., Huang B.L., Wagar E.A., McCabe E.R. Bacterial species identification after DNA amplification with a universal primer pair. Molecular Genetics and Metabolism. 1999;66(3):205–211. doi: 10.1006/mgme.1998.2795. [DOI] [PubMed] [Google Scholar]
- McGall G.H., Barone A.D., Diggelmann M., Fodor S.P.A., Gentalen E., Nga N. The efficiency of light-directed synthesis of DNA arrays on glass substrates. Journal of the American Chemical Society. 1997;119:5081–5090. [Google Scholar]
- Mengelle C., Mansuy J.M., Prere M.F., Grouteau E., Claudet I., Kamar N., et al. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clinical Microbiology and Infection. 2013;19(10):E458–E465. doi: 10.1111/1469-0691.12255. [DOI] [PubMed] [Google Scholar]
- Metso L., Maki M., Tissari P., Remes V., Piiparinen P., Kirveskari J., et al. Efficacy of a novel PCR- and microarray-based method in diagnosis of a prosthetic joint infection. Acta Orthopaedica. 2014;85(2):165–170. doi: 10.3109/17453674.2014.889978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijs W., De Vreese K., Devos A., Pottel H., Valgaeren A., Evans C., et al. Evaluation of a commercial line probe assay for identification of mycobacterium species from liquid and solid culture. European Journal of Clinical Microbiology & Infectious Diseases. 2002;21(11):794–802. doi: 10.1007/s10096-002-0825-y. [DOI] [PubMed] [Google Scholar]
- Miller M.B., Tang Y.W. Basic concepts of microarrays and potential applications in clinical microbiology. Clinical Microbiology Reviews. 2009;22(4):611–633. doi: 10.1128/CMR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure R., Espanol M., Tudo G., Vicente E., Coll P., Gonzalez-Martin J., et al. Characterization of the embB gene in Mycobacterium tuberculosis isolates from Barcelona and rapid detection of main mutations related to ethambutol resistance using a low-density DNA array. The Journal of Antimicrobial Chemotherapy. 2014;69(4):947–954. doi: 10.1093/jac/dkt448. [DOI] [PubMed] [Google Scholar]
- Naas T., Cuzon G., Bogaerts P., Glupczynski Y., Nordmann P. Evaluation of a DNA microarray (Check-MDR CT102) for rapid detection of TEM, SHV, and CTX-M extended-spectrum beta-lactamases and of KPC, OXA-48, VIM, IMP, and NDM-1 carbapenemases. Journal of Clinical Microbiology. 2011;49(4):1608–1613. doi: 10.1128/JCM.02607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Cuzon G., Truong H., Bernabeu S., Nordmann P. Evaluation of a DNA microarray, the check-points ESBL/KPC array, for rapid detection of TEM, SHV, and CTX-M extended-spectrum beta-lactamases and KPC carbapenemases. Antimicrobial Agents and Chemotherapy. 2010;54(8):3086–3092. doi: 10.1128/AAC.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah R., Khan G.Y., Miller S.A., Brooks G.F. Evaluation of a new-generation line-probe assay that detects 5′ untranslated and core regions to genotype and subtype hepatitis C virus. American Journal of Clinical Pathology. 2007;128(2):300–304. doi: 10.1309/RBAAC0HJFWRJB1HD. [DOI] [PubMed] [Google Scholar]
- Nasr Esfahani B., Rezaei Yazdi H., Moghim S., Ghasemian Safaei H., Zarkesh Esfahani H. Rapid and accurate identification of Mycobacterium tuberculosis complex and common non-tuberculous mycobacteria by multiplex real-time PCR targeting different housekeeping genes. Current Microbiology. 2012;65(5):493–499. doi: 10.1007/s00284-012-0188-2. [DOI] [PubMed] [Google Scholar]
- Navidad J.F., Griswold D.J., Gradus M.S., Bhattacharyya S. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. Journal of Clinical Microbiology. 2013;51(9):3018–3024. doi: 10.1128/JCM.00896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negoro E., Iwasaki H., Tai K., Ikegaya S., Takagi K., Kishi S., et al. Utility of PCR amplification and DNA microarray hybridization of 16S rDNA for rapid diagnosis of bacteremia associated with hematological diseases. International Journal of Infectious Diseases. 2013;17(4):e271–e276. doi: 10.1016/j.ijid.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Neonakis I.K., Gitti Z., Krambovitis E., Spandidos D.A. Molecular diagnostic tools in mycobacteriology. Journal of Microbiological Methods. 2008;75(1):1–11. doi: 10.1016/j.mimet.2008.05.023. [DOI] [PubMed] [Google Scholar]
- Ngamskulrungroj P., Price J., Sorrell T., Perfect J.R., Meyer W. Cryptococcus gattii virulence composite: Candidate genes revealed by microarray analysis of high and less virulent Vancouver island outbreak strains. PLoS One. 2011;6(1):e16076. doi: 10.1371/journal.pone.0016076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palka-Santini M., Cleven B.E., Eichinger L., Kronke M., Krut O. Large scale multiplex PCR improves pathogen detection by DNA microarrays. BMC Microbiology. 2009;9:1. doi: 10.1186/1471-2180-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palka-Santini M., Putzfeld S., Cleven B.E., Kronke M., Krut O. Rapid identification, virulence analysis and resistance profiling of Staphylococcus aureus by gene segment-based DNA microarrays: Application to blood culture post-processing. Journal of Microbiological Methods. 2007;68(3):468–477. doi: 10.1016/j.mimet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Palmieri M., Alessi E., Conoci S., Marchi M., Panvini G. In: Microfluidics, bioMEMs, and medical microsystems VI. Wang W., Vauchier C., editors. Vol. 6886. 2008. Developments of the in-check platform for diagnostic applications; p. 14. (Proceedings of the SPIE). 688602. [Google Scholar]
- Park S., Kang Y., Kim D.G., Kim E.C., Park S.S., Seong M.W. Comparison of the analytical and clinical performances of Abbott RealTime High Risk HPV, Hybrid Capture 2, and DNA Chip assays in gynecology patients. Diagnostic Microbiology and Infectious Disease. 2013;76(4):432–436. doi: 10.1016/j.diagmicrobio.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Park J.C., Kim J.M., Kwon O.J., Lee K.R., Chai Y.G., Oh H.B. Development and clinical evaluation of a microarray for hepatitis C virus genotyping. Journal of Virological Methods. 2010;163(2):269–275. doi: 10.1016/j.jviromet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Pasko C., Hicke B., Dunn J., Jaeckel H., Nieuwlandt D., Weed D., et al. Staph ID/R: A rapid method for determining staphylococcus species identity and detecting the mecA gene directly from positive blood culture. Journal of Clinical Microbiology. 2012;50(3):810–817. doi: 10.1128/JCM.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin L.F., de los D.S.-D.R.M., Sanchez I., Hernandez J., Gutierrez-Rios R.M., Lopez-Martinez I., et al. PhyloFlu, a DNA microarray for determining the phylogenetic origin of influenza A virus gene segments and the genomic fingerprint of viral strains. Journal of Clinical Microbiology. 2014;52(3):803–813. doi: 10.1128/JCM.03134-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease A.C., Solas D., Sullivan E.J., Cronin M.T., Holmes C.P., Fodor S.P. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(11):5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter H., Berggrav K., Thomas P., Pfeifer Y., Witte W., Templeton K., et al. Direct detection and genotyping of Klebsiella pneumoniae carbapenemases from urine by use of a new DNA microarray test. Journal of Clinical Microbiology. 2012;50(12):3990–3998. doi: 10.1128/JCM.00990-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowitch E.B., O'Neill S.S., Miller M.B. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. Journal of Clinical Microbiology. 2013;51(5):1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafi W., Chandramuki A., Mani R., Satishchandra P., Shankar S.K. Rapid diagnosis of acute bacterial meningitis: Role of a broad range 16S rRNA polymerase chain reaction. The Journal of Emergency Medicine. 2010;38(2):225–230. doi: 10.1016/j.jemermed.2008.02.053. [DOI] [PubMed] [Google Scholar]
- Rieg S., Jonas D., Kaasch A.J., Porzelius C., Peyerl-Hoffmann G., Theilacker C., et al. Microarray-based genotyping and clinical outcomes of Staphylococcus aureus bloodstream infection: An exploratory study. PLoS One. 2013;8(8):e71259. doi: 10.1371/journal.pone.0071259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringuet H., Akoua-Koffi C., Honore S., Varnerot A., Vincent V., Berche P., et al. hsp65 sequencing for identification of rapidly growing mycobacteria. Journal of Clinical Microbiology. 1999;37(3):852–857. doi: 10.1128/jcm.37.3.852-857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruettger A., Nieter J., Skrypnyk A., Engelmann I., Ziegler A., Moser I., et al. Rapid spoligotyping of Mycobacterium tuberculosis complex bacteria by use of a microarray system with automatic data processing and assignment. Journal of Clinical Microbiology. 2012;50(7):2492–2495. doi: 10.1128/JCM.00442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero P., McMillen T., Tang Y.W., Babady N.E. Evaluation of the BioFire FilmArray respiratory panel and the GenMark eSensor respiratory viral panel on lower respiratory tract specimens. Journal of Clinical Microbiology. 2014;52(1):288–290. doi: 10.1128/JCM.02787-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel L.P., Tibbetts R.J., Agotesku A., Fey M., Hensley R., Meier F.A. Evaluation of a microarray-based assay for rapid identification of Gram-positive organisms and resistance markers in positive blood cultures. Journal of Clinical Microbiology. 2013;51(4):1188–1192. doi: 10.1128/JCM.02982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sango A., McCarter Y.S., Johnson D., Ferreira J., Guzman N., Jankowski C.A. Stewardship approach for optimizing antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Enterococcus species. Journal of Clinical Microbiology. 2013;51(12):4008–4011. doi: 10.1128/JCM.01951-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Takayanagi A., Nagao K., Tomatsu N., Fukui T., Kawaguchi M., et al. Simple PCR-based DNA microarray system to identify human pathogenic fungi in skin. Journal of Clinical Microbiology. 2010;48(7):2357–2364. doi: 10.1128/JCM.02185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanne M., Bodem J., Gerhold-Ay A., Jacob A., Fellenberg K., Krausslich H.G., et al. Genotypic resistance testing in HIV by arrayed primer extension. Analytical and Bioanalytical Chemistry. 2008;391(5):1661–1669. doi: 10.1007/s00216-007-1775-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenzel J. Clinical relevance of new diagnostic methods for bloodstream infections. International Journal of Antimicrobial Agents. 2007;30(Suppl. 1):S2–S6. doi: 10.1016/j.ijantimicag.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Shang S., Chen G., Wu Y., Du L., Zhao Z. Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatric Research. 2005;58(1):143–148. doi: 10.1203/01.PDR.0000169580.64191.8B. [DOI] [PubMed] [Google Scholar]
- Shen H., Shi W., Wang J., Wang M., Li J., Zhang C., et al. Development of a new resequencing pathogen microarray based assay for detection of broad-spectrum respiratory tract viruses in patients with community-acquired pneumonia. PLoS One. 2013;8(9):e75704. doi: 10.1371/journal.pone.0075704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Gunther J., Rebeles J. Genotyping human papillomaviruses: Development and evaluation of a comprehensive DNA microarray. Gynecologic Oncology. 2013;128(3):433–441. doi: 10.1016/j.ygyno.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G., Wen S.Y., Chen S.H., Wang S.Q. Fabrication and optimization of the multiplex PCR-based oligonucleotide microarray for detection of Neisseria gonorrhoeae, Chlamydia trachomatis and Ureaplasma urealyticum. Journal of Microbiological Methods. 2005;62(2):245–256. doi: 10.1016/j.mimet.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Sibley C.D., Peirano G., Church D.L. Molecular methods for pathogen and microbial community detection and characterization: Current and potential application in diagnostic microbiology. Infection, Genetics and Evolution. 2012;12(3):505–521. doi: 10.1016/j.meegid.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes E.A., Patel C., Sung W.K., Lee C.W., Loh K.H., Lucero M., et al. Pathogen chip for respiratory tract infections. Journal of Clinical Microbiology. 2013;51(3):945–953. doi: 10.1128/JCM.02317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Gasson S., Green R.D., Yue Y., Nelson C., Blattner F., Sussman M.R., et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nature Biotechnology. 1999;17(10):974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- Soini H., Skurnik M., Liippo K., Tala E., Viljanen M.K. Detection and identification of mycobacteria by amplification of a segment of the gene coding for the 32-kilodalton protein. Journal of Clinical Microbiology. 1992;30(8):2025–2028. doi: 10.1128/jcm.30.8.2025-2028.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski R.G., Tu E., Butler W.F., O'Connell J.P., Heller M.J. Rapid determination of single base mismatch mutations in DNA hybrids by direct electric field control. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(4):1119–1123. doi: 10.1073/pnas.94.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence R.P., Wright V., Ala-Aldeen D.A., Turner D.P., Wooldridge K.G., James R. Validation of virulence and epidemiology DNA microarray for identification and characterization of Staphylococcus aureus isolates. Journal of Clinical Microbiology. 2008;46(5):1620–1627. doi: 10.1128/JCM.02453-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess B., Seifarth W., Hummel M., Frank O., Fabarius A., Zheng C., et al. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. Journal of Clinical Microbiology. 2007;45(11):3743–3753. doi: 10.1128/JCM.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommenger B., Schmidt C., Werner G., Roessle-Lorch B., Bachmann T.T., Witte W. DNA microarray for the detection of therapeutically relevant antibiotic resistance determinants in clinical isolates of Staphylococcus aureus. Molecular and Cellular Probes. 2007;21(3):161–170. doi: 10.1016/j.mcp.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Sullivan K.V., Deburger B., Roundtree S.S., Ventrola C.A., Blecker-Shelly D.L., Mortensen J.E. Pediatric multicenter evaluation of the Verigene gram-negative blood culture test for rapid detection of inpatient bacteremia involving gram-negative organisms, extended-spectrum beta-lactamases, and carbapenemases. Journal of Clinical Microbiology. 2014;52(7):2416–2421. doi: 10.1128/JCM.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland A., Thomas M., Brandon R.A., Brandon R.B., Lipman J., Tang B., et al. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Critical Care. 2011;15(3):R149. doi: 10.1186/cc10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.D., Briley D., Nguyen Q., Long K., Iannone M.A., Li M.S., et al. Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. Biotechniques. 2001;30(3):661–666. doi: 10.2144/01303dd04. 668–669. [DOI] [PubMed] [Google Scholar]
- Thierry D., Brisson-Noel A., Vincent-Levy-Frebault V., Nguyen S., Guesdon J.L., Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. Journal of Clinical Microbiology. 1990;28(12):2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry S., Hamidjaja R.A., Girault G., Lofstrom C., Ruuls R., Sylviane D. A multiplex bead-based suspension array assay for interrogation of phylogenetically informative single nucleotide polymorphisms for Bacillus anthracis. Journal of Microbiological Methods. 2013;95(3):357–365. doi: 10.1016/j.mimet.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Thissen J.B., McLoughlin K., Gardner S., Gu P., Mabery S., Slezak T., et al. Analysis of sensitivity and rapid hybridization of a multiplexed Microbial Detection Microarray. Journal of Virological Methods. 2014;201:73–78. doi: 10.1016/j.jviromet.2014.01.024. [DOI] [PubMed] [Google Scholar]
- Tobler N.E., Pfunder M., Herzog K., Frey J.E., Altwegg M. Rapid detection and species identification of Mycobacterium spp. using real-time PCR and DNA-microarray. Journal of Microbiological Methods. 2006;66(1):116–124. doi: 10.1016/j.mimet.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Tortoli E., Marcelli F. Use of the INNO LiPA Rif.TB for detection of Mycobacterium tuberculosis DNA directly in clinical specimens and for simultaneous determination of rifampin susceptibility. European Journal of Clinical Microbiology & Infectious Diseases. 2007;26(1):51–55. doi: 10.1007/s10096-006-0240-x. [DOI] [PubMed] [Google Scholar]
- Tortoli E., Mariottini A., Mazzarelli G. Evaluation of INNO-LiPA MYCOBACTERIA v2: Improved reverse hybridization multiple DNA probe assay for mycobacterial identification. Journal of Clinical Microbiology. 2003;41(9):4418–4420. doi: 10.1128/JCM.41.9.4418-4420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.F., Sammons L.R., Zhang X., Suffis S.D., Su Q., Myers T.G., et al. Microarray and molecular analyses of the azole resistance mechanism in Candida glabrata oropharyngeal isolates. Antimicrobial Agents and Chemotherapy. 2010;54(8):3308–3317. doi: 10.1128/AAC.00535-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenemans J., Overdevest I.T., Snelders E., Willemsen I., Hendriks Y., Adesokan A., et al. Next-generation sequencing for typing and detection of resistance genes: Performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. Journal of Clinical Microbiology. 2014;52(7):2454–2460. doi: 10.1128/JCM.00313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao B., Gao Q., Sun Y., Liu P., Feng L., et al. Detection of Enterobacter sakazakii and other pathogens associated with infant formula powder by use of a DNA microarray. Journal of Clinical Microbiology. 2009;47(10):3178–3184. doi: 10.1128/JCM.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., et al. Microarray-based detection and genotyping of viral pathogens. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Orlandi P.A., Stenger D.A. Simultaneous detection of four human pathogenic microsporidian species from clinical samples by oligonucleotide microarray. Journal of Clinical Microbiology. 2005;43(8):4121–4128. doi: 10.1128/JCM.43.8.4121-4128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wang M., Kong F., Gilbert G.L., Cao B., Wang L., et al. Development of a DNA microarray to identify the Streptococcus pneumoniae serotypes contained in the 23-valent pneumococcal polysaccharide vaccine and closely related serotypes. Journal of Microbiological Methods. 2007;68(1):128–136. doi: 10.1016/j.mimet.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Weile J., Schmid R.D., Bachmann T.T., Susa M., Knabbe C. DNA microarray for genotyping multidrug-resistant Pseudomonas aeruginosa clinical isolates. Diagnostic Microbiology and Infectious Disease. 2007;59(3):325–338. doi: 10.1016/j.diagmicrobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Wiesinger-Mayr H., Jordana-Lluch E., Martro E., Schoenthaler S., Noehammer C. Establishment of a semi-automated pathogen DNA isolation from whole blood and comparison with commercially available kits. Journal of Microbiological Methods. 2011;85(3):206–213. doi: 10.1016/j.mimet.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Wiesinger-Mayr H., Vierlinger K., Pichler R., Kriegner A., Hirschl A.M., Presterl E., et al. Identification of human pathogens isolated from blood using microarray hybridisation and signal pattern recognition. BMC Microbiology. 2007;7:78. doi: 10.1186/1471-2180-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witney A.A., Marsden G.L., Holden M.T., Stabler R.A., Husain S.E., Vass J.K., et al. Design, validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Applied and Environmental Microbiology. 2005;71(11):7504–7514. doi: 10.1128/AEM.71.11.7504-7514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright T.C., Jr., Stoler M.H., Behrens C.M., Apple R., Derion T., Wright T.L. The ATHENA human papillomavirus study: Design, methods, and baseline results. American Journal of Obstetrics and Gynecology. 2012;206(1):46.e1–46.e11. doi: 10.1016/j.ajog.2011.07.024. [DOI] [PubMed] [Google Scholar]
- Xiao-Ping K., Yong-Qiang L., Qing-Ge S., Hong L., Qing-Yu Z., Yin-Hui Y. Development of a consensus microarray method for identification of some highly pathogenic viruses. Journal of Medical Virology. 2009;81(11):1945–1950. doi: 10.1002/jmv.21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhang J., Li M., Cao Y., Xu Z., Cao Y., et al. DNA microarray analysis of fluconazole resistance in a laboratory Candida albicans strain. Acta Biochimica et Biophysica Sinica. 2008;40(12):1048–1060. doi: 10.1111/j.1745-7270.2008.00483.x. [DOI] [PubMed] [Google Scholar]
- Yao C., Zhu T., Li Y., Zhang L., Zhang B., Huang J., et al. Detection of rpoB, katG and inhA gene mutations in Mycobacterium tuberculosis clinical isolates from Chongqing as determined by microarray. Clinical Microbiology and Infection. 2010;16(11):1639–1643. doi: 10.1111/j.1469-0691.2010.03267.x. [DOI] [PubMed] [Google Scholar]
- Ye F., Li M.S., Taylor J.D., Nguyen Q., Colton H.M., Casey W.M., et al. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Human Mutation. 2001;17(4):305–316. doi: 10.1002/humu.28. [DOI] [PubMed] [Google Scholar]
- Yoo S.M., Choi J.Y., Yun J.K., Choi J.K., Shin S.Y., Lee K., et al. DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Molecular and Cellular Probes. 2010;24(1):44–52. doi: 10.1016/j.mcp.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Yoo S.M., Lee S.Y., Chang K.H., Yoo S.Y., Yoo N.C., Keum K.C., et al. High-throughput identification of clinically important bacterial pathogens using DNA microarray. Molecular and Cellular Probes. 2009;23(3–4):171–177. doi: 10.1016/j.mcp.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Yu X., Susa M., Knabbe C., Schmid R.D., Bachmann T.T. Development and validation of a diagnostic DNA microarray to detect quinolone-resistant Escherichia coli among clinical isolates. Journal of Clinical Microbiology. 2004;42(9):4083–4091. doi: 10.1128/JCM.42.9.4083-4091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Susa M., Weile J., Knabbe C., Schmid R.D., Bachmann T.T. Rapid and sensitive detection of fluoroquinolone-resistant Escherichia coli from urine samples using a genotyping DNA microarray. International Journal of Medical Microbiology. 2007;297(6):417–429. doi: 10.1016/j.ijmm.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Yue J., Shi W., Xie J., Li Y., Zeng E., Liang L., et al. Detection of rifampin-resistant Mycobacterium tuberculosis strains by using a specialized oligonucleotide microarray. Diagnostic Microbiology and Infectious Disease. 2004;48(1):47–54. doi: 10.1016/j.diagmicrobio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Zhang G., Cai F., Zhou Z., DeVos J., Wagar N., Diallo K., et al. Simultaneous detection of major drug resistance mutations in the protease and reverse transcriptase genes for HIV-1 subtype C by use of a multiplex allele-specific assay. Journal of Clinical Microbiology. 2013;51(11):3666–3674. doi: 10.1128/JCM.01669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Q., Wang D., Chen S., Wang S. Simultaneous detection of oseltamivir- and amantadine-resistant influenza by oligonucleotide microarray visualization. PLoS One. 2013;8(2):e57154. doi: 10.1371/journal.pone.0057154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Wen S., Liu Y., Li R., Zhong X., Feng L., et al. Development of a DNA microarray for detection and identification of Legionella pneumophila and ten other pathogens in drinking water. International Journal of Food Microbiology. 2011;145(1):293–300. doi: 10.1016/j.ijfoodmicro.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Zhu L.X., Zhang Z.W., Wang C., Yang H.W., Jiang D., Zhang Q., et al. Use of a DNA microarray for simultaneous detection of antibiotic resistance genes among staphylococcal clinical isolates. Journal of Clinical Microbiology. 2007;45(11):3514–3521. doi: 10.1128/JCM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimenkov D.V., Antonova O.V., Kuz'min A.V., Isaeva Y.D., Krylova L.Y., Popov S.A., et al. Detection of second-line drug resistance in Mycobacterium tuberculosis using oligonucleotide microarrays. BMC Infectious Diseases. 2013;13:240. doi: 10.1186/1471-2334-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]


