Highlights
-
•
Clinical specificity of Panther Fusion® is between 96 %–100 %, compared to LDT.
-
•
Clinical sensitivity Panther Fusion® is between 71.9 %–100 %, compared to LDT.
-
•
Overall linear regression showed good correlations between LDT and Panther Fusion® for all viruses, except RV and PIV-4.
-
•
The Panther Fusion® provides a random-access system with continuous loading and shorter sample-to-answer times compared to LDT.
Keywords: Panther fusion®, Respiratory viruses, Real-time PCR, Rapid diagnostics, Sample-to-answer
Abstract
Background
Respiratory tract infections are among the most common infections during winter season. Rapid diagnostics is required for clinical decision making regarding isolation of patients and appropriate therapy.
Objectives
The aim of this study was to evaluate the analytical and clinical performance characteristics of the Panther Fusion® respiratory panel using published laboratory-developed real-time PCR assays (LDT).
Study design
Analytical sensitivity of Panther Fusion® Flu A/B/RSV was assessed by testing dilutions of cell culture isolates. Clinical performance assessment included the complete Panther Fusion® respiratory panel (Flu-A/B/RSV, PIV 1-4 and AdV/hMPV/RV) and consisted of a retrospective and a prospective study-arm. The retrospective evaluation included 201, stored (−80 °C) samples collected between February 2006 and January 2017. Prospective evaluation was performed on 1045 unselected pretreated respiratory tract samples from patients presented to our hospital between November 2017 and May 2018.
Results
Analytical sensitivity was generally slightly lower for the Panther Fusion® assays. Clinical specificity and sensitivity was between 96 %–100 % and 71.9 %–100 %, respectively. Discrepant results were found in 146 samples of which 88 samples tested LDT positive / Panther Fusion® negative and 58 samples were LDT negative / Panther Fusion® positive. A total of ten discrepant samples with Ct-values <30 were sequenced to confirm the presence of 7 RV-C not-detected by LDT and 1 RV-A and 2 ADV-2 not detected by Panther Fusion®.
Conclusions
The Panther Fusion® provides a random-access system with continuous loading and much shorter sample-to-answer times compared to LDT, albeit with a slightly less clinical sensitivity compared to the LDT.
1. Background
During the winter season, viral respiratory tract infections are among the most common infections and can be severe in children, elderly, and immunocompromised patients, often leading to hospitalization. Fast and reliable laboratory testing is essential for patient management [[1], [2], [3]]. Many laboratories have developed multiplex qRT-PCR assays to detect respiratory viruses, but more recently also commercial PCR assays have become available. Some of these assays can be used at point-of-care (POCT) and can consist of a limited panel (e.g. Influenza A and B viruses) [[4], [5], [6]] or a comprehensive panel detecting a large number of viruses and even atypical respiratory bacteria [[7], [8], [9]]. In contrast to some POCT respiratory virus PCR assays, which can be fast and easy to perform, most laboratory developed tests (LDT) require highly qualified personnel to perform and interpret the results, much longer times to result (up to ∼6 h) and more hands-on time because of, among others, separated nucleic acid extraction, amplification, analysis and QC.
2. Objectives
We have evaluated the Panther Fusion® and accessory respiratory virus panels. Panther Fusion® is an automated random access system for molecular detection with continuous loading, a turnaround time of 2.5 h and a throughput of up to 120 respiratory tract samples within 8.5 h.
3. Study design
3.1. Analytical performance evaluation
Analytical sensitivity and repeatability was performed using a log10 dilution series of cell culture isolates for influenza A virus (FluA/H1N1/Netherlands/202/95), influenza B virus (FluB/Yamagata/Netherlands/138), respiratory syncytial virus(RSV)-A and RSV-B. Dilution series were prepared, aliquoted and stored at −80 °C until used. Each dilution was tested in 20 replicates in both the Panther Fusion® FLU A/B/RSV assay and a routine laboratory developed automated real-time RT-PCR (LDT) using Aurora FLOW (Roche, Almere, the Netherlands) as described before [4,10,11]. Relative analytical sensitivity was determined by probit analysis with 95 % positivity using IBM SPSS 24. Competitive interference for the FLU A/B/RSV assay was assessed using pairs of target viruses at different concentrations; a range of 0.5log10 serial dilutions were run to determine the sensitivity of these targets in the presence of a high amount of influenza A/B or RSV A/B. Repeatability of each assay was assessed using five replicates of (positive) process controls, containing corresponding virus. To determine the stability of samples in Panther specimens lysis tubes (STM), one replicate of each process control, was tested after 1 day and 6 weeks at 4 °C.
3.2. Clinical performance evaluation
Clinical performance was evaluated both retrospectively and prospectively. For retrospective evaluation stored (−80 °C), 201 pretreated respiratory tract samples collected between February 2006 and January 2017 were selected: This included 25 known LDT-positive samples each for FluA, FluB and RSV, 20 samples for each of the parainfluenza viruses (PIV 1–4) and human metapneumovirus (hMPV), and 40 for human rhinovirus (RV) as well as for adenovirus (AdV). Some of these samples were known to be positive for multiple targets. All samples were tested on the complete Panther Fusion® respiratory panel (Panther Fusion® FluA/B/RSV, Paraflu 1–4, AdV/hMPV/RV) and were retested in LDT to confirm the original results. Prospective evaluation was performed on unselected pretreated respiratory tract samples from patients presented to our hospital between November 2017 and May 2018, with suspicion of infection with respiratory virus. Pretreatment of sputa (SP), Broncho alveolar lavage (BAL) and nasal washes (NW) was performed as described before [4,10]. Briefly, pretreatment consisted of a 7-times dilution step in Dulbecco’s modified Eagle’s Medium (DMEM; Lonza Biowittaker, Vervier, Belgium) including 7.5 % NaHCO3 (Lonza Biowittaker, Vervier, Belgium) 1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Lonza Biowhittaker), 10 % penicillin/ streptomycin (Greiner Bio-one, Alphen a/d Rijn, the Netherlands), 5 % amphotericin B (Bristol-Meyers Squibb, Woerden, the Netherlands), and 40 % foetal bovine serum (Lonza Biowhittaker). After 5 min centrifugation at 3000 rpm (Hettich, Rotina 380, Beun de Ronde, Abcoude, the Netherlands), supernatant was stored at −80 °C or directly used for nucleic acid testing. 500 μl (pretreated) sample was added to the Panther specimens lysis tubes, containing 710 μl specimen transport media (STM), on the same day as the routine LDT assays [4,10,11] were performed. STMs were stored at 4 °C (maximum of 48 h), until Panther Fusion® assays were performed as described by the manufacturer.
3.3. Sanger sequencing
Samples with discrepant results and a Ct value <30 were repeated in both Panther Fusion® and LDT. Confirmed discrepancies were analyzed by Sanger sequencing. For RV a 549 bp fragment of the VP4/VP2-gene [12,13], and for AdV a 600bp fragment of the hexon gene [14], was amplified and sequenced using the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and a 3130xl genetic analyzer (Applied Biosystems). Sequences were analyzed using SeqMan Pro lasergene 10 software (DNA star) and genotypes were determined using NCBI blastn database.
3.4. Statistical analysis
Statistical analysis was performed using IBM SPSS v2.1. 95 % Confidence intervals for sample proportion were calculated using the Wilson method, Epitools (http://epitools.ausvet.com.au).
4. Results
4.1. Analytical performance evaluation
Relative Analytical sensitivity was determined by probit analyses at 95 % hitrate. Difference in relative sensitivity between Panther Fusion® and LDT was 0.53log10, 1.06log10, 0.36log10, 0.96log10, for FluA, FluB, RSV-A and RSV-B, respectively. The presence of two viruses in various concentrations was tested to establish the multiplex capacity of the assay. A high amount (Ct17) of influenza A virus resulted in an undetected internal control and a slightly less analytical sensitivity of the RSV-A and B viruses (Table 1 ). In contrast, high amounts of influenza B virus (Ct16) and RSV-A and B (Ct18-19) have no effect on the sensitivity.
Table 1.
Competitive interference within the FLUA/B/RSV assay.
| Single target |
FLU A H+ (Ct 17) |
FLU B H+ (Ct16) |
RSV-A H+ (Ct18) |
RSV-B H+ (Ct19) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dilution | FLUA (Ct) |
FLUB (Ct) |
RSVA (Ct) |
RSVB (Ct) |
IC# (Ct) |
FLUB (Ct) |
RSVA (Ct) |
RSVB (Ct) |
IC# (Ct) |
FLUA (Ct) | RSVA (Ct) | RSVB (Ct) | IC# (Ct) |
FLUA (Ct) | FLUB (Ct) | IC# (Ct) |
FLUA (Ct) | FL-B (Ct) | IC# (Ct) |
| 1 | 21.0 | 19.7 | 22.8 | 24.0 | <36^ | 19.2 | 23.4 | 23.1 | – | 20.9 | 23.9 | 23..4 | >39^ | 21.0 | 19.3 | – | 21.0 | 19.7 | >39^ |
| 2 | 23.9 | 22.0 | 26.3 | 27.4 | <36 | 21.7 | 27.0 | 28.6 | – | 23.3 | 27.1 | 28.0 | <36^ | 23.3 | 22.1 | – | 23.3 | 22.2 | >39^ |
| 3 | 24.7 | 22.9 | 27.6 | 28.3 | <36 | 22.8 | 28.4 | 30.4 | – | 24.2 | 27.9 | 28.9 | <36^ | 24.4 | 22.9 | – | 24.2 | 23.1 | >39^ |
| 4 | 27.3 | 25.5 | 30.4 | 31.0 | <36 | 25.4 | 32.6 | 34.3 | – | 26.7 | 30.1 | 31.8 | <36^ | 27.0 | 26.1 | >36 | 27.3 | 25.8 | <36 |
| 5 | 28.2 | 27.0 | 31.1 | 32.4 | <36 | 27.5 | 35.2 | 36.7 | – | 28.2 | 31.2 | 32.3 | <36^ | 28.1 | 26.6 | >36 | 28.4 | 27.1 | <36 |
| 6 | 30.0 | 29.4 | 33.0 | 33.2 | <36 | 32.8 | 36.3 | 40.6 | – | 30.2 | 33.8 | 35.3 | <36 | 30.2 | 29.2 | >36 | 31.1 | 29.7 | <36 |
| 7 | 31.7 | 30.2 | 34.8 | 35.3 | <36 | 34.9 | 38.2 | 40.7 | – | 31.1 | 35.1 | 36.1 | <36 | 31.5 | 30.5 | <36 | 32.1 | 30.3 | <36 |
| 8 | 34.6 | 31.5 | 37.1 | 37.2 | <36 | 39.8 | 40.6 | – | – | 34.2 | 36.8 | 40.0 | <36 | 36.4 | 35.1 | <36 | 35.0 | 34.0 | <36 |
| 9 | 36.1 | 33.5 | 37.9 | 37.7 | <36 | 40.8 | 44.1 | – | – | 36.1 | 39.6 | 39.7 | <36 | 36.6 | 34.1 | <36 | 35.6 | 33.0 | <36 |
| 10 | 36.6 | 35.4 | 41.5 | 40.4 | <36 | 41.8 | – | – | – | 36.0 | 39.1 | 42.3 | <36 | – | 39.2 | <36 | 40.4 | 36.0 | <36 |
#Average IC in single target samples = Ct 32.9 (32.1–34.6), IC Ct>40 was considered negative, ^IC FLU-A >40 or negative.
Repeatability was determined by calculating %coefficient of variation (CV) of five replicates of the positive process controls. All replicates for all targets tested positive, with a mean Ct-value of 32.3 (31.2–35.2) and %CV < 4.78 %. Measurements after 6 weeks at 4 °C were within 1.2 Ct-value difference compared to the initial results for all viruses except for PIV2 (Ct-value difference of 3.5).
4.2. Clinical performance evaluation
A total of 1246 respiratory tract samples (sample type distribution, Table 2 ) from 977 patients were collected, of which 201 retrospectively and 1045 prospectively. The retrospective study existed of 159 nasal washes (NW), 17 throat swabs (TS), 9 BAL and 16 sputa (SP). Of these samples 11.4 % was positive for FluA, 12.4 % FluB, 14.4 % RSV, 8 % PIV-1, 10 % PIV-2, 10.4 % PIV-3, 9 % PIV-4, 17.9 % AdV, 10 % hMPV, 19.4 % HRV and in both LDT and Panther Fusion®. Mean Ct-values ranged from 23.3–30.0 for LDT and from 26.5–31.8 for Panther Fusion®. The prospective study consisted of 679 throat swabs, 137 nasal washes, 112 sputa, 68 nasopharynx swabs, 47 BAL, 1 saliva and 1 pleural fluid. In this study-arm, 5.5 % was positive for FluA, 9.7 % FluB, 6.9 % RSV, 1 % PIV-1, 0.3 % PIV-2, 1.3 % PIV-3, 0.5 % PIV-4, 0.9 % AdV, 3.7 % hMPV and 11.5 % RV in LDT as well as on the Panther Fusion®. Mean Ct values ranged from 24.5–31.1 for LDT and from 28.4–36.3 for Panther Fusion®.
Table 2.
Sample type distribution including both (retro and prospective) study arms.
| BAL |
NS |
NW |
SP |
TS |
SA |
PF |
Total |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 56 | % | n = 68 | % | n = 296 | % | n = 128 | % | n = 696 | % | n = 1 | % | n = 1 | % | n = 1246 | % | |
| AdV | 1 | 1,8 | 0 | 0,0 | 29 | 9,8 | 4 | 3,1 | 11 | 1,6 | 0 | 0,0 | 0 | 0,0 | 45 | 3,6 |
| hMPV | 5 | 8,9 | 7 | 10,3 | 16 | 5,4 | 4 | 3,1 | 27 | 3,9 | 0 | 0,0 | 0 | 0,0 | 59 | 4,7 |
| FLUA | 2 | 3,6 | 5 | 7,4 | 26 | 8,8 | 8 | 6,3 | 40 | 5,7 | 0 | 0,0 | 0 | 0,0 | 81 | 6,5 |
| FLUB | 5 | 8,9 | 5 | 7,4 | 26 | 8,8 | 13 | 10,2 | 79 | 11,4 | 0 | 0,0 | 0 | 0,0 | 128 | 10,3 |
| RSV | 3 | 5,4 | 4 | 5,9 | 40 | 13,5 | 10 | 7,8 | 42 | 6,0 | 0 | 0,0 | 0 | 0,0 | 99 | 7,9 |
| PIV1 | 2 | 3,6 | 0 | 0,0 | 16 | 5,4 | 1 | 0,8 | 8 | 1,1 | 0 | 0,0 | 0 | 0,0 | 27 | 2,2 |
| PIV2 | 1 | 1,8 | 0 | 0,0 | 17 | 5,7 | 1 | 0,8 | 4 | 0,6 | 0 | 0,0 | 0 | 0,0 | 23 | 1,8 |
| PIV3 | 0 | 0,0 | 0 | 0,0 | 17 | 5,7 | 5 | 3,9 | 13 | 1,9 | 0 | 0,0 | 0 | 0,0 | 35 | 2,8 |
| PIV4 | 0 | 0,0 | 0 | 0,0 | 17 | 5,7 | 2 | 1,6 | 4 | 0,6 | 0 | 0,0 | 0 | 0,0 | 23 | 1,8 |
| RV | 6 | 10,7 | 8 | 11,8 | 74 | 25,0 | 23 | 18,0 | 49 | 7,0 | 0 | 0,0 | 0 | 0,0 | 160 | 12,8 |
| NVD | 30 | 53,6 | 32 | 47,1 | 59 | 19,9 | 56 | 43,8 | 387 | 55,6 | 1 | 100,0 | 1 | 100,0 | 566 | 45,4 |
Samples were considered positive if both LDT and Panther Fusion® results were positive. NS = nasopharynx swabs; BAL = Broncho-Alveolar Lavages; TS = throat swabs; SP = sputa; NW = nasal washes; SA = saliva, PF = pleural fluid; NVD = No Virus Detected; n = number.
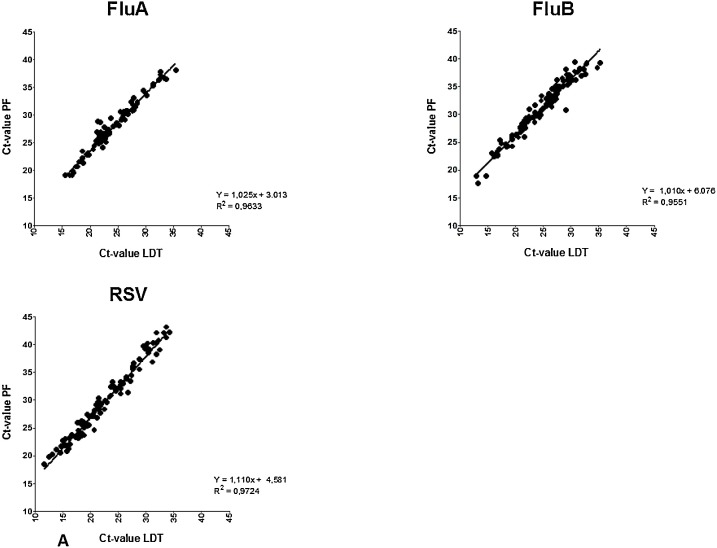
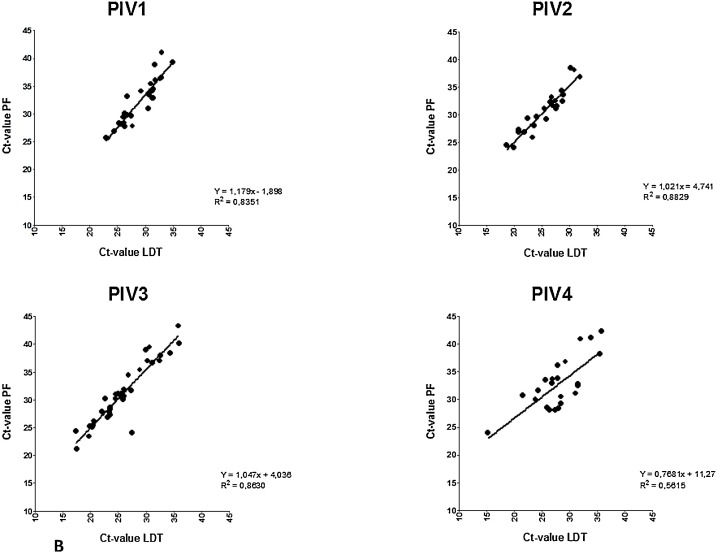
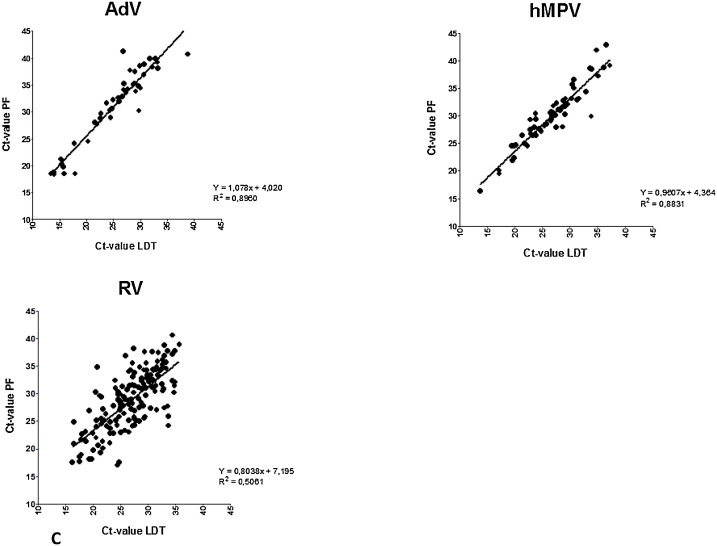
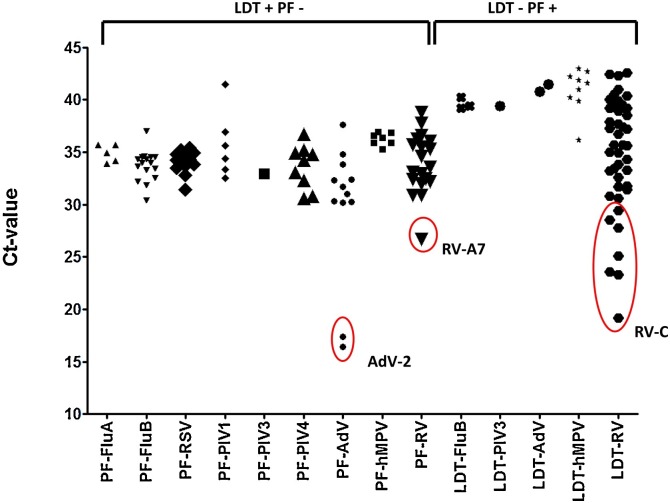
Clinical sensitivity and specificity was determined using all retrospective and prospective samples, with the LDT as the gold standard (Table 3 ). This resulted in a specificity of >99 % for all viruses except RV (96 %). Sensitivity of the Panther Fusion® varied from 71.9 %–100 %. Fig. 1 shows the distribution of paired LDT and Panther Fusion® Ct-values. Only samples positive in both assays were included (n = 680). Deming regression analysis shows a good correlation (slope 0.9607–1.179, R2>0.83) between LDT and Panther Fusion® for most viruses, except for PIV-4 (slope = 0.7681, R2 = 0.56) and RV (slope = 0.8038 R2 = 0.51). Average differences in Ct-value between Panther Fusion® and LDT varied from 3.3–7.1; lower Ct-values were generated by LDT, except for RV, where 28 % of the lower Ct-values were generated by Panther Fusion®. PIV-4 positive samples appear to be divided in two subgroups, samples above the linear regression line have a comparable Ct-value between LDT and Panther Fusion® with an average Ct-value difference of 1.3 (0.0–2.7), were samples below this line have an average Ct-value difference of 7.4 (5.9–9.2). Discrepant results were found for all viruses, except PIV-2 (Fig. 2 ). A total of 88 samples were positive in LDT but negative in the Panther Fusion® assay. Most of these samples exhibited high Ct-values in the LDT (26 with Ct>35; 59 with Ct 30–35; 3 with Ct<30). Conversely 58 samples were positive in the Panther Fusion® but negative in LDT, again, mostly at high Ct-values (38 with Ct>35; 13 with Ct 30–35; 7 with Ct<30). Discrepant samples with a Ct <30 were further investigated by Sanger sequencing. This included 2 AdV (LDT Ct-value 16.4 and 17.4) and 1 RV (LDT Ct-value 26.8) negative by Panther Fusion®; Ct), and 7 RV samples negative by LDT (Panther Fusion® Ct-value 19.2–29.4). All RV discrepant samples negative by LDT were typed as rhino virus type C. The discrepant samples negative by the Panther Fusion® were typed as AdV-2 and RV-A7. All other discrepant samples, with Ct > 30, were considered discrepant due to small differences in sensitivity between the assays and were not further investigated by sequencing.
Table 3.
Clinical sensitivity and specificity of Panther Fusion® respiratory panel compared to LDT.
| Sensitivity |
Specificity |
|||||
|---|---|---|---|---|---|---|
| N | % | 95 % CI | N | % | 95 % CI | |
| FLUA | 81/86 | 94.2 | [87.1–97.5] | 1162/1162 | 100.0 | [99.7–100.0] |
| FLUB | 128/144 | 88.9 | [82.7–93.0] | 1101/1104 | 99.7 | [99.2–99.9] |
| RSV | 99/112 | 88.4 | [81.1–93.1] | 1136/1136 | 100.0 | [99.7–100.0] |
| PIV-1 | 27/33 | 81.8 | [65.6–91.4] | 1215/1215 | 100.0 | [99.7–100.0] |
| PIV-2 | 23/23 | 100.0 | [85.7–100.0] | 1225/1225 | 100.0 | [99.7–100.0] |
| PIV-3 | 35/36 | 97.2 | [85.8–99.5] | 1211/1212 | 99.9 | [99.5–100.0] |
| PIV-4 | 23/32 | 71.9 | [54.6–84.4] | 1216/1216 | 100.0 | [99.7–100.0] |
| AdV | 45/57 | 78.9 | [66.7–87.5] | 1189/1191 | 99.8 | [99.4–100.0] |
| RV | 160/179 | 89.4 | [84.0–93.1] | 1026/1069 | 96.0 | [94.6–97.9] |
| hMPV | 59/66 | 89.4 | [79.7–94.8] | 1173/1182 | 99.2 | [98.6–99.6] |
Fig. 1.
Correlation plot of paired positive laboratory developed test (LDT) and Panther Fusion® (PF) Ct-values for Flu A/B/RSV (A), Paraflu 1–4 (B) and AdV/hMPV/RV (C) samples. Slopes show a significant deviation from zero for all viruses.
Fig. 2.
Scatter plot of discrepant results; samples with a Ct-value <30 were further investigated by sequencing and are indicated by circles.
5. Discussion
In this study, we evaluated the performance of the Panther Fusion® Flu A/B/RSV, Paraflu 1–4, AdV/hMPV/RV assays and compared this against a well-validated LDT assays. The relative analytical sensitivity of the Panther Fusion® system was slightly less for Flu A/B and RSV compared to the LDT, which corresponded with a slightly less clinical sensitivity compared to the LDT. Two recently published studies [15,16] have already evaluated Panther Fusion® assays. Banerjee et al. assessed the FluA/B and RSV assay against five other sample-to-answer tests and a LDT. However, this study only included retrospective samples, with Panther Fusion® low (<30) Ct-values, which might not give sufficient information about sensitivity. Sam et al. compared Panther Fusion® in a prospective study with the e-Plex (Genmark) and Lyra (Quidel) assays. Samples were considered consensus positive if 2 out of 3 assays gave positive results. Sample results were further divided in nasopharynx swabs and lower respiratory tract samples(LTR). Overall they showed a PPV of 100 % and NPV between 98.4 %–100 %, however, as no Ct-values, or other indications of quantity have been mentioned, sensitivity is hard to assess. Our study shows a Panther Fusion® specificity of >96 % and sensitivity of > 88 % for all viruses except PIV-4 (sensitivity 71.9 %) and AdV (sensitivity 78.9 %) compared to LDT. In addition, for Panther Fusion® Sam et al., found presumed false positive results for hMPV in 7 samples. In our study we also found 9 hMPV-positive samples in Panther Fusion® (Ct > 36.2) that were negative in our LDT. Amplification curves were re-evaluated and all exhibited low-fluorescence, which could result in ambiguous interpretation. It is not uncommon that multiplex syndromic PCR panels are less sensitive compared to well-established LDT assays [17,18]. These syndromic panels often are dependent on uniformed nucleic acid isolation and PCR procedures, which are a trade-off between sensitivity and consolidation.
Linear regression showed good correlations between LDT and Panther Fusion® for all viruses, except RV and PIV-4. These findings could be explained by the large sequence diversity of RV. In addition, the RV LDT assay was not designed to detect all rhinovirus type C strains efficiently, which also contributes to the lower correlation. The number of PIV-4 positive samples in both assays was low (n = 23) and appear to be divided in two different subgroups. Whether this can be attributed to genetic differences between PIV-4A and PIV-4B strains and potential primer/probe mismatches could not be investigated. Primer and probe sequences are unknown, since commercial companies do not tend to share these data. Although for AdV a good correlation between both assays was demonstrated, Panther Fusion® missed 2 high positive AdV-2 samples in the retrospective study. Most likely this was not due to mismatch of primer/probe for AdV-2 as other AdV-2 samples included in the study were detected properly. Why these two AdV-2 samples with low Ct values were missed in the Panther Fusion® remains unclear.
The Panther Fusion® respiratory assays are officially only CE-IVD approved for nasopharyngeal swabs. However, in our diagnostic setting the most common respiratory tract samples are throat swabs, sputa, BAL and nasal washings. Therefore, those materials were added to the study. Sputa, BAL and nasal washings were pretreated according to our diagnostic standard protocol, direct use of these materials has not been tested. All internal controls where within the assay limits, according to the Panther Fusion® software. However, the software only declares a result as invalid if the internal control is negative and no other virus is tested positive. As a consequence a certain degree of PCR inhibition will be tolerated by the Panther Fusion®. In specific cases laboratories could formulate additional cut-off values for the internal control, but these would have to be monitored and interpreted by the user. In addition, in case of for instance a high positive influenza A virus result the internal control may be negative, presumably by competition, but the result will not be rejected by the Panther Fusion® software. Theoretically, co-infections with for instance influenza B virus or RSV at low amounts of virus could inadvertently be reported as negative (Table 1) due to PCR inhibition, that is not monitored correctly.
The Panther Fusion® respiratory panel does not include coronaviruses, enterovirus and bocavirus. Other syndromic respiratory panels such as ePlex (GenMark), Biofire® FilmArray® (bioMérieux), and xTAG RVP FASTv2 (Luminex) do. These panels do not distinguish between rhino- or enteroviruses but ePlex and FilmArray® do have quicker turnaround times of about 60 min, whereas the turnaround time for xTAG is about 6 h and requires more hands-on time [9]. Although the first Panther Fusion® results are produced after approximately 2.5 h, after every 15 min five additional, complete panel results are generated. Panther Fusion® also provides an open channel on which LDT tests can be adapted and run simultaneously with the respiratory panel (and other CE-IVD assays). Furthermore, from one extraction a total of three PCR reactions can be performed, allowing efficient use of samples with limited volume.
In conclusion, although slightly less sensitive as compared to an optimized LDT assay, Panther Fusion® and accessory respiratory virus panels offers a reliable, fully automated system with a minimum hands-on time, ease of use and short sample–to-answer times.
Ethical approval
The study was approved by the Medical Ethical Commission of the Erasmus MC (MEC-2016-702).
CRediT authorship contribution statement
Jolanda J.C. Voermans: Writing - original draft, Formal analysis, Validation. Daphne G.J.C. Mulders: Investigation, Validation, Writing - review & editing. Suzan D. Pas: Conceptualization, Funding acquisition, Writing - review & editing. Marion P.G. Koopmans: Funding acquisition, Writing - review & editing. Annemiek A. van der Eijk: Resources, Writing - review & editing. Richard Molenkamp: Writing - review & editing, Supervision.
Declaration of Competing Interest
This study was supported by a research grant from Hologic Deutschland GmbH.
References
- 1.Templeton K.E., Scheltinga S.A., Beersma M.F., Kroes A.C., Claas E.C. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etemadi M.R., Jalilian F.A., Othman N., Lye M.S., Ansari S., Yubbu P. Diversity of respiratory viruses detected among hospitalized children with acute lower respiratory tract infections at Hospital Serdang, Malaysia. J. Virol. Methods. 2019;269:1–6. doi: 10.1016/j.jviromet.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J., Mao N.Y., Zhang C., Yang M.J., Wang M., Xu W.B. The development of a GeXP-based multiplex reverse transcription-PCR assay for simultaneous detection of sixteen human respiratory virus types/subtypes. BMC Infect. Dis. 2012;12:189. doi: 10.1186/1471-2334-12-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voermans J.J., Seven-Deniz S., Fraaij P.L., van der Eijk A.A., Koopmans M.P., Pas S.D. Performance evaluation of a rapid molecular diagnostic, MultiCode based, sample-to-answer assay for the simultaneous detection of Influenza A, B and respiratory syncytial viruses. J. Clin. Virol. 2016;85:65–70. doi: 10.1016/j.jcv.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Lowe C.F., Leung V., Karakas L., Merrick L., Lawson T., Romney M.G. Targeted management of influenza A/B outbreaks incorporating the cobas((R)) Influenza A/B & RSV into the virology laboratory. J. Hosp. Infect. 2019;101:38–41. doi: 10.1016/j.jhin.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Hardick J., Dugas A., Goheen J., Rothman R., Gaydos C. Prospective comparison of RT-PCR/ESI-MS to prodesse ProFlu plus and Cepheid GenXpert for the detection of influenza a and B viruses. J. Virol. Methods. 2015;214:43–45. doi: 10.1016/j.jviromet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babady N.E. The FilmArray(R) respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev. Mol. Diagn. 2013;13:779–788. doi: 10.1586/14737159.2013.848794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babady N.E., England M.R., Jurcic Smith K.L., He T., Wijetunge D.S., Tang Y.W. Multicenter evaluation of the ePlex respiratory pathogen panel for the detection of viral and bacterial respiratory tract pathogens in nasopharyngeal swabs. J. Clin. Microbiol. 2018:56. doi: 10.1128/JCM.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popowitch E.B., O’Neill S.S., Miller M.B. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J. Clin. Microbiol. 2013;51:1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoek R.A., Paats M.S., Pas S.D., Bakker M., Hoogsteden H.C., Boucher C.A. Incidence of viral respiratory pathogens causing exacerbations in adult cystic fibrosis patients. Scand. J. Infect. Dis. 2013;45:65–69. doi: 10.3109/00365548.2012.708942. [DOI] [PubMed] [Google Scholar]
- 11.Dewhurst-Maridor G., Simonet V., Bornand J.E., Nicod L.P., Pache J.C. Development of a quantitative TaqMan RT-PCR for respiratory syncytial virus. J. Virol. Methods. 2004;120:41–49. doi: 10.1016/j.jviromet.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Rahamat-Langendoen J., Riezebos-Brilman A., Borger R., van der Heide R., Brandenburg A., Scholvinck E. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J. Clin. Virol. 2011;52:103–106. doi: 10.1016/j.jcv.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 13.Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 14.Sarantis H., Johnson G., Brown M., Petric M., Tellier R. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 2004;42:3963–3969. doi: 10.1128/JCM.42.9.3963-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sam S.S., Caliendo A.M., Ingersoll J., Abdul-Ali D., Hill C.E., Kraft C.S. Evaluation of performance characteristics of panther fusion assays for detection of respiratory viruses from nasopharyngeal and lower respiratory tract specimens. J. Clin. Microbiol. 2018:56. doi: 10.1128/JCM.00787-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee D., Kanwar N., Hassan F., Essmyer C., Selvarangan R. Comparison of six sample-to-Answer influenza A/B and respiratory syncytial virus nucleic acid amplification assays using respiratory specimens from children. J. Clin. Microbiol. 2018:56. doi: 10.1128/JCM.00930-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramanan P., Bryson A.L., Binnicker M.J., Pritt B.S., Patel R. Syndromic panel-based testing in clinical microbiology. Clin. Microbiol. Rev. 2018:31. doi: 10.1128/CMR.00024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruning A.H.L., Leeflang M.M.G., Vos J., Spijker R., de Jong M.D., Wolthers K.C. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin. Infect. Dis. 2017;65:1026–1032. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]