Abstract
Background
Human Epstein-Barr virus-transformed lymphoblastoid cell lines (LCLs) have been thought to be a useful model system for pharmacogenomics studies. The purpose of this study was to determine the effect of EBV transformation on gene expression changes by dexamethasone (Dex) in LCLs and primary B cells (PBCs) derived from the same individuals.
Patients and methods
We prepared LCLs and purified PBCs from the same six male donors participating the Childhood Asthma Management Program clinical trial, and compared mRNA profiles after 6 hours incubation with Dex (10−6 M) or sham buffer. We assessed differential expression and put the list of differentially expressed genes into the web interface of ConsensusPathDB to find the pathway-level interpretation of our genes specified. As a supplementary analysis, we looked at the expression of the Dex-regulated (inducing or repressing) genes in treatment-naïve PBCs and LCLs (pre Dex-treatment) from the GSE30916 dataset.
Results
By hierarchical clustering, we found clustering of probes by cell types but not by individuals irrespective of Dex-treatment. We observed that the Dex-regulated genes significantly overlapped in PBCs and LCLs. In addition, the expression of these genes showed significant correlations between treatment-naïve PBCs and LCLs. Common genes showing significantly decreased expressions by the Dex-treatment in both cells were enriched in immune responses and pro-inflammatory signaling pathways.
Conclusion
Taken together, these results suggest the use of LCLs are representative of primary biologic effects of corticosteroids treatment.
Keywords: Steroid, Gene expression, Lymphoblastoid cell lines, B cell
Introduction
In searching for the genetic variants associated with drug responses, human Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines (LCLs) has been thought to be a useful model system. However, EBV transformation itself can alter gene expression [1], and thus it is controversial that gene regulation in LCLs recapitulates that of untransformed primary B cells (PBCs). An examination of matched PBCs and LCLs derived from the same donors showed that the expression profiles of more than half of the studied genes were affected by the EBV transformation [2]. In addition, it has been reported that only 9.8% of expression quantitative trait loci identified in LCLs were observed in PBCs [3]. While these changes have been noted in association with baseline gene expression, this model system has also been prominently featured in relation to pharmacogenomics studies linking drug treatment response within the cells to drug treatment in individuals [4–6]. Taken together, for a pharmacogenomics model, we need to understand gene expression differences in LCLs compared with PBCs in the context of response to a particular drug.
Corticosteroids are widely used to treat many inflammatory and immunologic diseases which act by inducing or repressing a wide range of genes [7]. Recently it was reported that genes involved in cholesterol metabolism were similarly regulated by statin between LCLs and PBCs from the same donors [8]. However, in this study, the expression of genes implicated in cell cycle, apoptosis and alternative splicing differed between two cells and authors suggested that drug effects on these pathways may be affected in LCLs [8]. As mentioned before, corticosteroids act by various signaling pathways related with apoptosis, immune reaction, inflammation and so on [7,9]. Therefore we need to evaluate whether gene expression changes in LCLs by corticosteroids are similar with those observed in PBCs to use LCLs as a model system for the pharmacogenomics study of corticosteroid. The purpose of this study was to determine the effect of EBV transformation on gene expression changes by dexamethasone (Dex) in LCLs and PBCs derived from the same individuals.
Patients and methods
Patients and B cell preparation
Blood of six male subjects participating the Childhood Asthma Management Program (CAMP) clinical trial was analyzed. The CAMP clinical trial enrolled 1,041 children aged 5-12 years with mild-to-moderate asthma, who were randomly assigned to treatment with budesonide, nedocromil, or placebo and followed for a mean of 4.8 years [10]. This study was approved by the Institutional Review Board of the corresponding institution and informed consent was obtained from all study participants. Demographics of donors are shown in the online supplementary material (Table S1, Supplemental Digital Content 1, http://links.lww.com/FPC/B339). Peripheral blood mononuclear cells (PBMCs) were isolated from 10 ml of blood using IsoPrep (Robbins Scientific, CA) and transformed by EBV as described previously [11]. Viable cells remaining after 6 weeks were considered to be LCLs. PBMCs were isolated from the remaining 50 ml of blood using Lymphoprep (Alere Technologies, Oslo, Norway), and B cells were specifically isolated from PBMCs using the B cell Negative Isolation kit (Invitrogen, MA) according to the manufacturer’s protocol. Aliquots of B-cells were incubated with FITC-conjugated CD19 fluorescent antibodies and the proportion of CD19-positive cells (B cells only) was determined by FACS analysis (BD FACS Calibur, BD Bioscience, MA) of 10,000-gated events. Only preparations with > 90% B cells purity were used for further analysis.
Gene expression and statistical analysis
Each cell line culture was split into two equal parts. One part was treated with 10−6M Dex and the other part was sham treated. After 6 hours, expression levels were measured using the Illumina HumanRef8 v2 BeadChip (Illumina, San Diego, CA). Each pair of the Dex-treated and sham-treated arrays was in the same batch. We then did vst transformation and quantile normalization to reduce the effects of technical noises and to make the distribution of expression level for each array closer to normal distribution. After filtering, we evaluated 21,175 gene probes (17,370 unique genes). The robust multi-array average algorithm from the affy package in R statistical software (http://cran.at.r-project.org/) was employed to convert probe-level data into expression measures. Then a non-specific intensity-based filtering was done using genefilter package in Bioconductor statistical software (http://bioconductor.org/) and finally 7,780 probes (36.7%) presenting 6,833 genes were forwarded for the differentially expressed genes (DEGs) analysis. The samr package in R statistical software was employed to identify DEGs between the Dex-treated and sham-treated LCLs and between the Dex-treated and sham-treated PBCs. A minimum fold change (expression value of Dex-treated sample/expression value of sham-treated sample) =1 and a false discovery rate (FDR) < 0.05 were used as the cut-off criteria. The delta value for LCLs = 0.7 and the delta value for PBCs = 0.45 were selected respectively based on the delta table and FDR values. We put the list of DEGs into the web interface of ConsensusPathDB (http://cpdb.molgen.mpg.de) to find the pathway-level interpretation of our genes specified. ConsensusPathDB is a meta-database that integrates different types of functional interactions from heterogeneous interaction data resources [12].
Supplementary analysis using Gene Expression Omnibus database
Although we were not able to find another gene expression dataset of the Dex-treated PBCs and LCLs, we looked at the expression of the Dex-regulated genes (genes which showed significant differential expressions in response to the Dex-treatment in the primary analysis) in treatment-naïve PBCs and LCLs (thereby mimicking pre Dex-treatment state). If expression of the Dex-regulated genes showed significant correlation between treatment-naïve PBCs and LCLs, we thought that it would be lends an additional evidence supporting our conclusion. For this purpose, we used the GSE30916 dataset which was generated from a study to investigate variability and consistency in gene expression profiles between five of the most common post venipuncture methods of cell and RNA isolation (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30916) [13]. They measured global gene expression profiles of samples from the same six individuals using the Illumina human-6 v2.0 expression BeadChip (Illumina). We utilized EBV-transformed LCL and CD19-specific B-cell datasets in the present study.
Results
Hierarchical clustering of gene expressions (6 Dex- and sham-treated PBCs and 6 Dex- and sham-treated LCLs) showed a distinct clustering by cell type irrespective of treatment status (Fig. 1). Plotting of gene expressions between the Dex-treated PBCs and Dex-treated LCLs and between the sham-treated PBCs and sham-treated LCLs showed good correlations (Fig. S1, Supplemental Digital Content 1, http://links.lww.com/FPC/B339). Comparison of gene expression between the Dex-treated and sham-treated cells identified 46 transcripts (42 genes) significantly differentially expressed (q < 0.05, Table 1) in PBCs and 107 transcripts (99 genes) in LCLs (q < 0.05, Table 1). Of these, 20 genes showed increased expressions responding to the Dex-treatment in both PBCs and LCLs and this overlap was statistically significant (P = 3.4 × 10−17 and odds ratio = 1258.4, Fisher’s exact test) (Fig. 2A). Among 9 genes which showed decreased expression responding to the Dex-treatment in PBCs, 8 genes were significantly overlapped with those identified in LCLs (P = 1.3 × 10−36 and odds ratio = 345.2, Fisher’s exact test) (Fig. 2B). SAM plots of gene expression in the Dex-treated cells compared to the sham-treated cells were provided in Figure S2, Supplemental Digital Content 1, http://links.lww.com/FPC/B339. These overlapped genes showed significant correlations of expression changes between PBCs and LCLs in response to the Dex-treatment (Fig. 3). Table 2 shows enriched pathways from the common genes showing significantly differential expressions between the Dex-treated and sham-treated cells in both PBCs and LCLs. A total of 32 enriched pathways involving transcriptional regulation and immune response were identified from the common genes with significant decreases in expression by the Dex-treatment in both cells. Results of enrichment analysis using genes showing significant expression changes by the Dex-treatment in either PBCs LCLs were provided in the online supplementary materials (Table S2, Supplemental Digital Content 1, http://links.lww.com/FPC/B339).
Figure 1. Clustering of primary B cells and lymphoblastoid cell lines by individual’s gene expression.
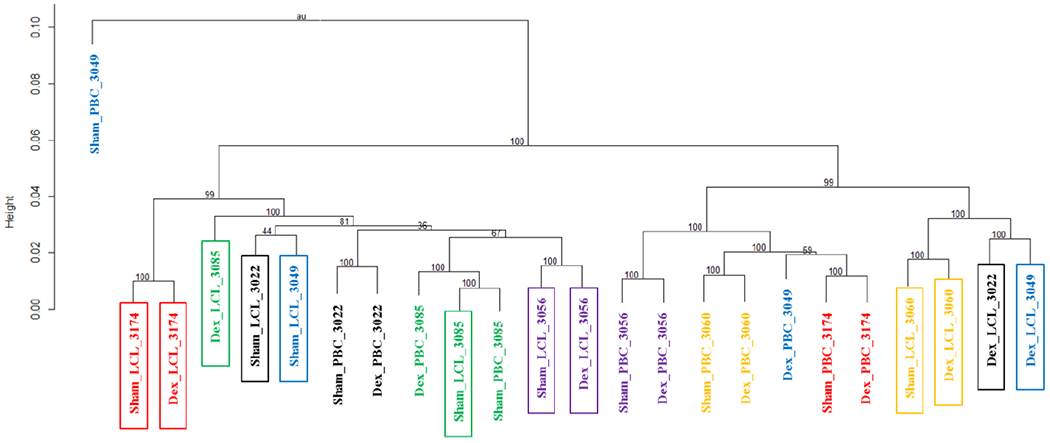
Individuals are designated with treatment_cell_subject’s ID. Boxed individuals indicate lymphosblastoid cells. Numbers on the branches are boot strap support (n = 1,000). Dex, Dexamethasone; PBC, Primary B cell; LCL, Lymphoblastoid cell line
Table 1.
Genes showing significantly differential expressions between dexamethasone-treated and sham-treated cells
| Primary B cells | Lymphoblastoid cell lines | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Increased expression | Decreased expression | Increased expression | Decreased expression | ||||||||
| Gene | Log2FC | Q value (%) | Gene | Log2FC | Q value (%) | Gene | Log2FC | Q value (%) | Gene | Log2FC | Q value (%) |
| TSC22D3 | 1.183 | 0 | LTA | −1.105 | 0 | TSC22D3 | 1.186 | 0 | LTA | −1.115 | 0 |
| SAP30 | 1.132 | 0 | EGR2 | −1.072 | 0 | SAP30 | 1.172 | 0 | PEA15 | −1.08 | 0 |
| DDIT4 | 1.128 | 0 | LYSMD2 | −1.068 | 0 | DDIT4 | 1.128 | 0 | PRMT1 | −1.069 | 0 |
| FKBP5 | 1.118 | 0 | PEA15 | −1.056 | 0 | TXNIP | 1.112 | 0 | SAMSN1 | −1.068 | 0 |
| SMAP2 | 1.107 | 0 | HMOX1 | −1.073 | 3.39 | FKBP5 | 1.102 | 0 | CD83 | −1.054 | 0 |
| CCND3 | 1.092 | 0 | LPXN | −1.058 | 4.82 | SMIM3 | 1.101 | 0 | SLAMF7 | −1.053 | 0 |
| BLK | 1.08 | 0 | BCL2A1 | −1.042 | 4.82 | PSCDBP | 1.09 | 0 | NUS1P1 | −1.05 | 0 |
| SMIM3 | 1.067 | 0 | CSK | −1.041 | 4.82 | CASP6 | 1.082 | 0 | LPXN | −1.043 | 0 |
| SOCS1 | 1.063 | 0 | CYBASC3 | −1.032 | 4.82 | CLIP1 | 1.08 | 0 | P2RY10 | −1.042 | 0 |
| CNDP2 | 1.06 | 0 | P4HA1 | 1.08 | 0 | PRDX1 | −1.038 | 0 | |||
| H1FX | 1.054 | 0 | TOX2 | 1.077 | 0 | OBFC2A | −1.034 | 0 | |||
| CASP6 | 1.051 | 0 | SMAP2 | 1.076 | 0 | TRAF1 | −1.029 | 0 | |||
| TIPARP | 1.049 | 0 | SLC16A3 | 1.063 | 0 | NHP2 | −1.028 | 0 | |||
| P4HA1 | 1.045 | 0 | CDCP1 | 1.06 | 0 | PLEK | −1.081 | 1.5 | |||
| VIL2 | 1.044 | 0 | CNDP2 | 1.059 | 0 | GAR1 | −1.054 | 1.5 | |||
| TXNIP | 1.145 | 3.19 | BIN1 | 1.058 | 0 | CSK | −1.053 | 1.5 | |||
| DUSP1 | 1.099 | 3.19 | S100A10 | 1.055 | 0 | DDX21 | −1.043 | 1.5 | |||
| BTG1 | 1.065 | 3.19 | CCDC6 | 1.055 | 0 | BCL2A1 | −1.058 | 3.24 | |||
| THOP1 | 1.059 | 3.19 | CCND3 | 1.052 | 0 | CD40 | −1.056 | 3.24 | |||
| TOX2 | 1.058 | 3.19 | AP3S1 | 1.05 | 0 | EGR2 | −1.054 | 3.24 | |||
| SEMA4D | 1.055 | 3.19 | THOP1 | 1.049 | 0 | MS4A1 | −1.047 | 3.24 | |||
| DSTN | 1.054 | 3.19 | FGD2 | 1.049 | 0 | HSPD1P4 | −1.045 | 3.24 | |||
| ETS1 | 1.053 | 3.19 | BTLA | 1.049 | 0 | LOC148430 | −1.044 | 3.24 | |||
| MXD4 | 1.051 | 3.19 | AIDA | 1.043 | 0 | NOP16 | −1.04 | 3.24 | |||
| SORT1 | 1.047 | 3.19 | HOST2 | 1.043 | 0 | TIMM23 | −1.03 | 3.24 | |||
| CSDA | 1.046 | 3.19 | STRBP | 1.04 | 0 | PRELID1 | −1.029 | 3.24 | |||
| PLP2 | 1.038 | 3.19 | PTPN1 | 1.039 | 0 | APEX1 | −1.028 | 3.24 | |||
| IQGAP2 | 1.037 | 3.19 | IQGAP2 | 1.037 | 0 | HMOX1 | −1.111 | 4.18 | |||
| KLF2 | 1.059 | 4.82 | CCDC102A | 1.035 | 0 | DNAJA1 | −1.073 | 4.18 | |||
| BIN1 | 1.046 | 4.82 | COQ8A | 1.031 | 0 | LYSMD2 | −1.067 | 4.18 | |||
| CDCP1 | 1.04 | 4.82 | FOXN3 | 1.027 | 0 | ACADM | −1.058 | 4.18 | |||
| CHSY1 | 1.035 | 4.82 | SYCP3 | 1.069 | 1.26 | SPIB | −1.052 | 4.18 | |||
| MAP2K1 | 1.034 | 4.82 | DSTN | 1.066 | 1.26 | HSP90AB1 | −1.052 | 4.18 | |||
| SORT1 | 1.066 | 1.26 | TLR7 | −1.048 | 4.18 | ||||||
| CHPT1 | 1.065 | 1.26 | GADD45B | −1.047 | 4.18 | ||||||
| HACD2 | 1.062 | 1.26 | ADO | −1.045 | 4.18 | ||||||
| EZR | 1.056 | 1.26 | MTX2 | −1.043 | 4.18 | ||||||
| STUM | 1.052 | 1.26 | DYNLL1 | −1.04 | 4.18 | ||||||
| RASSF2 | 1.051 | 1.26 | AGPAT5 | −1.039 | 4.18 | ||||||
| KLF2 | 1.042 | 1.26 | SQSTM1 | −1.038 | 4.18 | ||||||
| INIP | 1.037 | 1.26 | POLR1C | −1.037 | 4.18 | ||||||
| CFLAR | 1.035 | 1.26 | DKC1 | −1.036 | 4.18 | ||||||
| TMEM243 | 1.034 | 1.26 | HSPD1 | −1.035 | 4.18 | ||||||
| VKORC1 | 1.024 | 1.26 | IFIH1 | −1.035 | 4.18 | ||||||
| TIPARP | 1.076 | 3.24 | HVCN1 | −1.034 | 4.18 | ||||||
| VAMP5 | 1.064 | 3.24 | ZNF267 | −1.031 | 4.18 | ||||||
| SLC44A2 | 1.056 | 3.24 | YRDC | −1.03 | 4.18 | ||||||
| RASSF4 | 1.039 | 3.24 | ADAR | −1.029 | 4.18 | ||||||
| SLC15A4 | 1.046 | 4.18 | SMARCC1 | −1.024 | 4.18 | ||||||
| MCUB | 1.045 | 4.18 | |||||||||
FC, fold change. Bold denotes the common genes showing significantly differential expressions between dexamethasone- and sham-treated cells in both primary B cells and lymphoblastoid cell lines
Figure 2. Overlap analysis of the dexamethasone-regulated genes in primary B cells and lymphoblastoid cell lines.
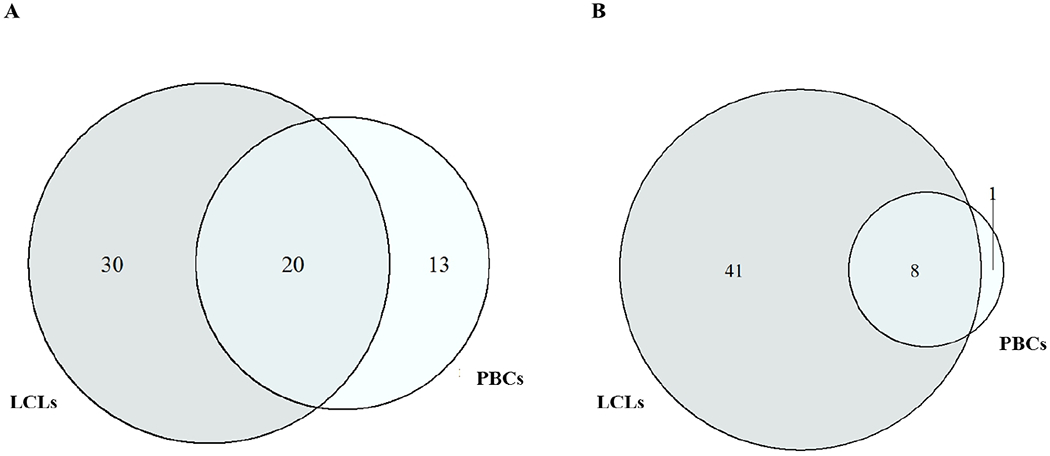
A. Number of genes showing increased expressions
B. Number of genes showing decreased expressions
Figure 3. Correlation of gene expression changes by dexamethasone in paired primary B cells and lymphoblastoid cell lines.
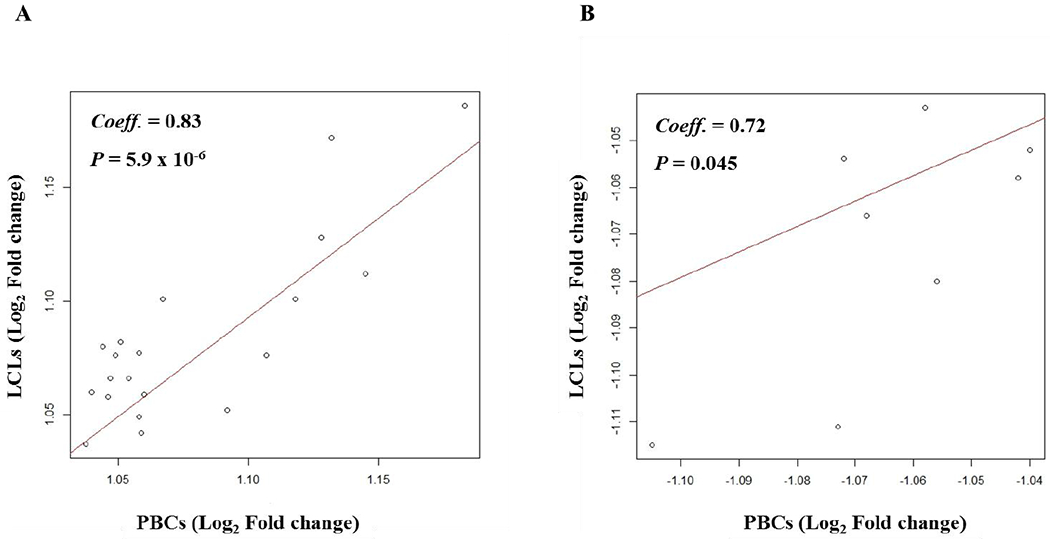
A. Genes showing increased expressions
B. Genes showing decreased expressions
Dex, Dexamethasone; PBC, Primary B cell; LCL, Lymphoblastoid cell line; Coeff, Pearson’s correlation coefficient
Table 2.
Enriched pathways obtained from the common genes showing significantly differential expressions between the dexamethasone-treated and sham-treated cells in both primary B cells and lymphoblastoid cell lines
| p-value | q-value | Pathway | Source | Overlapped genes† |
|---|---|---|---|---|
| Decreased expression | ||||
| 0.00053655 | 0.01007031 | IL4-mediated signaling events | PID | EGR2; LTA |
| 0.00060452 | 0.01007031 | B Cell Receptor Signaling Pathway | PID | CSK; BCL2A1 |
| 0.00107213 | 0.01007031 | Apoptosis Modulation and Signaling | Wikipathways | PEA15; BCL2A1 |
| 0.00111892 | 0.01007031 | NF-kappa B signaling pathway - Homo sapiens | KEGG | LTA; BCL2A1 |
| 0.00175505 | 0.01353898 | Cytokine Signaling in Immune system | Reactome | PEA15; LTA; CSK |
| 0.0049051 | 0.01362912 | RAF/MAP kinase cascade | Reactome | PEA15; CSK |
| 0.0049051 | 0.01362912 | SHC1 events in EGFR signaling | Reactome | PEA15; CSK |
| 0.0049051 | 0.01362912 | SOS-mediated signaling | Reactome | PEA15; CSK |
| 0.0049051 | 0.01362912 | GRB2 events in EGFR signaling | Reactome | PEA15; CSK |
| 0.00509938 | 0.01362912 | Signaling to p38 via RIT and RIN | Reactome | PEA15; CSK |
| 0.00509938 | 0.01362912 | ARMS-mediated activation | Reactome | PEA15; CSK |
| 0.0051485 | 0.01362912 | Frs2-mediated activation | Reactome | PEA15; CSK |
| 0.00519785 | 0.01362912 | MAPK1/MAPK3 signaling | Reactome | PEA15; CSK |
| 0.00524742 | 0.01362912 | Prolonged ERK activation events | Reactome | PEA15; CSK |
| 0.00529721 | 0.01362912 | Signaling by Leptin | Reactome | PEA15; CSK |
| 0.00534722 | 0.01362912 | Signaling to RAS | Reactome | PEA15; CSK |
| 0.00539745 | 0.01362912 | Interleukin receptor SHC signaling | Reactome | PEA15; CSK |
| 0.00554948 | 0.01362912 | VEGFR2 mediated cell proliferation | Reactome | PEA15; CSK |
| 0.0057035 | 0.01362912 | Signaling to ERKs | Reactome | PEA15; CSK |
| 0.00575528 | 0.01362912 | IL2 signaling | Reactome | PEA15; CSK |
| 0.00585949 | 0.01362912 | RET signaling | Reactome | PEA15; CSK |
| 0.00623116 | 0.01362912 | IL3, IL5 and GM-CSF signaling | Reactome | PEA15; CSK |
| 0.00650319 | 0.01362912 | NCAM signaling for neurite out-growth | Reactome | PEA15; CSK |
| 0.00700651 | 0.01362912 | MAPK family signaling cascades | Reactome | PEA15; CSK |
| 0.0075273 | 0.01362912 | IRS-mediated signaling | Reactome | PEA15; CSK |
| 0.00770476 | 0.01362912 | Insulin receptor signaling cascade | Reactome | PEA15; CSK |
| 0.00776433 | 0.01362912 | IRS-related events triggered by IGF1R | Reactome | PEA15; CSK |
| 0.00776433 | 0.01362912 | IGF1R signaling cascade | Reactome | PEA15; CSK |
| 0.00782413 | 0.01362912 | Signaling by Type 1 Insulin-like Growth Factor 1 Receptor | Reactome | PEA15; CSK |
| 0.00812628 | 0.0137131 | HTLV-I infection - Homo sapiens | KEGG | EGR2; LTA |
| 0.00925778 | 0.0151491 | Signaling by Insulin receptor | Reactome | PEA15; CSK |
| 0.00958414 | 0.01522188 | VEGFA-VEGFR2 Pathway | Reactome | PEA15; CSK |
| Increased expression | ||||
| 0.0019247 | 0.02265025 | Arginine and proline metabolism - Homo sapiens | KEGG | P4HA1; CNDP2 |
| 0.0038069 | 0.02265025 | TNFalpha | NetPath | TXNIP; FKBP5; IQGAP2 |
| 0.0039971 | 0.02265025 | p75(NTR)-mediated signaling | PID | SORT1; CASP6 |
| 0.00621013 | 0.02639305 | Retinoblastoma in Cancer | Wikipathways | CCND3; SAP30 |
Input options: 13 data bases, minimum overlap with input list = 2 and p-value cutoff = 0.01;
Genes overlapped between the input set and the data base set
Among 113 Dex-regulated genes, 102 were available for interrogation in GSE30916 (Table S3, Supplemental Digital Content 1, http://links.lww.com/FPC/B339). The expressions of the Dex-regulated genes showed significant correlations between treatment-naïve PBCs and LCLs in each matched pair of subjects (P < 2.2 × 10−22) (Fig. 4). Only two genes (DUSP1 and SOCS1) showed significant differential expressions between PBCs and LCLs (q < 0.05) (Table S3, Supplemental Digital Content 1, http://links.lww.com/FPC/B339).
Figure 4.
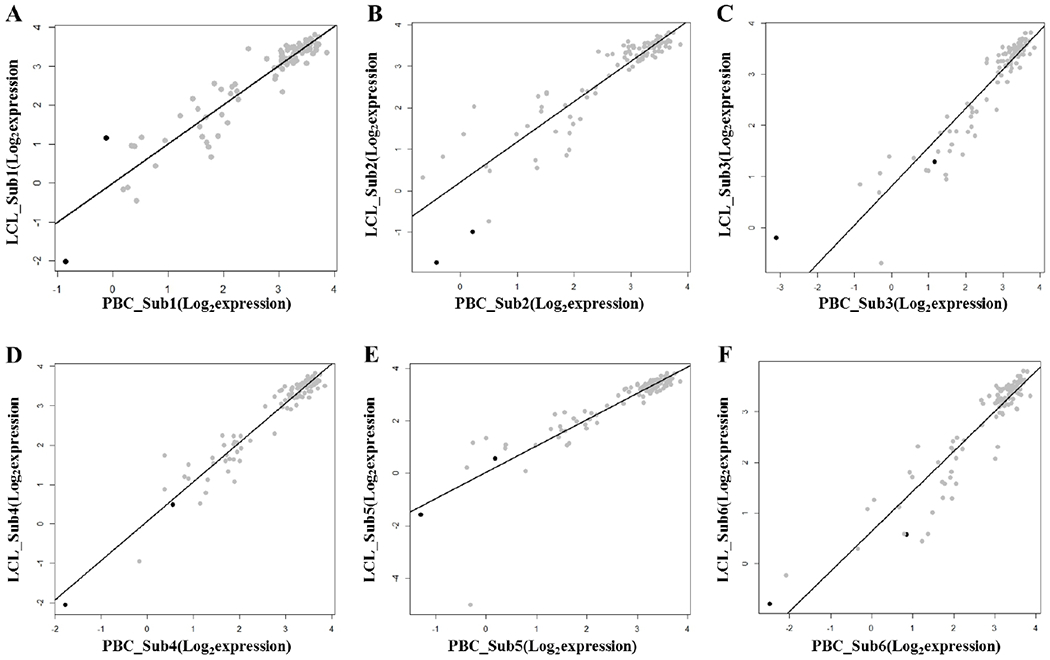
Correlation of the dexamethasone-regulated gene expressions between primary B cells and lymphoblastoid cell lines in the GSE30916 dataset
A. Subject 1
B. Subject 2
C. Subject 3
D. Subject 4
E. Subject 5
F. Subject 6
Black dots represent two genes showing significantly differential expressions between PBCs and LCLs in GSE30916 (FDR q < 0,05).
Discussion
In this study, we aimed to assess the effect of EBV transformation of B cells on transcriptional response to corticosteroids (dexamethasone), a widely prescribed class of anti-inflammatory drugs. Although previous reports have shown that LCLs are a useful model for the study of cholesterol metabolism and statin response in vitro [8,14,15], it is totally unknown that LCLs can recapitulate the naturally occurring gene expression changes in PBCs in response to corticosteroids. As well known, corticosteroids have a complicated mode of action and thus affect a bunch of genes. By hierarchical clustering, we found clustering of probes by cell types but not by individuals irrespective of the Dex-treatment. As we used a subset of probes in our analysis, we cannot say that our results represent the genome-wide effects of EBV transformation. However, these findings suggest that EBV transformation may affect natural genetic or epigenetic control of transcription. In this study, we observed that the Dex-regulated (inducing or repressing) genes significantly overlapped in PBCs and LCLs. Common genes showing significantly decreased expressions by the Dex-treatment in both cells were enriched in immune responses and pro-inflammatory signaling pathways. Taken together, these results suggest the use of LCLs would be suitable for the study of repressing effects on immune and inflammatory reactions by corticosteroids.
Corticosteroids regulate gene expressions in several ways. They suppress the multiple inflammatory genes that are activated in chronic inflammatory diseases and activate transcription of anti-inflammatory genes at higher concentrations [7]. Using common genes showing significantly increased expressions by the Dex-treatment in both PBCs and LCLs, we identified only 4 pathways in common, none of which classically related with corticosteroids action. In contrast, we found that diverse pathways were enriched in genes showing significantly increased expressions by the Dex-treatment in PBCs only. These findings may underscore some of innate differences between two cellular states and explain in part why some genes did not overlap. Taken together, LCLs may not be a useful model system for the study of corticosteroids effects on gene activations.
EBV transformation ultimately leads to cellular proliferation including the activation of the NFkB signaling pathway, in part, by mimicking CD40 activation [16]. As expected, the enriched pathways from genes showing significantly decreased expressions by the Dex-treatment in LCLs only included Telomere Extension By Telomerase pathway, NF-kappa B signaling pathway, and CD40/CD40L signaling pathway. This LCLs’ activated state may also implicate that they are a suitable system for studying the immunorepressive effects of corticosteroids. Moreover, some regulatory variants that affect corticosteroids response in LCLs may be shared with other cell types, as observed for baseline expression [17–19].
A small number of subject enrolled and a lack of replication analysis were limitations of our study. In prior work [20], we had noted over 5,000 differentially expressed genes following differential expression in a large number of LCLs. Therefore, it is highly likely that the subset of overlapping genes would increase with an increased sample size. As mentioned before, we were not able to find another gene expression dataset to replicate our findings, we performed a supplementary analysis using gene expression profiles of the treatment-naïve (pre Dex-treatment) PBCs and LCLs. Resultantly, we noticed that the expression of the Dex-regulated genes obtained from our analysis was significantly correlated between two cells. In addition to our observation that the Dex-regulated genes significantly overlapped between the Dex-treated PBCs and LCLs, the expression of these genes showed significant correlations between treatment-naïve PBCs and LCLs. This finding lends an additional evidence supporting our conclusion.
Conclusion
In conclusion, we observed that the Dex-regulated (inducing or repressing) genes significantly overlapped in PBCs and LCLs and found that use of LCLs for the study of repressing effects on immune and inflammatory reactions by corticosteroids would be suitable by gene set enrichment analysis.
Supplementary Material
Acknowledgments
Funding
NIH R01 HL092197, U01 HL065899, and R01 HL127332
Footnotes
Conflicts of interest
The authors declare that they have no competing interests relevant to this study.
References
- 1.Carter KL, Cahir-McFarland E, Kieff E. Epstein-barr virus-induced changes in B-lymphocyte gene expression. J Virol 2002; 76:10427–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caliskan M, Cusanovich DA, Ober C, Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum Mol Genet 2011; 20:1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet 2012; 44:502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Huang RS, Dolan ME. Cell-based models for discovery of pharmacogenomic markers of anticancer agent toxicity. Trends Cancer Res 2008; 4:1–13. [PMC free article] [PubMed] [Google Scholar]
- 5.Niu N, Wang L. In vitro human cell line models to predict clinical response to anticancer drugs. Pharmacogenomics 2015; 16:273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K, Bolotin E, Theusch E, Huang H, Medina MW, Krauss RM. Prediction of LDL cholesterol response to statin using transcriptomic and genetic variation. Genome Biol 2014; 15:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes PJ. How corticosteroids control inflammation: Quintiles Prize Lecture 2005. Br J Pharmacol 2006; 148:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolotin E, Armendariz A, Kim K, Heo SJ, Boffelli D, Tantisira K, et al. Statin-induced changes in gene expression in EBV-transformed and native B-cells. Hum Mol Genet 2014; 23:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruver-Yates AL, Cidlowski JA. Tissue-Specific Actions of Glucocorticoids on Apoptosis: A Double-Edged Sword. Cells 2013; 2:202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials 1999; 20:91–120. [PubMed] [Google Scholar]
- 11.Pressman S, Rotter JI. Epstein-Barr virus transformation of cryopreserved lymphocytes: prolonged experience with technique. Am J Hum Genet 1991; 49:467. [PMC free article] [PubMed] [Google Scholar]
- 12.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc 2016; 11:1889–1907. [DOI] [PubMed] [Google Scholar]
- 13.Min JL, Barrett A, Watts T, Pettersson FH, Lockstone HE, Lindgren CM, et al. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics 2010; 11:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min JL, Barrett A, Watts T, Pettersson FH, Lockstone HE, Lindgren CM, et al. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics 2010; 11:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangravite LM, Engelhardt BE, Medina MW, Smith JD, Brown CD, Chasman DI, et al. A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature 2013; 502:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham JP, Arcipowski KM, Bishop GA. Differential B-lymphocyte regulation by CD40 and its viral mimic, latent membrane protein 1. Immunol Rev 2010; 237:226–248. [DOI] [PubMed] [Google Scholar]
- 17.Bullaughey K, Chavarria CI, Coop G, Gilad Y. Expression quantitative trait loci detected in cell lines are often present in primary tissues. Hum Mol Genet 2009; 18:4296–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, et al. GR. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet 2010; 87:779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet 2011; 7:e1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu W, Rogers AJ, Damask A, Raby BA, Klanderman BJ, Duan QL, et al. Pharmacogenomics: novel loci identification via integrating gene differential analysis and eQTL analysis. Hum Mol Genet 2014; 23:5017–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


