Abstract
Background
A novel influenza A virus, subtype A/H1N1v emerged in April 2009 and caused the first influenza pandemic of the 21st century. Reliable detection and differentiation from seasonal influenza viruses is mandatory for appropriate case management as well as public health.
Objectives
To develop and technically validate a novel one-step real-time RT-PCR assay which can be used for influenza A virus screening and subtyping of A/H1N1v in a singleplex fashion. To assess the clinical performance of a novel commercial influenza RT-PCR kit based on the in-house version.
Study design
A real-time RT-PCR assay targeting the matrix gene of influenza A viruses was developed and validated using in vitro transcribed RNA derived from influenza A/H1N1v, A/H1N1 and A/H3N2 virus as well as plaque-quantified influenza A/H1N1v, A/H1N1 and A/H3N2 virus samples. After validation of the in-house version the commercial RealStar kit was used to assess the clinical performance and specificity on a panel of influenza viruses including A/H1N1v, A/H1N1, swine A/H1N1, A/H3N2, avian A/H5N1 as well as patient specimens.
Results
The lower limit of detection of the in-house version was 2149, 1376 and 2994 RNA copies/ml for A/H1N1v, A/H1N1 and A/H3N2, respectively. The RealStar kit displayed 100% sensitivity and specificity and could reliably discriminate influenza A viruses from A/H1N1v. No cross reaction with swine A/H1N1 and A/H1N2 was observed with the RealStar A/H1N1v specific probe.
Conclusion
Both assays demonstrated high sensitivity and specificity and might assist in the diagnosis of suspected influenza cases.
Keywords: Influenza A virus, Pandemic A/H1N1v virus, Commercial real-time RT-PCR
1. Background
In April 2009 a novel influenza A virus, subtype A/H1N1v emerged and caused the first influenza pandemic of the 21st century.1 Although predominance of A/H1N1v is likely during the upcoming influenza seasons, co-circulation of seasonal influenza A subtypes may occur.2 Definite laboratory diagnosis remains crucial for individual patient management as well as epidemiological surveillance. Most testing algorithms include universal screening assays for influenza A virus and, in case of positive results, type-specific differentiation by separate assays.3, 4 Due to the cumbersome work flow and high costs, multiplex assays have been proposed that can simultaneously screen for A/H1N1v and the former seasonal viruses.5 However, multiplexing of PCR reactions generally reduces their sensitivity and reliability. In addition, these assays have so far not been available in a uniform, quality-controlled, and ready-to-use format that can easily be adopted by clinical laboratories.
The aims of this study were twofold. We developed a matrix gene-based real-time RT-PCR that allows for detecting seasonal influenza A and simultaneously A/H1N1v. This assay was not a multiplex assay, but used only a single amplicon. Discrimination of the respective viruses is accomplished by use of two different fluorescent probes. In a second step the assay was transformed into a commercially available, homogenous test kit format and evaluated on a comprehensive panel of clinical specimens. The commercial assay used the same oligonucleotides as the in-house version and included an internal control system to monitor possible inhibitory substances or procedural failures.
2. Objectives
To develop and validate a singleplex real-time RT-PCR assay for rapid detection and simultaneous differentiation of influenza A from A/H1N1v virus. To compare the limit of detection of the in-house assay with a commercial assay, based on the in-house version. To evaluate this commercial kit on a panel of clinical samples.
3. Study design
Upon alignment of 373 contemporary sequences of segment 7 of various influenza A virus subtypes primer and probe combinations were selected manually using procedures as described.6 A universal primer pair was chosen to amplify an 87 bp amplicon and two specific hydrolysis probes to discriminate influenza A virus and A/H1N1v (Fig. 1 ). Reaction conditions are outlined in Table 1 . Based on these same oligonucleotides, the RealStar influenza RT-PCR version 1.0 (astra diagnostics, Hamburg, Germany) kit was developed using proprietary technology. This commercial assay contained 10 μl RNA sample in a total reaction volume of 25 μl. An internal heterologous control system was included to monitor possible inhibitory effects of the sample matrix. It comprises an exogenously spiked RNA at low copy numbers. The RealStar kit was performed on an ABI Prism 7500 real-time cycler (Applied Biosystems, Weiterstadt, Germany).
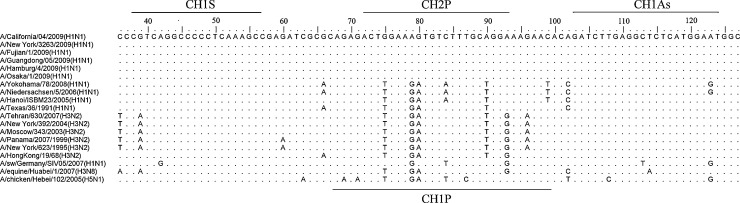
Fig. 1.
Alignment of segment 7 of selected influenza A viruses. Nucleotide alignment of randomly selected influenza A viruses including representative strains from the current pandemic. Dots are representing identities to the head sequence whereas capital letters indicate deviations. Numbering is according to reference sequence A/California/04/2009 (H1N1) (GenBank accession number FJ966085).
Table 1.
Oligonucleotides for singleplex real-time RT-PCR assay.
| Oligonucleotide name | Purposea | Sequence and label, 5′ → 3′ | Position (Acc. No.)b |
|---|---|---|---|
| CH1S1 | Universal forward primer | CGTCAGGCCTCCTCAAAGC | 37–55 (FJ966085) |
| CH1As | Universal reverse primer | ATTCCATGAGAGCCTCAAGATC | 123–102 (FJ966085) |
| CH1P | Detection probe, influenza A viruses | YAK-AGAGACTTGAAGATGTATTTGCTGGGAAGAAT-BHQ1 | 77–108 (CY028100) |
| CH2P | Detection probe, A/H1N1v | FAM-TCCTGCAAAGACACTTTCCAGT-BHQ1 | 92–71 (FJ966085) |
All oligonucleotides were used in the following assay: 20 μL reaction volume, 3 μL of RNA extract (Viral RNA Mini Kit, QIAGEN, Hilden, Germany), OneStep RT-PCR Kit (Qiagen), 240 nmol/L each primer, 120 nmol/L each probe. Cycling at 50 °C for 15 min, 95 °C for 15 min, 45 cycles each at 95 °C for 10 s and 60 °C for 30 s, LightCycler 2.0 (Roche, Mannheim, Germany).
Acc. No., GenBank accession number.
The majority of clinical specimens were nasopharyngeal swabs in universal transport medium (Copan, Brescia, Italy) and a few broncho-alveolar lavage fluids. RNA was extracted by the viral RNA mini kit (140 μl input and 60 μl elution volume; Qiagen, Hilden, Germany).
To determine the lower limit of detection (LOD) of the in-house assay plaque-quantified influenza A/H1N1/Texas/91, A/H3N2/HK/1/68 and A/H1N1v/FR/09 cell culture supernatants as well as in vitro RNA transcripts of partial segment 7 were spiked into viral transport medium and extracted and processed as described.3 For influenza A/H1N1/Texas/91 and A/H3N2/HK/1/68 partial fragments of the matrix gene were amplified using primers H1S (5′-AGTCTTCTAACCGAGGTCGAAACGT-3′) and H1As (5′-GTCTACGCTGCAGTCCTCGCTCACTGG-3′) for A/H1N1 and H3S (5′-GATGAGCCTTCTAACCGAGGTCGA-3′) and H3As (5′-GAACGTTATCTCCCTCTTAAGTTTCC-3′) for A/H3N2, respectively. In-vitro RNA transcripts were constructed as described.3 Cross-reactivity of the in-house assay was tested on a panel of 30 respiratory samples containing adenovirus (n = 1), respiratory syncytial virus-A (n = 8), respiratory syncytial virus-B (n = 2), human coronaviruses OC43 (n = 2), 229E (n = 2) and NL63 (n = 1), human metapneumovirus (n = 2), parainfluenzavirus 3 (n = 4), parainfluenzavirus 4 (n = 1) and entero-/rhinoviruses (n = 7). An additional 20 samples negative for any respiratory virus was also analyzed by the in-house assay. 55 stored influenza A virus positive patient samples including 20 A/H1N1v samples were re-analyzed by the in-house assay only. Specificity was also tested on 22 A/H1N1v and 27 seasonal influenza A PCR positive samples. Additionally the RealStar kit was evaluated on a panel of influenza A virus cell culture supernatants including A/H5N1, A/H3N3, A/H3N8, A/H3N2, A/H1N1, A/H1N1v, classical swine influenza A/H1N1 and A/H1N2, and on a panel of 126 previously quantified A/H1N1v patient specimens.7
4. Results
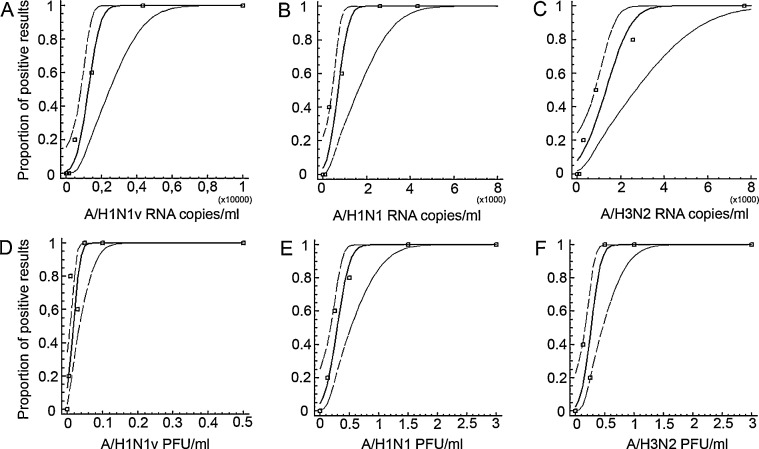
In a first step we used in vitro transcribed RNA to assess the linear range of the in-house assay. Tenfold serial dilutions were tested in triplicates. A linear relationship was observed from 4 × 101 to at least 4 × 107 copies per reaction for influenza A/H1N1v, A/H1N1 and A/H3N2, respectively (data not shown). PCR efficiency was calculated to be 1.8 for A/H1N1v, 1.9 for A/H1N1 and 1.9 for A/H3N2, respectively according to the PCR amplification formula E = 10(1/slope); E being the PCR efficiency. Next in vitro transcribed RNA was used to determine the limit of detection (LOD) using probit analysis.3 Replicate testing of each analyte concentration yielded a 95% LOD of 2149 [95% CI 1506–5170], 1323 [95% CI 881–3875] and 2858 [95% CI 1926–6852] RNA copies/ml for influenza A/H1N1v, seasonal influenza A/H1N1 and A/H3N2, respectively (Fig. 2 ). To assess the LOD with whole virion cell culture supernatants containing plaque-quantified influenza A virus were spiked at different concentrations into universal transport medium and processed as described above. The 95% LOD was 0.04 [95% CI 0.03–0.1], 0.5 [95% CI 0.4–1.7] and 0.5 [95% CI 0.4–1.3] PFU/ml for A/H1N1v, A/H1N1 and A/H3N2, respectively (Fig. 2). No cross reactivity was observed on a panel of clinical samples containing respiratory viruses other than influenza A virus. Another 20 respiratory samples which tested negative by PCR for respiratory viruses yielded no positive results in the in-house assay. In addition 22 A/H1N1v and 27 seasonal influenza A virus containing original patient samples were correctly identified (Table 2 ). In both influenza A virus panels a range of different RNA target concentrations was used with cycle threshold (C t) values from 21.7 to >40. Accordingly 55 stored influenza A virus positive patient samples including 20 A/H1N1v samples yielded positive results by the in-house assay.
Fig. 2.
Probit analysis of influenza A/H1N1v, A/H1N1 and A/H3N2 detection by novel in-house real-time RT-PCR. Probit regression analysis to determine the LOD. X-axis: Different concentrations of in vitro transcribed RNA copies per ml of A/H1N1v, A/H1N1 and A/H3N2 (A–C) as well as plaque-quantified A/H1N1v, A/H1N1 and A/H3N2 (D–E) were tested by real-time RT-PCR in 5 replicate reactions per datum point. Y-axis: Depicted are the observed proportion of positive test results in parallel experiments (square datum point), as well as the derived predicted proportion of positive results at a given input concentration of RNA. Center line denotes the prediction, thin broken border lines are 95% confidence intervals.
Table 2.
Cycle threshold (Ct) values for 27 patient samples containing seasonal influenza A virus and 22 patient samples containing A/H1N1v as assessed by the in-house RT-PCR version, the RealStar kit and reference assays for A/H1N1v3 (based on the hemagglutinin gene) and general influenza A3 (based on the matrix gene), respectively.
|
Ct value |
|||
|---|---|---|---|
| Sample | In-house RT-PCR | RealStar kit | Reference assay |
| Seasonal influenza A virus | |||
| 1 | 29.4 | 27.3 | 29.1 |
| 2 | 29.7 | 25.9 | 28.5 |
| 3 | 34 | 27.7 | 31.5 |
| 4 | 26.8 | 23.3 | 25.7 |
| 5 | 23.8 | 19.3 | 21.7 |
| 6 | 30.6 | 25.9 | 29.6 |
| 7 | 27.6 | 24.1 | 26.2 |
| 8 | 31.6 | 26.6 | 30.2 |
| 9 | 33.9 | 30.0 | 32.6 |
| 10 | 37.9 | >40 | 36.9 |
| 11 | 35.6 | 36.9 | 33.6 |
| 12 | 34.9 | 28.8 | 32.8 |
| 13 | 33.8 | 33.1 | 32.8 |
| 14 | 24.9 | 21.1 | 24.4 |
| 15 | 24.7 | 21.9 | 23.8 |
| 16 | >40 | >40 | 38.8 |
| 17 | 26.3 | 22.8 | 24.9 |
| 18 | 28.7 | 23.6 | 27.5 |
| 19 | 33.7 | 28.6 | 32.9 |
| 20 | 26.1 | 22.6 | 25.9 |
| 21 | 37.5 | >40 | 37.2 |
| 22 | 39.7 | >40 | 38.6 |
| 23 | 29.7 | 24.3 | 28.5 |
| 24 | 33.9 | 35.1 | 33.6 |
| 25 | 29.8 | 24.5 | 29.5 |
| 26 | 36.5 | 32.9 | 35.3 |
| 27 | 28.7 | 23.9 | 27.9 |
| Pandemic A/H1N1v virus | |||
| 1 | 32.9 | 27.2 | 31 |
| 2 | 32.2 | 28.2 | 30.5 |
| 3 | 35.7 | 34.8 | 35.3 |
| 4 | 28.7 | 22.7 | 26.9 |
| 5 | 34.7 | 26.7 | 27.8 |
| 6 | 29 | 22.8 | 34.8 |
| 7 | 36.3 | 29 | 32.8 |
| 8 | 34.9 | 28.3 | 25 |
| 9 | 27.8 | 25.1 | >40 |
| 10 | 30.3 | 20.4 | 32.1 |
| 11 | 33.5 | 27.3 | 31.3 |
| 12 | 33.3 | 27 | 27.3 |
| 13 | 29 | 23.7 | 32.6 |
| 14 | 34.8 | 28 | 34 |
| 15 | 35.7 | 30.3 | 30 |
| 16 | 31.5 | 25.1 | 30.7 |
| 17 | 32.4 | 26.1 | 36.7 |
| 18 | 32.5 | 26.1 | 32.9 |
| 19 | 34.7 | 28.5 | 30.9 |
| 20 | 31.1 | 25.4 | 29.3 |
| 21 | 30.8 | 25.5 | 31.8 |
| 22 | 33.3 | 27.5 | 27.7 |
The LOD of the RealStar kit was assessed using the plaque-quantified influenza A/H1N1, A/H3N2 and A/H1N1v virus. Tenfold limiting dilution series were tested in triplicates. At least 1–10 PFU/ml were detectable by both assays rendering them equally sensitive (data not shown). Correspondingly the LOD was determined using in vitro transcribed RNA as above. A 95% LOD of 380 [95% CI 285–998], 586 [95% CI 427–1300] and 1399 [95% CI 1006–2831] RNA copies/ml for influenza A/H1N1v, seasonal influenza A/H1N1 and A/H3N2 was determined, respectively. A panel of different influenza A virus subtypes including swine A/H1N1 and A/H1N2 yielded no fluorescent signal by the A/H1N1v-specific probe but was positive by the universal influenza A virus probe. Furthermore all 27 seasonal influenza A and 22 A/H1N1v positive samples were correctly identified by the RealStar assay (Table 2). Similar to the in-house version even low concentrated samples were detectable. Of note, C t values obtained by the RealStar kit were in general lower than those obtained by the in-house version and the reference assays which might in part reflect the higher sample input volume of the kit. Finally, 126 A/H1N1v positive patient samples were re-evaluated by the RealStar kit and all yielded concordant results.7 C t values for the A/H1N1v specific RT-PCR ranged from 22.5 to 42 (mean 32.8) and for the RealStar kit from 17.9 to 38.5 (mean 27.4), respectively (p < 0.0001, paired t-test). The internal control yielded a valid reaction in 126/126 samples demonstrating the robustness of the assay.
5. Discussion
The described matrix gene-based assay simultaneously detects influenza A viruses and differentiates A/H1N1v in a singleplex fashion with uniformly good overall sensitivity. Analytical sensitivity of the in-house version was comparable to published singleplex RT-PCR assays.3, 8 For the first time the LOD for each of the contemporary human influenza A viruses was thoroughly assessed from a technical standpoint. In addition we describe the evaluation of one of the first commercial test kits specifically designed to meet the demands of the current pandemic. The RealStar kit rendered to be even more sensitive than the in-house assay most likely attributed to the higher sample input volume and equally specific. Particularly on a panel of original A/H1N1v patient samples the RealStar kit showed superior performance to a widely used real-time RT-PCR.3 Detection of low concentrated samples may become important to, e.g. monitor virus concentrations in patients treated with oseltamivir.9
In the midst of an epidemic rapid provision of ready-to-use test kits has a significant impact on the performance of laboratories.10, 11 Although A/H1N1v in house diagnostic tests were designed and implemented with outstanding rapidity frontline laboratories may lack the capacity to implement these assays.11 Availability of ready-to-use systems might therefore assist laboratories in patient management and surveillance. Combined with its rapidity, internal control system and quality controlled reagents this approach might be useful for general public health laboratories.
In conclusion both assays have demonstrated their usefulness in the current pandemic and might assist laboratories to provide results in a timely, standardized and cost effective manner.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We are grateful to Berit Hollmann and Ulrike Reber for excellent technical assistance. We would like to thank Dr. Ralf Dürrwald (IDT Biologika, Dessau-Rosslau, Germany) for providing swine influenza A viruses. This work was supported by BMBF (contract #01ES0830).
Contributor Information
Marcus Panning, Email: marcus.panning@uniklinik-freiburg.de.
Christian Drosten, Email: drosten@virology-bonn.de.
References
- 1.Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 2.Kelly H.A., Grant K.A., Williams S., Fielding J., Smith D. Epidemiological characteristics of pandemic influenza H1N1 2009 and seasonal influenza infection. Med J Aust. 2009;191:146–149. doi: 10.5694/j.1326-5377.2009.tb02723.x. [DOI] [PubMed] [Google Scholar]
- 3.Panning M., Eickmann M., Landt O., Monazahian M., Olschlager S., Baumgarte S. Detection of influenza A(H1N1)v virus by real-time RT-PCR. Euro Surveill. 2009:14. [PubMed] [Google Scholar]
- 4.Carr M.J., Gunson R., Maclean A., Coughlan S., Fitzgerald M., Scully M. Development of a real-time RT-PCR for the detection of swine-lineage influenza A (H1N1) virus infections. J Clin Virol. 2009;45:196–199. doi: 10.1016/j.jcv.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunson R., Maclean A., Davies E., Bennett S., Miller R., Carman W.F. Development of a multiplex real-time RT-PCR that allows universal detection of influenza A viruses and simultaneous typing of influenza A/H1N1/2009 virus. J Virol Methods. 2010;163:258–261. doi: 10.1016/j.jviromet.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panning M., Laue T., Olschlager S., Eickmann M., Becker S., Raith S. Diagnostic reverse-transcription polymerase chain reaction kit for filoviruses based on the strain collections of all European biosafety level 4 laboratories. J Infect Dis. 2007;196(Suppl. 2):S199–204. doi: 10.1086/520600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drexler J.F., Helmer A., Kirberg H., Reber U., Panning M., Muller M. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis. 2009;15:1662–1664. doi: 10.3201/eid1510.091186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panning M., Hess M., Fischer W., Grywna K., Pfeffer M., Drosten C. Performance of the RealStar Chikungunya virus real-time reverse transcription-PCR kit. J Clin Microbiol. 2009;47:3014–3016. doi: 10.1128/JCM.01024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.To K.K., Chan K.H., Li I.W., Tsang T.Y., Tse H., Chan J.F. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J Med Virol. 2010;82:1–7. doi: 10.1002/jmv.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panning M., Charrel R.N., Mantke O.D., Landt O., Niedrig M., Drosten C. Coordinated implementation of chikungunya virus reverse transcription-PCR. Emerg Infect Dis. 2009;15:469–471. doi: 10.3201/eid1503.081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drosten C., Doerr H.W., Lim W., Stohr K., Niedrig M. SARS molecular detection external quality assurance. Emerg Infect Dis. 2004;10:2200–2203. doi: 10.3201/eid1012.040416. [DOI] [PMC free article] [PubMed] [Google Scholar]