Highlights
-
•
New sequencing technologies increase our knowledge regarding the composition of the human virome.
-
•
There are beneficial and detrimental viruses.
-
•
The human virome can impact upon health at body surfaces (skin or gut) and within tissues.
-
•
Some animal viruses are transmitted via the oral route by consumption of food.
Keywords: next-generation sequencing, virus, metagenomics, virome, microbiome
Abstract
The human virome is the viral component of the microbiome. Its composition, and interindividual and temporal variability are not precisely known. Its impact on human health has received less attention than that of the bacterial microbiome, but is likely to be equally important, both in homeostasis and disease. Here we review the recent advances in this field and the questions that arise in the context of our rapidly increasing knowledge regarding the composition and function of the human virome. With the ever-extending use of next-generation sequencing (NGS) on a variety of clinical samples, rapid progress on the composition of the human virome and its impact upon human health are to be expected in the coming years.
The human virome – the viral component of the human microbiome
The human virome is defined as the viral component of the human microbiome, itself defined as the microbial communities of the various niches of the human body. This is indeed a broad definition, and we have excluded from this review endogenous human retroviruses that do not produce replication-competent particles able to ensure horizontal transmission, despite recent results showing that replication-competent retroviruses do in fact emerge in immunodeficient mice, in a process that is dependent on the intestinal microbiota [1]. We have also excluded viruses of bacteria (bacteriophages).
The history of human virology has been dominated by the discovery of pathogenic viruses as ultrafilterable infectious particles able to multiply in cultured cells. A second wave of discovery came with the development of molecular techniques which enabled the discovery of major human viral pathogens that were not cultivable in vitro (e.g., hepatitis C virus, HCV [2]; hepatitis E virus, HEV [3]; and human herpesvirus 8, HHV8 [4]). Nobel prizes have been delivered to virus hunters [5] in recognition of their contribution to health and science. Currently, the power of new molecular techniques has entirely modified the landscape: although numerous new viruses have been identified, determining their association to disease, if any, will remain a challenging task and a focus for future clinical and pathogenesis investigations. In fact, it is increasingly apparent that viral pathogens are only salient members of a larger group of viruses associated with humans that are not directly linked to disease, analogous to the bacterial component of the microbiome. In particular, viruses abound at the body interfaces with the external environment, on the skin, and in the mucosa. By contrast, the presence of viruses in the blood and organ parenchyma of healthy people tends to be the exception than the rule, although a limited number of viral species establish persistent, albeit mostly silent, systemic infections, more often in immunocompromised individuals (Figure 1 ). Here we review the current knowledge on the human virome at body barriers and systemic compartments, as well as its impact upon human health, both in homeostasis and disease.
Figure 1.
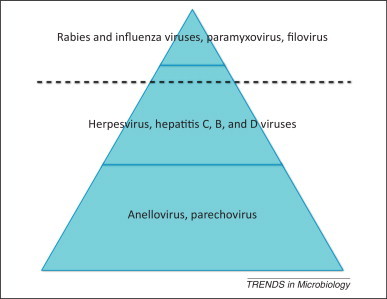
The iceberg of viral infections. The rate at which people show overt disease symptoms as a proportion of those infected can be high (rabies and influenza viruses, paramyxovirus, and filovirus; tip of iceberg), medium (herpesvirus, hepatitis C, B, and D viruses), or low (anellovirus and parechovirus; base of iceberg). In the history of virus discovery attention has progressively shifted from the tip to the base of the iceberg. The volume below the dashed line corresponds to the asymptomatic infections.
New tools reveal the unexpected complexity of the human virome
The history of medicine teaches us that pathogen discovery has been, until recently, a very difficult, time-consuming and low-throughput task, limited by technical hurdles, and requiring the coordination of clinicians and basic scientists. Moreover, this activity has long been mainly hypothesis-driven, similarly to many other research activities. Recently, the development of DNA arrays 6, 7 and new high-throughput DNA sequencing techniques (also known as NGS) have paved the way for quick, unbiased, and sensitive discovery and characterization of pathogens. In addition to these paradigm-shifting technological improvements, global networks dedicated to surveillance of animal and human infectious diseases have been set up, both at the regional and global scale, providing access to highly valuable clinical samples, which is key for pathogen discovery. Moreover, clusters of cases of infectious diseases of unknown origin are identified and pinpointed by diffusion lists such as ProMed (www.promedmail.org) on an almost daily basis. Finally, there is a growing list of microbes associated with chronic conditions including cancers [8], suggesting that increased efforts in microbial investigation of clinical conditions of unknown etiology will be decisive for future personalized diagnostics and therapeutics.
Analysis of the recent literature shows a very rapid increase in NGS usage in the field of environmental microbiology, physiology, and pathogenesis. Recently, very broad and sensitive methodologies have been developed to detect and characterize previously unknown or variant viruses. Technological pipelines for pathogen discovery that create an interface between clinicians, virologists, molecular biologists, and bioinformaticians can uncover viruses referenced in databases with a threshold of sensitivity equivalent to that of quantitative PCR, and are now becoming even more sensitive than PCR with the increasing depth of sequencing 9, 10, 11. This type of pipeline also allows the acquisition and assembly of de novo full-length genomes from biological samples, and therefore the discovery of new viruses even when very distant from known viruses (see [12] for recent review). These tools have begun to reveal the existence and composition of the human virome and unravel its intrinsic complexity and interindividual variability in healthy individuals. Even so, the precise composition and potential impact on health of the human virome at the body surfaces (such as the skin and gut) and within tissues largely remains to be determined.
Viruses at body barriers
Skin
Skin is the most obvious body barrier. The presence of a complex ecosystem at the cutaneous barrier plays a well-established role in preventing adherence and invasion of pathogens, which involves competition between commensal species and their colonization of favorable ecological niches. Similarly to the skin microbiome, the skin virome is composed of both resident and transient viruses. Cutaneous β and γ human papillomaviruses (β- and γ-HPVs) are commonly present on the superficial layers of the skin in most individuals 13, 14. Several studies have described the interindividual and temporal genetic diversity of cutaneous HPV, and new human virus species belonging to the Polyomaviridae family have recently been described, including the tumorigenic Merkel's virus [15].
Unbiased analysis of viral DNA sequences by NGS at the surface of the skin revealed three predominant families [Papillomaviridae (β- and γ-papillomaviruses), Polyomaviridae, and Circoviridae] [16]. This provides definitive evidence for the asymptomatic carriage of numerous HPV strains (up to 17 different strains on individual skin samples). Furthermore, 13 new γ-HPV strains have been identified, which suggests that the diversity of the resident cutaneous γ-HPV group might actually be larger than previously described [16], and their complete inventory thus remains to be established. Merkel cell polyomavirus (MCPyV), human polyomavirus 6 (HPyV6), human polyomavirus 7 (HPyV7), and human polyomavirus 9 (HPyV9) are newly discovered skin-tropic polyomaviruses [17], which indicate that chronic carriage and shedding at the surface of healthy skin is also a hallmark of human polyomaviruses with cutaneous tropism.
Numerous Circoviridae members are also present at the skin surface, mainly belonging to the Cyclovirus genus. Cycloviruses mainly infect animals with cross-species transmission appearing likely. They have also been detected in the feces of primates including humans, suggesting that the relevance of the detection of cycloviruses on the skin remains to be established 18, 19.
Gut
Persistent or intermittent shedding of enteric viruses from healthy people is well established. For example, human enterovirus (HEV) [20] and parechovirus (HPeV) [21] are excreted by a large fraction of children under the age of five without any evidence of association with disease. A 1 year NGS longitudinal study of the stool of two healthy infant siblings in samples taken at 1 week intervals demonstrated that viruses were continuously excreted [22]. The most frequently observed viruses, in decreasing order, were anellovirus (Torque teno viruses, TTV, but also TT-like mini viruses, TTMV), picobirnavirus, and HPeV types 1 and 6. Bocavirus (HBoV-1), adenovirus groups C and F, Aichi virus, astroviruses, and rotavirus were less frequently detected. Surprisingly, other enteric viruses such as noroviruses, coronaviruses, cardioviruses, cosaviruses, saliviruses, and sapoviruses were not detected, although they are frequently detected in stool, indicating that the results of this survey only provide a first indication of the composition of the gut virome, which would benefit from the study of larger samples. Some viruses, including adenoviruses, anelloviruses, picobirnaviruses, parechoviruses, and human bocavirus, were shed for months. These viruses are more likely to represent a significant portion of the normal human virome, owing to their ability to establish persistent infections.
The gut virome is not only composed of fully adapted human viruses: an intriguing feature is the presence of some animal viruses that can be transmitted by the oral route by consumption of contaminated food, and whose kinship to the human virome remains unclear. The first is HEV, which is typically responsible for acute hepatitis in humans. Genotypes 1 and 2 are human-specific but types 3 and 4 have a reservoir in pigs [23]. Although HEV infection is considered to be silent in pigs, this is difficult to ascertain because the main target of HEV in humans are the elderly with previous liver injury and this is not a clinical situation that can be observed in pig husbandry. Furthermore, even in humans, the morbidity rate of HEV is low, which may preclude the identification of disease in pigs. Acute infection is the rule in humans, and the virus can be seen as a typical zoonotic virus. Nevertheless, long-term chronic infections have been described in immunocompromised patients, highlighting the importance of the host immune status on virome composition and its potentially pathogenic properties 24, 25. Also, some genotypes (1 and 2) are circulating between humans, which suggests that genotypes 3 and 4, which very frequently infect humans from the animal reservoir (up to 50% of the population is antibody-positive in some areas [26]), might be adapting to humans. The situation is even less clear for gyroviruses. Within the Circoviridae family, only the chicken anemia virus (CAV) was previously known as a member of this genus until recently. The first gyrovirus in humans (HGyV), a member of the Circoviridae family, was initially found by NGS at the surface of the skin of healthy people [27]. A very similar virus, AGV2, was found a few months later in chickens, and then a third gyrovirus was detected in human stool, in which CAV and HGyV/AGV2 were also identified [28]. A fourth gyrovirus, GyV4, was also found in China in chickens and in human stool [29]. Some reports [29] reveal a high frequency of these four gyroviruses in human stool, possibly owing to cross-species transmission of gyroviruses and their replication in humans, or alternatively to passive transit of animal viruses via food intake, as has been observed for plant viruses [30]. Within the Circoviridae family the cycloviruses constitute a new genus, and are found in the feces of humans and other animal species, posing similar questions as gyroviruses [31].
Systemic compartment
In healthy individuals there seem to be a paucity of viruses in the plasma, with the exception of anelloviruses. Although anelloviruses are so far considered to be non-pathogenic, they cause persistent viremia and can be identified in more than 70% of the general population worldwide [32]. Recently, plasma samples from febrile and afebrile children under the age of 3 years were compared using PCR and NGS [10]. Plasma from afebrile children contained no viral reads other than anelloviruses, whereas sequences from other viral species were recovered from approximately 70% of samples from children with unexplained fever. Virus reads obtained by NGS were on average fivefold more abundant in samples from febrile children than from afebrile children, and enterovirus/rhinoviruses and roseoloviruses were particularly abundant. Herpesviridae, including herpes simplex virus types 1 and 2 (HSV1 and HSV2), cytomegalovirus (CMV), Epstein–Barr virus (EBV), and HHV types 6 to 8, are well-characterized human viruses. Their prevalence in human plasma is high and increases with age. PCR surveys in healthy, febrile and/or immunosuppressed patients have shown that each of these viruses can be detected in human tissues, including blood samples. CMV, EBV, HHV6, and HHV7 are leukotropic viruses and their corresponding viral DNA sequences can be readily detected from circulating human leucocytes [33]. The determination of the significance and imputability of such findings requires careful analysis that integrates the immune status of the host, comorbidities, and concomitant symptoms, as well as the sensitivity of the detection technique. Also, the latent stage of cell-associated viral DNA should be distinguished from productive cycles leading to the production of virus particles. Under homeostatic conditions, these viruses could be considered to be a non-pathogenic component of the human virome. Nevertheless, most of these viruses can be associated with either acute disease at the time of primary infection or with active viral replication upon immunosuppression (Figure 2 ), and either directly or indirectly with pathologic conditions such as lymphoma or sarcoma (EBV and HHV8), encephalitis (HSV, CMV, EBV, and HHV6), or digestive symptoms (CMV and HHV6). It is also likely that the interplay between the host, these viruses, and the microbiome translates into medically significant outcomes, such as inflammatory conditions 34, 35. The case of HHV6 merits particular attention because the genome of this virus can be chromosomally integrated and transmitted vertically. The significance for health of this chromosomal integration remains unclear [36].
Figure 2.
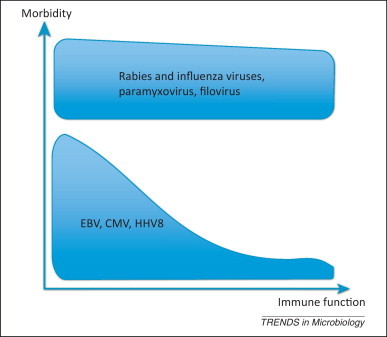
Morbidity rates (likelihood that a person infected with a particular virus will suffer disease) as a function of host immune competence. Abbreviations: CMV, cytomegalovirus; EBV, Epstein–Barr virus; HHV8, human herpesvirus 8.
Of note, some viruses that persistently infect the gut can also be transiently or more persistently present in the blood. A pathogenic strain of simian immunodeficiency virus (SIV), as opposed to a non-pathogenic strain, has recently been described to expand the enteric virome [37] in nonhuman primates. It was suggested that immunosuppression results in an increased level of enteric viral infection that promotes and accelerates acquired immune deficiency syndrome (AIDS) via the modification of the intestinal mucosa and the induction of systemic immune activation. In support of the relationship between immune suppression and expansion of the enteric virome, evidence of HGyV was found in the blood of three of 100 solid organ transplant recipients and in one HIV-infected person, and circulating HGyV was detected transiently in longitudinal plasma samples [38]. HGyV was also detected in 3 of 352 (0.85%) plasma samples from healthy persons [39]. This illustrates that the immune system has an impact upon virome composition. Future studies should focus on studying the impact of virome changes as an indicator of immunosuppression and as a causal factor for disease progression and even occurrence.
Virome and health
It has often been assumed that perfectly adapted viruses are clinically silent, having found an ideal equilibrium with their host to avoid their counter-selection, thereby leading to viral persistence and favoring transmission. However, in some cases, symptoms such as cough, diarrhea, and aggressiveness (for example, in animal rabies increased biting behavior facilitates increased virus transmission) increase the dissemination of the virus. In view of the number of new orphan viruses that are discovered, the question arises as to whether some viruses have actually been selected because they provide a selective advantage to their hosts. For example, some viruses could participate in physiological processes, as is well established for commensal bacteria. The formation of a syncytium layer at the maternal–fetal interface has been demonstrated in primates, rodents, lagomorphs, carnivores, and recently in ruminants [40], and is governed by genes that now encode syncytin protein but that were captured millions of years ago from retroviral genes encoding envelope glycoproteins. Most papillomaviruses are considered to be innocent bystanders of the skin, with no significant tissue damage resulting from their replication and chronic asymptomatic shedding. Nonetheless, it has been speculated that β-HPV could be a human symbiont and actively participate in the proliferation of keratinocytes during wound healing. Local inflammation might transiently decrease the expression of members of the EVER/ZnT-1 complex (which maintains cellular zinc homeostasis) together with barrier efficacy against β-HPV infection [41].
Some viral infections also seem to confer better fitness to infected individuals in the context of other infections. The GBV-C (GB virus type C), also known as hepatitis G virus, is a lymphotropic virus classified in the Flaviviridae family. Similarly to the closely related GBV-A and GB-V viruses of New World primates, it is distantly related to a major human pathogen, HCV. GBV-C is frequently found in humans, with viremia detected in approximately 1–4% of healthy donors in developed countries but in up to 20% in some developing regions. GBV-C is not responsible for any known disease but, conversely, a set of studies associate survival in HIV-infected individuals with GBV-C infection. These epidemiological results are experimentally supported by the observation of an inhibition of HIV replication by GBV-C, which is demonstrable in T cell cultures, although the underlying mechanisms are only partly understood. GBV-C infection is thought to alter the expression of HIV entry receptors, inhibit HIV replication, enhance innate immune responses, polarize cytokines towards a T helper 1 (Th1) profile, protect CD4+ T cells from apoptosis, decrease lymphocyte activation, and interfere with IL-2-induced CD4+ T cell expansion [42].
It has been shown that latent infection by mouse herpesviruses highly similar to human EBV and CMV activates the innate immune response and protects mice against distant microorganisms including bacteria [43]. It is tempting to speculate that this beneficial effect of infection might extend to human herpesviruses. Among the possible mechanisms, latent herpesvirus infection was sufficient to arm natural killer (NK) cells [44]. Even if such protection against acute infections was shown to be relatively short-lived [45], host fitness could be improved when there is a high risk of infection. How fitness is increased and balanced with the rare complications associated with these viruses, such as EBV-malignancies and CMV infections in immunocompromised patients, together with implications regarding the benefit of vaccination against herpesviruses, has been the subject of discussion [46]. Moreover, fitness may vary as a function of the environment and also over time. Beneficial viruses may have turned into enemies in modern times because of longer lifespans and the introduction of new therapies such as immunosuppressive drugs (e.g., leading to possible long-term deleterious effects of persistent viruses on the immune system and cell survival).
It is noteworthy that gyroviruses harbor an apoptin gene, which encodes a protein that has the rare quality of being specifically cytotoxic for cancer cells. Indeed, natural infection could be of benefit in controlling the development of tumor cells [47]. Because these viruses have been found in human stool and at the surface of the skin, they could potentially be of benefit in controlling the early amplification of transformed cells. For example, the expression of polyomavirus large T is sufficient to render cells susceptible to apoptosis upon expression of apoptin. The biological significance of these observations, particularly in light of the many new polyomaviruses found at the surface of the skin or in stool, merits further studies.
Persistence of commensal viruses and immune responses
For viruses to be part of the human virome they must be able to persist in humans. Because the mechanisms underlying viral persistence constitute a broad topic out of the scope of this review, we will highlight here only two specific situations that are applicable to the human virome. First, as more and more viruses have been described on the skin, it is becoming more important to understand viral persistence on the skin. Second, the persistence of anellovirus in blood is a particularly intriguing example.
As described above, papillomaviruses are very frequently found at the surface of the skin and high seropositivity (90%) to at least one HPV type has been reported (reviewed in [48]). Papillomaviruses can persist in the epidermis at a stage of that resembles latency [49] in which gene expression is limited and the immune system is confronted with little exposure to structural proteins that are expressed by late genes in fully differentiated keratinocytes. The innate response to HPV is poorly understood. The virus capsid and DNA are potential pathogen-associated molecular patterns (PAMPs), and HPV interferes with Toll-like receptor and interferon signal-transduction pathways. Control of HPV is associated with a cell-mediated response to early HPV antigens. Keratinocytes can act as non-professional antigen-presenting cells able to present peptides in association with class I and II major histocompatibility complex (MHC) molecules, to secrete proinflammatory cytokines and chemokines, and to activate CD4+ and CD8+ memory T cells. The major professional antigen-presenting cells in the epidermis are the Langerhans cells (LC). In tissues infected with β-HPV types, LCs are associated with a high numbers of regulatory T cells (Treg), which have immunosuppressive properties and are thought to limit the immune response to self-antigens. T cells present in the epidermis are predominantly memory T cells, which have shown limited ability to control infections localized to peripheral tissues. Nevertheless, a distinct tissue-resident memory (TRM) cell subset, which is recruited during the immune response, remains localized in the periphery. It was recently suggested that the TRM cell subset could control viruses that are dependent on initial local replication before systemic infection 50, 51. Therefore HPV must be able to evade their surveillance.
TTVs are circular, single-stranded (ss) DNA viruses classified in the Anelloviridae family together with TTMV, and TT midi virus (TTMDV). Their situation is very unusual because viremia can be identified in nearly all individuals and antibodies against the capsid protein VP1 can be detected in nearly 100% of the population [52]. Viremia is controlled by the innate [53] and adaptive immune response, and people under therapeutic immune suppression exhibit increased virus load [54]. One of the most trivial explanations of immune escape in healthy people is the variability of the virus. In contrast to the double-stranded DNA HPVs and HPyVs, the ssDNA anelloviruses are genetically highly variable [55]. For example, according to the last edition of the International Committee on Taxonomy of Viruses (ICTV) list (v2, 2011; http://ictvonline.org/), there are at least 29 species of TTV which cluster in 5 genogroups, 12 species of TTMV, and 15 species of TTMDV. Children followed for 3 years showed seroconversion against most of the genogroups, with only 5–7% of the children remaining seronegative. Moreover, children who were initially seropositive for a single strain seroconverted later against multiple genogroups. It therefore remains possible that TTV persistence at least partly results from multiple reinfections with different non-cross-reactive species. This shows that sequential infections by different genogroups occur, and that antibodies against a given TTV genotype are not cross-protective against others. The viral genome length is very short (the TTV genome comprises a 1.2 kb non-coding region and a 2.6 kb coding region). It remains unclear how such a level of intra- and inter-genogroup variability is maintained within such a small genome, allowing evasion of the neutralizing antibody response while maintaining interactions with cell receptors.
Viruses able to establish long-term persistence within their host represent a subpopulation of the virome that is particularly fascinating. First, in most cases, our understanding of the mechanisms of persistence remains limited. Second, it remains questionable if such a permanent stimulation of the immune system is favorable or deleterious because it may favor autoimmunity and/or immunosenescence [56].
Concluding remarks
The human virome expands as knowledge increases. Emerging acute human infections are generally of animal origin, and wildlife is rich in viruses that could exploit epidemiological circumstances to cross species barriers into humans [57]. A typology of zoonotic viruses has been proposed according to their capacity to disseminate in human populations, and dissemination might require initial rounds of replication in humans before fully adapted strains emerge [58]. It can also be anticipated that novel viruses with roles in chronic diseases such as cancers still remain to be discovered, in the same way as Merkel polyomavirus and HHV8 have been implicated in Merkel's disease and Kaposi sarcoma, respectively. For example, childhood leukemia has long been suspected to be linked to viruses and, in particular, to polyomaviruses [59]. Conversely, the sensitivity of NGS is so high that negative results have high predictive value, and will at last allow elimination of specific viruses suspected to have a role in particular diseases with a reasonable degree of confidence [60]. It can be anticipated that assessing the virome and bacteriome of patients by broad-range techniques such as NGS (referred to as the microbioscope) will be of interest in medical diagnosis and patient management, and give highly valuable information to adapt treatments such as antimicrobial, anti-inflammatory, and immunosuppressive therapies.
Acknowledgments
We thank Jennifer Richardson for critical reading of the manuscript.
References
- 1.Young G.R. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houghton M. Discovery of the hepatitis C virus. Liver Int. 2009;29(Suppl. 1):82–88. doi: 10.1111/j.1478-3231.2008.01925.x. [DOI] [PubMed] [Google Scholar]
- 3.Reyes G.R. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 5.Weiss R.A. On viruses, discovery, and recognition. Cell. 2008;135:983–986. doi: 10.1016/j.cell.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen E.C. Using a pan-viral microarray assay (Virochip) to screen clinical samples for viral pathogens. JoVE. 2011;50 doi: 10.3791/2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthet N. Massively parallel pathogen identification using high-density microarrays. Microb. Biotechnol. 2008;1:79–86. doi: 10.1111/j.1751-7915.2007.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Martel C. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 9.Cheval J. Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J. Clin. Microbiol. 2011;49:3268–3275. doi: 10.1128/JCM.00850-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wylie K.M. Sequence analysis of the human virome in febrile and afebrile children. PLoS ONE. 2012;7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greninger A.L. A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS ONE. 2010;5:e13381. doi: 10.1371/journal.pone.0013381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu C.Y. Viral pathogen discovery. Curr. Opin. Microbiol. 2013 doi: 10.1016/j.mib.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonsson A. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J. Gen. Virol. 2003;84:1881–1886. doi: 10.1099/vir.0.18836-0. [DOI] [PubMed] [Google Scholar]
- 14.Chen A.C-H. Human papillomavirus type spectrum in normal skin of individuals with or without a history of frequent sun exposure. J. Gen. Virol. 2008;89:2891–2897. doi: 10.1099/vir.0.2008/003665-0. [DOI] [PubMed] [Google Scholar]
- 15.Dalianis T., Hirsch H.H. Human polyomaviruses in disease and cancer. Virology. 2013;437:63–72. doi: 10.1016/j.virol.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 16.Foulongne V. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feltkamp M.C.W. From Stockholm to Malawi: recent developments in studying human polyomaviruses. J. Gen. Virol. 2012;94:482–496. doi: 10.1099/vir.0.048462-0. [DOI] [PubMed] [Google Scholar]
- 18.Li L. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J. Gen. Virol. 2011;92:768–772. doi: 10.1099/vir.0.028704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 2010;84:1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witsø E. High prevalence of human enterovirus a infections in natural circulation of human enteroviruses. J. Clin. Microbiol. 2006;44:4095–4100. doi: 10.1128/JCM.00653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolehmainen P. Human parechoviruses are frequently detected in stool of healthy Finnish children. J. Clin. Virol. 2012;54:156–161. doi: 10.1016/j.jcv.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Kapusinszky B. Nearly constant shedding of diverse enteric viruses by two healthy infants. J. Clin. Microbiol. 2012;50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith D.B. Genetic variability and the classification of hepatitis E virus. J. Virol. 2013;87:4161–4169. doi: 10.1128/JVI.02762-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lhomme S. Hepatitis E virus quasispecies and the outcome of acute hepatitis E in solid-organ transplant patients. J. Virol. 2012;86:10006–10014. doi: 10.1128/JVI.01003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koning L. Clinical implications of chronic hepatitis E virus infection in heart transplant recipients. J. Heart Lung Transplant. 2013;32:78–85. doi: 10.1016/j.healun.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Kamar N. Hepatitis E. Lancet. 2012;379:2477–2488. doi: 10.1016/S0140-6736(11)61849-7. [DOI] [PubMed] [Google Scholar]
- 27.Sauvage V. Identification of the first human gyrovirus, a virus related to chicken anemia virus. J. Virol. 2011;85:7948–7950. doi: 10.1128/JVI.00639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan T.G. A third gyrovirus species in human faeces. J. Gen. Virol. 2012;93:1356–1361. doi: 10.1099/vir.0.041731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu D.K.W. Characterization of a novel gyrovirus in human stool and chicken meat. J. Clin. Virol. 2012;55:209–213. doi: 10.1016/j.jcv.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delwart E., Li L. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 2012;164:114–121. doi: 10.1016/j.virusres.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hino S., Miyata H. Torque teno virus (TTV): current status. Rev. Med. Virol. 2007;17:45–57. doi: 10.1002/rmv.524. [DOI] [PubMed] [Google Scholar]
- 33.Zumla A. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Mandell G.L., editor. Churchill Livingstone; 2000. pp. 1937–2022. [Google Scholar]
- 34.Cadwell K. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuss S.K. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellett P.E. Chromosomally integrated human herpesvirus 6: questions and answers. Rev. Med. Virol. 2012;22:144–155. doi: 10.1002/rmv.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Handley S.A. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maggi F. Human gyrovirus DNA in human blood, Italy. Emerg. Infect. Dis. 2012;18:956–959. doi: 10.3201/eid1806.120179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biagini P. Human gyrovirus in healthy blood donors, france. Emerg. Infect. Dis. 2013;19:1014–1015. doi: 10.3201/eid1906.130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornelis G. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E828–E837. doi: 10.1073/pnas.1215787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazarczyk M. The EVER proteins as a natural barrier against papillomaviruses: a new insight into the pathogenesis of human papillomavirus infections. Microbiol. Mol. Biol. Rev. 2009;73:348–370. doi: 10.1128/MMBR.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattarai N., Stapleton J.T. GB virus C: the good boy virus? Trends Microbiol. 2012;20:124–130. doi: 10.1016/j.tim.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barton E.S. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 44.White D.W. Latent herpesvirus infection arms NK cells. Blood. 2010;115:4377–4383. doi: 10.1182/blood-2009-09-245464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yager E.J. Gamma-herpesvirus-induced protection against bacterial infection is transient. Viral Immunol. 2009;22:67–72. doi: 10.1089/vim.2008.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barton E.S. Herpesvirus latency and symbiotic protection from bacterial Infection. Viral Immunol. 2009;22:3–4. doi: 10.1089/vim.2008.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Los M. Apoptin, a tumor-selective killer. Biochim. Biophys. Acta. 2009;1793:1335–1342. doi: 10.1016/j.bbamcr.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Antonsson A. Review: antibodies to cutaneous human papillomaviruses. J. Med. Virol. 2012;84:814–822. doi: 10.1002/jmv.23257. [DOI] [PubMed] [Google Scholar]
- 49.Hibma M.H. The immune response to papillomavirus during infection persistence and regression. Open Virol. J. 2012;6:241–248. doi: 10.2174/1874357901206010241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mackay L.K. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7037–7042. doi: 10.1073/pnas.1202288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang X. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ott C. Use of a TT virus ORF1 recombinant protein to detect anti-TT virus antibodies in human sera. J. Gen. Virol. 2000;81:2949–2958. doi: 10.1099/0022-1317-81-12-2949. [DOI] [PubMed] [Google Scholar]
- 53.Maggi F. Torque teno virus viremia load size in patients with selected congenital defects of innate immunity. Clin. Vaccine Immunol. 2011;18:692–694. doi: 10.1128/CVI.00466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moen E.M. Effect of immune modulation on TT virus (TTV) and TTV-like-mini-virus (TLMV) viremia. J. Med. Virol. 2003;70:177–182. doi: 10.1002/jmv.10356. [DOI] [PubMed] [Google Scholar]
- 55.Biagini P. Classification of TTV and related viruses (anelloviruses) Curr. Top. Microbiol. Immunol. 2009;331:21–33. doi: 10.1007/978-3-540-70972-5_2. [DOI] [PubMed] [Google Scholar]
- 56.Reddy J. Autoimmunity in viral myocarditis. Curr. Opin. Rheumatol. 2013;25:502–508. doi: 10.1097/BOR.0b013e3283620036. [DOI] [PubMed] [Google Scholar]
- 57.Morse S.S. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfe N.D. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zur Hausen H. Childhood leukemias and other hematopoietic malignancies: interdependence between an infectious event and chromosomal modifications. Int. J. Cancer. 2009;125:1764–1770. doi: 10.1002/ijc.24365. [DOI] [PubMed] [Google Scholar]
- 60.Dereure O. No evidence for viral sequences in mycosis fungoides and Sézary syndrome skin lesions: a high-throughput sequencing approach. J. Invest. Dermatol. 2013;133:853–855. doi: 10.1038/jid.2012.371. [DOI] [PubMed] [Google Scholar]


