Abstract
Computer-assisted technologies of the genomic structure, biological function, and evolution of viruses remain a largely neglected area of research. The attention of bioinformaticians to this challenging field is currently unsatisfying in respect to its medical and biological importance. The power of new genome sequencing technologies, associated with new tools to handle “big data”, provides unprecedented opportunities to address fundamental questions in virology. Here, we present an overview of the current technologies, challenges, and advantages of Next-Generation Sequencing (NGS) in relation to the field of virology. We present how viral sequences can be detected de novo out of current short-read NGS data. Furthermore, we discuss the challenges and applications of viral quasispecies and how secondary structures, commonly shaped by RNA viruses, can be computationally predicted. The phylogenetic analysis of viruses, as another ubiquitous field in virology, forms an essential element of describing viral epidemics and challenges current algorithms. Recently, the first specialized virus-bioinformatic organizations have been established. We need to bring together virologists and bioinformaticians and provide a platform for the implementation of interdisciplinary collaborative projects at local and international scales. Above all, there is an urgent need for dedicated software tools to tackle various challenges in virology.
Keywords: Virus sequence analysis, Bioinformatics, Virology, Software
“Big data” has been awarded to be the second-best Anglicism in 2014.a Although microorganisms and particularly viruses are tiny, the standard properties of big data apply: volume, variety, velocity, and veracity. The biodiversity of viruses with its coverage of multiple scales and its high complexity is a big challenge for algorithm and software development in the big data field (Beckstein et al., 2014).
Recently, we have started to explore the virus’ and host's genomes, transcriptomes, metabolome, proteome, and metagenome but also their phenotype, occurrence, and environment. Linking such raw heterogeneous data with current data, e.g., collected from social networks on cumulative occurrences of disease-carrying mosquitoes, is a challenging task. For example, such a task might be solved by combining geo-reference photos from mobile phones with an automatic determination software, allowing better decisions on overarching questions (Graham et al., 2011).
The storage of such data is essential and currently a computationally unsolved problem. Additionally, calculations on computational cluster machines have annual electricity costs of one third of its acquisition costs. Medical data are usually only semianonymous and therefore cannot be stored and computed in clouds.b In the future, we will need novel, qualitatively different computational methods and paradigms. We will witness the rapid extension of computational pan-genomics, a new subarea of research in bioinformatics. A prominent example for a computational paradigm shift is the transition from the representation of single reference genomes as strings to cloud-like representations as graphs (Marschall et al., 2016). Especially, viruses are notorious mutation machines. Therefore, a viral quasispecies is a cloud of viral haplotypes that surround a given master virus (Qin et al., 2012).
Interestingly, already the storage of simple linear viral genomes is complicated. For instance, although most viral genomes are stored in the NCBI, many virologists refuse to integrate their data due to the generality of the database: One of the first questions during the upload process is “What chromosome is this?” Therefore, virus-specific databases are necessary, however, only a few exist so far (Table 1 ), and a general database for all viruses needs to be urgently developed.
Table 1.
Virus-Specific Databases Besides the General NCBI Database
| Tool | Description | Ref. |
|---|---|---|
| ViPR | ViPR database integrates genomes and various other types of data for multiple virus families belonging to the Arenaviridae, Bunyaviridae, Caliciviridae, Coronaviridae, Flaviviridae, Filoviridae, Hepeviridae, Herpesviridae, Paramyxoviridae, Picornaviridae, Poxviridae, Reoviridae, Rhabdoviridae, and Togaviridae families. | Pickett et al. (2012) |
| EpiFluTM | GISAID EpiFluTM is the world's most complete collection of genetic sequence data of influenza viruses and related clinical and epidemiological data. EpiFluTM is tailored to the needs of influenza researchers from both the human and the veterinary fields. The data is publicly accessible but not public domain (GISAID does not remove nor waive any preexisting rights). | Shu and McCauley (2017) |
| HIV | The HIV database contains data on HIV genetic sequences and immunological epitopes. The website also provides an access to several tools that can be used for analysis and visualization. | Druce et al. (2016) |
| HCV | HCV is a comprehensive database of the hepatitis C virus (HCV). | Kuiken et al. (2005) |
| ViralZone | ViralZone is a web-resource from the Swiss Institute of Bioinformatics for all viral genus and families, providing general molecular and epidemiological information, along with virion and genome figures. Each virus or family page gives an easy access to UniProtKB/Swiss-Prot viral protein entries. | Hulo et al. (2011) |
| VVR | The virus variation resource (VVR) is a selection of web retrieval interfaces, analysis, and visualization tools for virus sequence datasets. | Hatcher et al. (2017) |
1. Next-Generation Sequencing
Next-Generation Sequencing (NGS) has dramatically increased the accessibility of genetic information, generating in only a few hours massive amounts of genome and transcriptome data that is rapidly changing the landscape of many life science disciplines (Goodwin et al., 2016). In April 2003, the complete human genome was announced and the project succeeded after spending $3-billion with a high-quality human reference genome (Schmutz et al., 2004). Although the assembly of such a huge genome is still a very challenging task, nowadays the sequencing can be done in just a few days and for only some thousands of dollars (Goodwin et al., 2016) by utilizing the still emerging NGS technologies.
In recent years, DNA sequencing (DNA-Seq) based on novel NGS technologies (Table 2 ) became the most sophisticated method for the sequencing of full genomes. A general DNA-Seq workflow starts with the library preparation including the fragmentation (chemically, physically) of the DNA molecules. After amplification and sequencing millions of short subsequences, so-called reads, are produced. In general, methods like Illumina and Ion Torrent produce reads with a length between 50 and 500 bp, depending on the setup and machine used (Goodwin et al., 2016). Next to that short read producing NGS technologies more and more long read NGS approaches are emerging. Very popular is the single-molecule real-time sequencing (SMRT) introduced by Pacific Biosciences (Rhoads and Au, 2015) (PacBio) producing reads with an average length of 15,000 bp and a maximum of >40,000 bases. However, PacBio produces only ∼50,000 reads per SMRT cell, whereas Illumina yields ∼180 million reads on one HiSeq2500 lane (Goodwin et al., 2016). It is clearly important to produce longer reads to improve the results of various analyzes like the de novo assembly of highly repetitive, large or fast mutating genomes.
Table 2.
Commonly Used Next-Generation Sequencing (NGS) Technologies and Their Major Specifications
| Platform | Length (bp) | Throughput | Number of Reads | Error | Cost Per Gb |
|---|---|---|---|---|---|
| Short-read NGS | |||||
| Sequencing by synthesis: SNA | |||||
| 454 Pyrosequencing | 400–1000 | 35–700 Mb | 0.1–1 M | 1%, indel | $10–40,000 |
| Ion Torrent | 200–400 | 100 Mb–15 Gb | 2–80 M | 1%, indel | $500–2000 |
| Sequencing by synthesis: CRT | |||||
| Illumina Solexa | 25–300 | 2–900 Gb | 10 M–4 B | 0.1%, subst. | $7–1000 |
| Qiagen GeneReader | 100 | NA | 10 M–4 B | 0.1%, subst. | NA |
| Sequencing by ligation | |||||
| SOLiD | 60–100 | 10–320 Gb | 700 M–1.4 B | 0.1%, AT bias | $100 |
| Long-read SMRT NGS | |||||
| Pacific BioSciences | up to 40 Kb | 0.5–7 Gb | ∼55 k | 13% (single) | $1000 |
| 1% (circular) | |||||
| Oxford Nanopore (MinION) | up to 200 Kb | up to 1.5 Gb | >100 k | 12%, indel | $750 |
Generally, NGS technologies can be divided in short-read and long-read approaches, depending on the length of the produced reads. SNA, single-nucleotide addition; CRT, cyclic reversible termination; SMRT, single-molecule real-time sequencing; indel, nucleotide insertion–deletion; subst., nucleotide substitution.
This table is mainly based on recent reviews Goodwin et al., 2016, Mardis, 2017.
Nanopore sequencing is another recent incumbent in the SMRT area: the way nanopore-based sequencing works is by pulling a nucleotide strand (DNA or RNA) through a kind of molecular channel isolated from a bacterium. While passing through the pore, the nucleotide sequence produces a small change in the applied voltage, which can be reinterpreted as the familiar sequence of the bases A, C, T/U, and G, including also modifications such as methylation (Jain et al., 2016). Because each pore produces its own signal, this technology can be highly parallelized. For example, with the current USB-sized MinION sequencer, 2048 pores are situated on a membrane of the size of a finger nail. The sequencer itself costs a fraction of the aforementioned ones. Furthermore, each pore's signal can be detected in real time (Gardy et al., 2015), allowing unprecedented speed and mobility in sequence-based diagnostics, as exemplary demonstrated in field trials during the 2014 Ebola outbreak (Quick et al., 2016). Furthermore, nanopore sequencing is currently the only technique that does (in theory) not technologically limit the potential read length, which means an entire viral genome can be sequenced in one part at an intact pore. No additional assembly step would be required. The current read length maximum is >900 Kbp (personal communication with N. Loman). The MinION's throughput has been shown to provide up to 15 Gb in 48 h with a protocol-dependent error rate of 5%–15%.
Besides the sequencing of genomic DNA, RNA sequencing (RNA-Seq) emerged as a powerful method for discovering, profiling, and quantifying RNA transcripts or viral RNA genomes (Mortazavi et al., 2008). However, with currently available short-read NGS techniques such as Illumina it is not possible to directly sequence RNA molecules—first the RNA must be reversely transcribed to complementary DNA (cDNA) for sequencing. Strikingly, nanopore just recently announced a sequencing kit that should allow for the direct sequencing of RNA molecules (and therefore also RNA viruses).
Importantly, within each NGS project one should consider the need and amount of replication, different protocols for molecule selection and library preparation, the achieved throughput and length of the reads and further specific parameters like strand-specificity and the insertion size between paired-end reads.
2. Detection of De Novo Viruses
Within the last decade numerous genomes of previously unknown viruses have been identified. However, it is still a challenging task to discriminate an outnumbered amount of viral sequences from the majority of host reads. Genome assemblers specifically designed for viral genomes are rare (Table 3 ) and cannot overcome an uneven or incomplete coverage of viral genomes.
Table 3.
De Novo Assembly Tools Suitable for the Assembly of Viral Genomes
| Tool | Description | Ref. |
|---|---|---|
| AV454 | AV454 is a de novo consensus assembler designed for small and nonrepetitive genomes sequenced at high depth. | Henn et al. (2012) |
| RIEMS | RIEMS is a software for the sensitive and reliable analysis of metagenomic datasets. | Scheuch et al. (2015) |
| V-FAT | V-FAT is a tool to perform automated computational finishing and annotation of de novo viral assemblies. | Charlebois et al. (n.d.) |
| VICUNA | VICUNA is a de novo assembly tool targeting populations with high mutation rates. | Yang et al. (2012) |
| VrAP | The VrAP (Viral Assembly Pipeline) is based on the genome assembler SPAdes (Bankevich et al., 2012) combined with an additional read correction and several filter steps. The pipeline classifies the contigs (contiguous sequences constructed from short reads) to distinguish host from viral sequences. VrAP can identify viruses without any sequence homology to known references. | Fricke et al. (2017) |
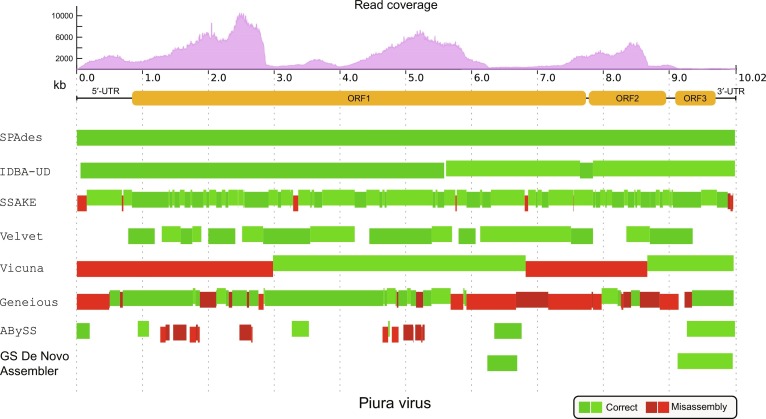
Many assembly tools and software suites have been developed for the complete genome assembly in general, such as Velvet (Zerbino and Birney, 2008), ABySS (Simpson et al., 2009), or Geneious (Kearse et al., 2012) (Fig. 1 ). These common tools often fail to assemble full viral genomes, due to a low and uneven read coverage (Peng et al., 2012), as well as repetitive elements in the viral UTR regions. However, algorithms developed for single-cell sequencing like SPAdes (Bankevich et al., 2012) or IDBA-UD (Peng et al., 2012) perform very well for tested samples and outperform assembly tools like VICUNA (Yang et al., 2012), especially designed for viral data (Fig. 1).
Fig. 1.
Comparison of eight assembly tools based on a sequenced C6/36 cell, infected with a Piura virus strain from Mexico. The figure depicts an alignment of de novo assembled contigs (rectangles) to the reference genome of Piura virus (KM249340.1). SPAdes assembles the full viral genome without any difficulties. All other assemblers fail to build a continuous single contig. Green—contigs that align correctly. Red—misassemblies. The different color shades are only for a better visualization of adjacent contigs. The alignment plot was created with Quast (Gurevich et al., 2013).
Figure adapted from Fricke, M., Zirkel, F., Drosten, C., Junglen, S., Marz, M., 2017. VrAP: full length de novo genome assembly of unknown RNA viruses (submitted for publication).
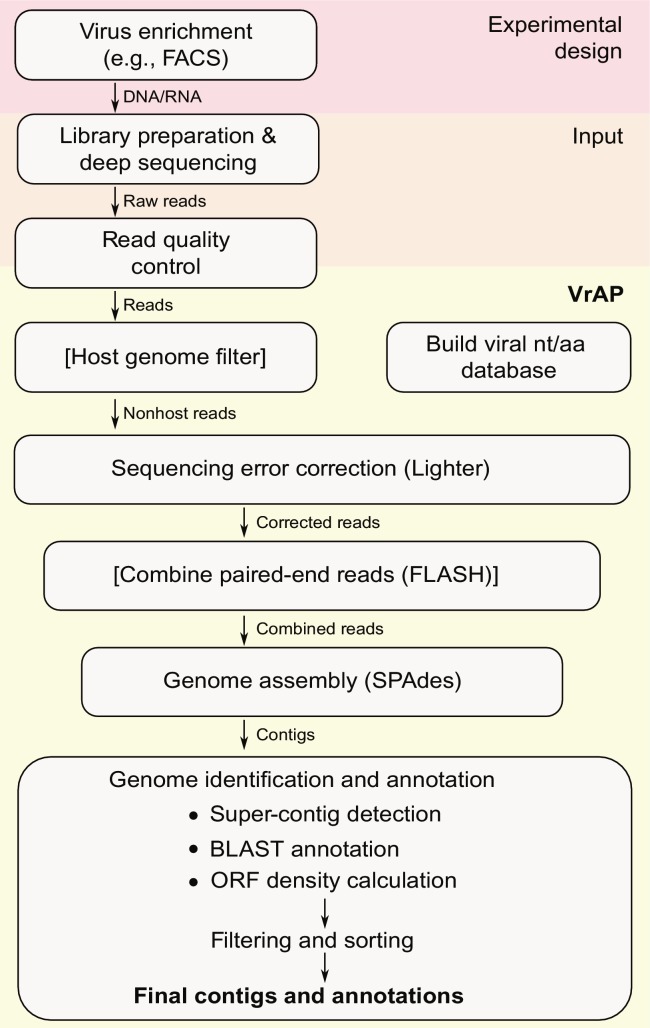
For an efficient viral de novo assembly we suggest enriching of the viruses by, e.g., ultracentrifugation or FACS prior to the library preparation step. After the sequencing, a standard read quality control should be conducted followed by a host genome filter step, if possible. Finally, the assembly step can be performed based on de Bruijn graphs or overlapping layout consensus (OLC) approaches. If possible, the usage of multiple k-mer values is recommended. The final assembly can be used for annotation and identification of contigs from viral origin. Fig. 2 shows the viral assembly workflow as used in the VrAP assembly pipeline (Fricke et al., 2017).
Fig. 2.
Workflow of the viral de novo assembly pipeline VrAP. The pipeline requires (preprocessed) reads as input. The output consists of final contigs and an annotation list. The pipeline combines multiple read corrections with SPAdes, a super-contig construction and a contig classification. VrAP comes as an easy to use command-line tool (http://www.rna.uni-jena.de/en/vrap/). All steps in square bracket are optional. FACS, fluorescent activated cell sorting.
Figure adapted from Fricke, M., Zirkel, F., Drosten, C., Junglen, S., Marz, M. 2017. VrAP: full length de novo genome assembly of unknown RNA viruses (submitted for publication).
3. Viral Quasispecies
The above described de novo assembly methods can reconstruct viral genomes. However, to yield a small number of contigs, the algorithms usually include a step that calls a consensus on a given sequence position. This consensus is implemented to reduce the noise in the raw assembly. However, in the context of viral haplotype variants, this step is misleading, because it effectively ignores low-frequency variants and technical errors (Marz et al., 2014).
To gain insights into viral haplotypes, the reads should be mapped either to a known reference genome or to the contigs that were generated during assembly. This “classification” can be used to infer the viral population structure of each individual species in the sample, thereby increasing the resolution of the diversity estimate. (Intrahost) viral populations consist of many related virions, generated by mutation, recombination, and selection. The resulting diversity is especially large for RNA viruses (Holmes, 2009). Even low-frequency variants can be of great interest, for example, because they may harbor drug resistance mutations (Barzon et al., 2011), facilitate immune escape (Luciani et al., 2012), or affect virulence (Töpfer et al., 2013). Estimating intrahost viral genetic diversity and reconstructing the individual haplotype sequences relies on both error correction and read assembly (Pulido-Tamayo et al., 2015). It can be performed on different spatial scales, including single sites of the genome (single-nucleotide-variant calling), small sliding windows (local reconstruction), or complete genomes (global reconstruction). Viral haplotype reconstruction tools can quantify viral diversity from NGS data (e.g., Beerenwinkel et al., 2012). It was shown that haplotypes differ enough, current NGS reads are not too short and the coverage is high enough to assemble accurate viral haplotype genomes (Zagordi et al., 2012). A common prerequisite for these tools is a high-quality alignment of the reads (e.g., Töpfer et al., 2014). However, tools exist that allow haplotype calling without a reference genome as presented in Gregor et al. (2016). Nevertheless, the short-read-based discovery of viral sequences in mixed samples remains challenging (Marschall et al., 2016) because most analysis steps are not easily automated and various technical or biological limitations exist (Fricke et al., 2017). There is a need for an integrated workflow combining the different processing steps in viral diversity studies to discover the underlying virus populations that can be used on a daily basis by clinicians and virologists. The advent of SMRT sequencing provides new opportunities. One of the main limitations of the past was the limited length of the sequenced nucleotide fragments. Currently, it is not possible to write cDNA longer than a few thousand of nucleotides (e.g., ∼2000 nucleotides for the wheat stripe rust pathogen (Ling et al., 2007)). However, even if the cDNA transcription would be no limiting factor, current short-read sequencing technologies such as Illumina are only able to sequence small fragments of several hundred nucleotides. Nanopore sequencing lifts these two constraints: it is now possible to sequence much longer fragments (as described above) and to sequence the RNA directly, without the need of a cDNA intermediate, advancing the detection of viral quasispecies.
4. Secondary Structures of RNA Viruses
RNA viruses are flanked by highly structured 5′- and 3′-untranslated regions (UTRs), which are indispensable for translation and replication of the viral genome Liu et al., 2009, Lohmann, 2013.
Standard RNA secondary structure prediction tools such as mfold and RNAfold (Table 4 ) are based on the calculation of the minimum free energy (MFE) and can fold reliably on small local windows of up to 300 nt. Secondary structures of larger genomic segments or interactions spanning larger regions, including pseudogenes, are still bioinformatically challenging. Foldings based on not only one but also multiple sequences are generally more reliable due to following the footsteps of evolution by compensatory mutations. Viruses usually come along with a high mutation rate and therefore with a bunch of similar sequences perfect for a large alignment and predicting secondary structures.
Table 4.
A Selection of Tools for the Detection of Secondary Structures in RNA Viruses
| Tool | Description | Alignment | Ref. |
|---|---|---|---|
| RNAfold | RNAfold is a tool to predict secondary structures of single stranded RNA or DNA sequences. | No | Gruber et al. (2008) |
| mfold | mfold is a web server that provides easy access to RNA and DNA folding and hybridization software. | No | Zuker (2003) |
| RNAalifold | RNAalifold is a tool for calculating secondary structures for a set of aligned RNAs. It is part of the Vienna RNA Package. | Yes | Hofacker (2007) |
| LocARNA | LocARNA is a multiple alignment tool based on the calculation of sequence and structure simultaneously. | Yes | Will et al. (2007) |
| LRIscan | LRIscan is a tool for the prediction of long-range interactions in full viral genomes based on a multiple genome alignment. LRIscan is able to find interactions spanning thousands of nucleotides. | Yes | Fricke and Marz (2016) |
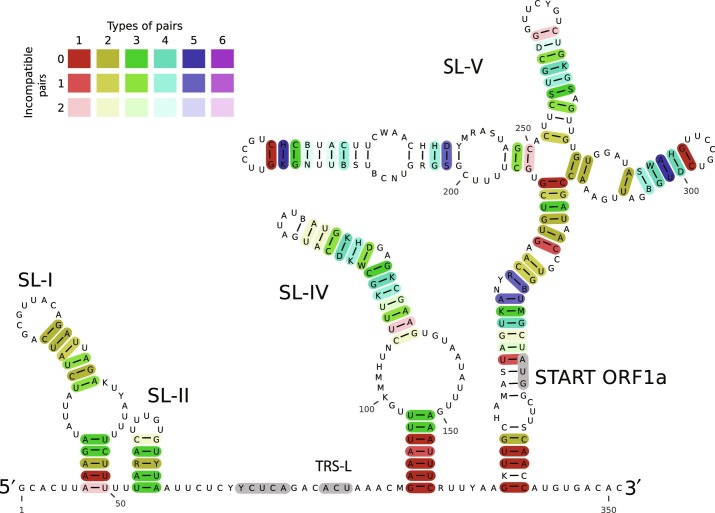
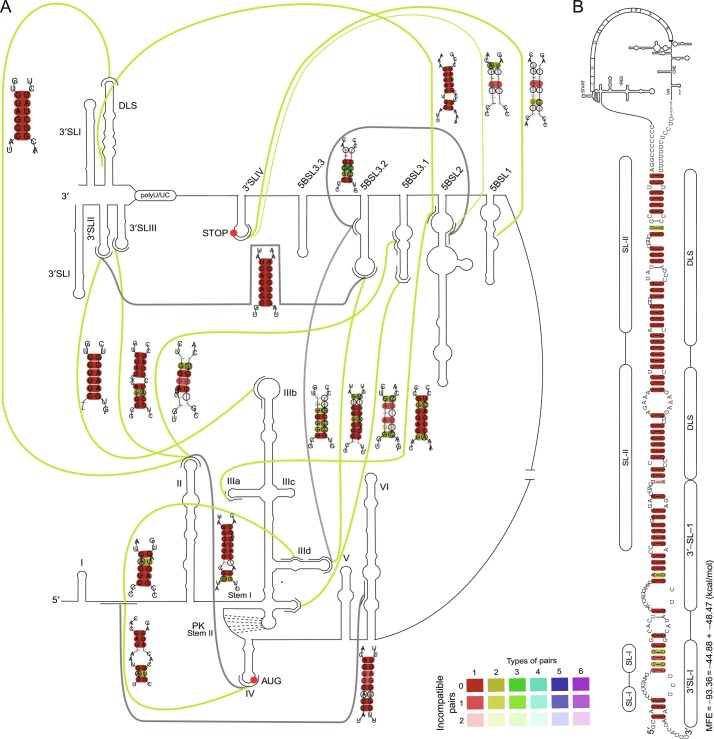
For example, LocARNA creates a multiple alignment based on sequence and structure simultaneously. Based on this tool larger genomic regions up to 800 nt can be reliable predicted as shown for coronaviruses (Fig. 3 ) (Madhugiri et al., 2014) and HCV (Fig. 4 ) (Fricke et al., 2015). Nowadays, long-range interactions (LRIs) are computationally predictable by tools such as LRIscan (Fricke and Marz, 2016), suggesting circularizations of viruses during replication.
Fig. 3.
Alignment-based secondary structure prediction of 5′ genome regions of alphacoronaviruses. The viruses included in this analysis represent all currently recognized species in the genus Alphacoronavirus. The alignment (not shown) was calculated by LocARNA (Will et al., 2007) and the structure by RNAalifold (Hofacker, 2007). The consensus sequence is represented using the IUPAC code. Colors are used to indicate conserved base pairs: from red (conservation of only one base pair type) to purple (all six base pair types are found); from dark (all sequences contain this base pair) to light colors (one or two sequences are unable to form this base pair). To refine the alignment, an anchor at the highly conserved core TRS-L was used.
Figure adapted from Madhugiri, R., Fricke, M., Marz, M., Ziebuhr, J., 2014. RNA structure analysis of alphacoronavirus terminal genome regions. Virus Res. 194, 76–89. doi:10. 1016/j.virusres.2014.10.001.
Fig. 4.
Long-range interactions in 5′-UTR, CRE, VR, and X-tail of HCV (Fricke et al., 2015). (A) Overview and possible interactions for all tested HCV sequences. Gray lines—known interactions derived from literature, validated by this analysis for all examined isolates; Green lines—novel interactions (based on new calculations). The detailed interactions are shown on the right side next to each corresponding interaction line. The leftmost interaction can be extended for a possible circularization of HCV. (B) Possible circularization of HCV. Interacting loops of SLII and DLS of the HCV plus-strand can be extended to at least 62 bp in all available 19 isolates.
5. Analysis of Transcriptomic Host Reactions to Viral Infections
The general workflow of a short-read RNA-Seq experiment involves: (1) the extraction of total RNA from a biological sample of interest, (2) the purification of the sample to enrich a certain type of RNA such as mRNAs or microRNAs, and (3) the preparation of a library ready for short-read NGS. The generation of the library may involve steps like the fragmentation of longer RNA molecules, followed by the reverse transcription of the RNA to cDNA, ligation of adapters to the 5′- and/or 3′-ends of the cDNA fragments and PCR amplification to enrich the library for correctly ligated cDNA fragments (Corney, 2013). The resulting reads from an RNA-Seq experiment can be used to estimate the abundances of certain transcripts within each sequenced sample. If different conditions are sequenced, the obtained transcript abundances can be further used to identify differential expressed genes. Before RNA-Seq came up, gene expression studies were performed with hybridization-based microarrays. Contrasting the microarray technology, RNA-Seq allows for the identification of novel transcripts and does not necessarily need a sequenced reference genome. Furthermore, RNA-Seq allows for the genome-wide analysis of transcripts at a single-nucleotide resolution and therefore includes the identification of single-nucleotide variants, gene fusions, allele-specific expression, and alternative splicing events (Corney, 2013).
However, besides all its advantages, RNA-Seq is still an expensive technology. Therefore, in most RNA-Seq studies the number of biological replicates is limited (only 3–5 replicates per condition are quite common) contrasting the comparative high number of genes that are simultaneously tested.
A typical RNA-Seq experiment, involving an eukaryotic cell line and involving two different conditions (untreated, infected), three time points and four biological replicates already results in the sequencing of 24 samples. The current Ensembl annotation of the human genome (v85) consists of 58,051 genes comprising 19,961 genes coding for proteins. In a differential gene expression study, all expressed genes can be compared between different conditions and time points, resulting in an overwhelming amount of data. Genes can be further analyzed for differential expressed isoforms and clustered according to their function. With a de novo gene prediction, one of the huge advantages of RNA-Seq in comparison to microarrays, an incomplete annotation can be further extended and even more genes are possibly involved. The use of different library preparation protocols can extend the complexity of such an RNA-Seq study even further.
Therefore, the statistical analysis of RNA-Seq data with the final goal to define significantly differential expressed genes is a challenging task. Especially, if a high number of reads originating from viral transcripts is involved, outshining the expression of host genes. Furthermore, the generation of a sensible number of biological replicates can be difficult when working with such deadly viruses like Ebola. The analysis can become even more complicated when no reference genome for mapping and quantification of the RNA-Seq reads is available. In this case, a de novo transcriptome assembly can be constructed and annotated from scratch.
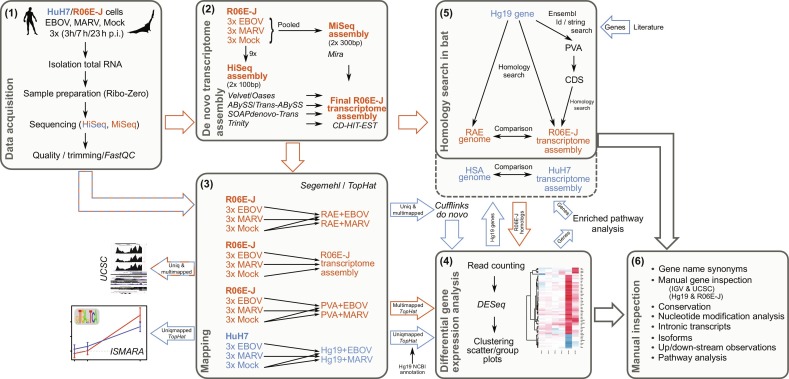
To tackle these difficulties, profoundly occurring when working with virus infected RNA-Seq data, different tools and parameter settings should be conducted and combined to achieve a comprehensive overview picture of the host's transcriptional reaction to a viral infection. An exemplary pipeline combining different tools for mapping and assembly and working on a genomic and transcriptomic context as well is given in Fig. 5 . The overall goal of the underlying study was to understand why bats can live with the Ebola virus, while humans suffer so much from this deadly infection.
Fig. 5.
Host–virus RNA-Seq methods pipeline for the detection of differential expressed genes (see text for details).
Modified from Hölzer , M., Krähling, V., Amman, F., Barth, E., Bernhart, S.H., Carmelo, V.A.O., Collatz, M., Doose, G., Eggenhofer, F., Ewald, J., Fallmann, J., Feldhahn, L.M., Fricke, M., Gebauer, J., Gruber, A.J., Hufsky, F., Indrischek, H., Kanton, S., Linde, J., Mostajo, N., Ochsenreiter, R., Riege, K., Rivarola- Duarte, L., Sahyoun, A.H., Saunders, S.J., Seemann, S.E., Tanzer, A., Vogel, B., Wehner, S., Wolfinger, M.T., Backofen, R., Gorodkin, J., Grosse, I., Hofacker, I., Hoffmann, S., Kaleta, C., Stadler, P.F., Becker, S., Marz, M., 2016. Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells. Sci. Rep. 6, 34589. doi:10.1038/srep34589.
In this study, performed by Hölzer et al. (2016), (1) total RNA from a human HuH7 cell line and a fruit bat cell line (R06E-J; Rousettus aegyptiacs) infected with either the Ebola or Marburg virus (EBOV, MARV) was harvested 3, 7, and 23 h postinfection, depleted of ribosomal RNA and sequenced on an Illumina HiSeq2500. The bat RNA was further pooled and additionally sequenced on an Illumina MiSeq system. Initial quality control and trimming of the raw data were conducted with FastQC (Andrews, 2010) and PRINSEQ (Schmieder and Edwards, 2011). (2) For bat RNA, a de novo transcriptome assembly was constructed by combining MiSeq and HiSeq data using Velvet/Oases Schulz et al., 2012, Zerbino and Birney, 2008, ABySS/Trans-ABySS Birol et al., 2009, Simpson et al., 2009, SOAPdenovo-Trans (Luo et al., 2012), Trinity (Grabherr et al., 2011), and Mira (Chevreux et al., 2004) with default parameters and multiple k-mer values, if possible. (3) The mapping of the RNA-Seq short-reads was performed for Mock-, EBOV-, and MARV-treated cells onto human/bat genomes and the bat transcriptome with Segemehl (Hoffmann et al., 2014) and TopHat (Kim et al., 2013). (4) A differential gene expression analysis was performed by counting uniquely mapped reads with HTSeq-count (Anders et al., 2015) and applying a DESeq (Love et al., 2014) analysis in R. The results were further used for clustering and scatter/group plot analyzes. (5) A homology search in bats was performed for all significantly differential expressed genes from (4) and for the genes assumed to be involved in the response to infection based on an enriched pathway analysis and the literature. The Rousettus aegyptiacus genome and coding sequences from Pteropus vampyrus, a closely related bat species, were used to validate but also to detect homologous sequences in the bat transcriptome. Detected homologs were employed for the differential gene expression analysis. (6) One huge advantage of this comprehensive study was the manual inspection of ∼7.5 % of the human genes. Each candidate gene was manually investigated in the IGV (Thorvaldsdóttir et al., 2013) and UCSC (Dreszer et al., 2012) browsers for the human and bat samples from all time points. Single-nucleotide modifications (differential SNPs, posttranscriptional modifications), intronic transcripts and regulators, alternative splicing and isoforms, as well as upstream and downstream transcript characteristics were described.
6. Viral Phylogeny/Cophylogeny
Phylogenetic analysis is a common method in virology, forming a crucial element of investigations describing viruses or viral epidemiology. Nevertheless, many characteristics of viruses pose distinct challenges for phylogenetics: (1) strong differences in evolution rates, (2) great potential for recombination and gene transfer, (3) evolutionary relationships between viruses and their hosts, (4) lack of physical “fossil records” of viruses, and (5) the abundance of genomic viral fossils as parts of ancient viral genomes that occur within the genomes of extant species.
Today, various phylogenetic tree-building methods such as MrBayes (Ronquist and Huelsenbeck, 2003), BEAST (Drummond et al., 2012), PhyloBayes (Lartillot et al., 2009), and RAxML (Stamatakis et al., 2008) exist. However, trees cannot represent complex evolutionary relations relevant for viruses such as horizontal gene transfer, interspecific recombination, or virus–host coevolution. Different types of phylogenetic networks were developed to represent such relations (e.g., Huson et al., 2011). However, there is still a high need for research on how to reconstruct such aspects of virus phylogeny.
Genomic evolution can be already observed over the course of years or even days due the fact that the short-term evolution rates of many viruses are so high. It is important that the phylogenetic methods can include the sampling dates of the sequences for analyzing short-term evolution as implemented in TipDate (Rambaut, 2000). Furthermore, spatial dispersal processes play an essential role, for example, the spatial distribution of a virus within the host's body (Bloomquist et al., 2010). Moreover, the evolutionary substitution rates of viruses can differ even for short-term evolutionary scenarios. One reason is that substitution rates reflect a complex product of mutation rate, generation time, effective population size, and fitness Jenkins et al., 2002, Sanjuan et al., 2010. Particularly in viruses, substitutions might be an artifact generated by polymerase errors and nucleotide modifications (Domingo and Holland, 1997). Thus, the classical assumption of a time-homogeneous substitution process used by different phylogeographic statistical inference methods does not hold and new approaches that can include varying evolutionary rates have been already introduced (e.g., Bielejec et al., 2014).
Another problem for viral “deep phylogeny” reconstruction is the genetic distance between viruses. The distance can be so large that reasonable alignments become impossible to calculate. To achieve biologically correct alignments, the development of advanced approaches would help, however, can only marginally alleviate the problem of saturated substitution processes. Including aspects such as genome organization or protein structure as phylogenetic characters could further improve viral alignments and phylogenies (Holmes, 2011).
Several ancient viruses have left parts of their genome (or other traces) in the genome of germ line cells of their hosts. Such parts, called endogenous viral elements (EVEs), have survived as nonfunctional, neutrally evolving pseudogenes, or even became fixed as functional. Most EVEs stem from retroviruses because they integrate into host genomes as part of their life cycle. For example, ∼8% of the human genome is derived from >100,000 retroviral fossils (Lander et al., 2001). However, in recent years, EVEs from many other viruses have been found (Horie and Tomonaga, 2011). Different programs have been developed to detect EVEs in complete genome sequences such as RepeatMasker (Smit et al., n.d.), LTR_STRUC (McCarthy and McDonald, 2003), and RetroTector (Sperber et al., 2009). Moreover, a combination of several of these programs seems very promising for the calculation of viral phylogenies (Lerat, 2010).
Withal, associations between viruses and their hosts can influence the phylogeny of both partners. A divergence of the host can also lead to a divergence of the virus (codivergence) and thus to a (local) congruence of both phylogenies. A match of the virus phylogeny with host evolutionary events at known dates can be used to adjust the virus phylogeny or corresponding molecular clocks (Sharp and Simmonds, 2011). The ability of viruses to switch their hosts can enable viruses to replicate and spread more efficiently. This process is commonly known as an epidemic and is observed in pathogenic viruses (Weiss, 2003). Owing to the advantages conferred by the conquest of new host territory, several researchers presume host switching as an elementary component of virus evolution that might initiate viral speciation (Kitchen et al., 2011). Attributed to the fact that virologists are highly interested in the reconstruction of the common history of viruses and their hosts, several bioinformatic tools have been developed for this purpose (de Vienne et al., 2013).
However, there is still a huge amount of research questions that need to be answered based on new computational methods. For example, the inclusion of biogeographic information, ecological traits, or preferential host switching are crucial tasks (Cuthill and Charleston, 2013). A better knowledge of the timing and underlying conditions of those processes might enable projections into the future and thereby contribute to tackle one of the major issues in today's infectious diseases research: the prediction and prevention of future pandemics and outbreaks.
7. Conclusions and Future Perspectives
It is essential to bundle the expertise's of virus bioinformatics to follow with larger steps the small footsteps that were already taken. There is an urgent need for novel and specialized tools that allow the efficient detection, assembly, and classification of already known and completely new viruses in a fast and reliable way.
One big step in this direction involves the establishment of research networks between experienced scientists to facilitate the exchange of knowledge and to speed-up the development of powerful tools. DiaMETA-net is a German network which focuses on metagenomics in infection medicine. The research groups within the network devote themselves to the very broad detection and characterization of pathogens (viruses, bacteria, parasites) by means of NGS. However, the first specialized virology-bioinformatics organization, the EVBC (European Virus Bioinformatics Center), has been established rather recently on March 2017, comprising up to now 100 members from over 50 research institutions distributed across 13 European countries.
The future of virus bioinformatics clearly depends on how fast we develop specific bioinformatical tools, take first steps to establish a useful virus-specific database, and help to establish joint research projects. We must initiate and coordinate ring trials, undergraduate courses, graduate summer schools, and courses for principal investigators.
Whereas the list of bioinformatical tools presented in this section is supposed to be incomplete, they should provide a good overview and starting point to dive even deeper into the computational analysis of viral sequences.
Footnotes
Cloud storage or cloud computing refers to shared computer processing resources and data on demand.
References
- Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc Available online at: [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzon L., Lavezzo E., Militello V., Toppo S., Palu G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011;12:7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstein C., Böcker S., Bogdan M., H. Bruelheide H.M.B., Denzler J., Dittrich P., Grosse I., Hinneburg A., König-Ries B., Löffler F., Marz M., Müller-Hannemann M., Winter M., Zimmermann W. Explorative analysis of heterogeneous, unstructured, and uncertain data: a computer science perspective on biodiversity research. In: Helfert M., Holzinger A., Belo O., Francalanci C., editors. SciTePress; 2014. pp. 251–257. (Proceedings of the 3rd International Conference on Data Management Technologies and Applications, DATA 2014, Vienna, Austria, August 29–31, 2014). [Google Scholar]
- Beerenwinkel N., Günthard H.F., Roth V., Metzner K.J. Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front. Microbiol. 2012;3:329. doi: 10.3389/fmicb.2012.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F., Lemey P., Baele G., Rambaut A., Suchard M.A. Inferring heterogeneous evolutionary processes through time: from sequence substitution to phylogeography. Syst. Biol. 2014;63(4):493–504. doi: 10.1093/sysbio/syu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I., Jackman S.D., Nielsen C.B., Qian J.Q., Varhol R., Stazyk G., Morin R.D., Zhao Y., Hirst M., Schein J.E., Horsman D.E., Connors J.M., Gascoyne R.D., Marra M.A., Jones S.J.M. De novo transcriptome assembly with ABySS. Bioinformatics (Oxford, England) 2009;25:2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- Bloomquist E.W., Lemey P., Suchard M.A. Three roads diverged? Routes to phylogeographic inference. Trends Ecol. Evol. 2010;25(11):626–632. doi: 10.1016/j.tree.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois, P., Yang, X., Newman, R., Henn, M., Zody, M., n.d. Vfat: A post-assembly pipeline for the finishing and annotation of viral genomesPlease provide year of publication for this reference, if available. https://www.broadinstitute.org/viral-genomics/v-fat (accessed 07.03.17).
- Chevreux B., Pfisterer T., Drescher B., Driesel A.J., Müller W.E., Wetter T., Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14(6):1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney D.C. RNA-seq using next generation sequencing. Mater Methods. 2013;3:203. [Google Scholar]
- Cuthill J.H., Charleston M.A. A simple model explains the dynamics of preferential host switching among mammal RNA viruses. Evolution. 2013;67:980–990. doi: 10.1111/evo.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne D.M., Refrégier G., López-Villavicencio M., Tellier A., Hood M.E., Giraud T. Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 2013;198:347–385. doi: 10.1111/nph.12150. [DOI] [PubMed] [Google Scholar]
- Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Dreszer T.R., Karolchik D., Zweig A.S., Hinrichs A.S., Raney B.J., Kuhn R.M., Meyer L.R., Wong M., Sloan C.A., Rosenbloom K.R., Roe G., Rhead B., Pohl A., Malladi V.S., Li C.H., Learned K., Kirkup V., Hsu F., Harte R.A., Guruvadoo L., Goldman M., Giardine B.M., Fujita P.A., Diekhans M., Cline M.S., Clawson H., Barber G.P., Haussler D., James Kent W. The UCSC genome browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druce M., Hulo C., Masson P., Sommer P., Xenarios I., Le Mercier P., De Oliveira T. Improving HIV proteome annotation: new features of BioAfrica HIV Proteomics Resource. Database (Oxford) 2016 doi: 10.1093/database/baw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke M., Marz M. Prediction of conserved long-range RNA-RNA interactions in full viral genomes. Bioinformatics (Oxford, England) 2016;32:2928–2935. doi: 10.1093/bioinformatics/btw323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke M., Dünnes N., Zayas M., Bartenschlager R., Niepmann M., Marz M. Conserved RNA secondary structures and long-range interactions in hepatitis C viruses. RNA. 2015;21(7):1219–1232. doi: 10.1261/rna.049338.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke M., Zirkel F., Drosten C., Junglen S., Marz M. VrAP: full length de novo genome assembly of unknown RNA viruses. 2017 submitted for publication. [Google Scholar]
- Gardy J., Loman N.J., Rambaut A. Real-time digital pathogen surveillance—the time is now. Genome Biol. 2015;16:155. doi: 10.1186/s13059-015-0726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham E.A., Henderson S., Schloss A. Using mobile phones to engage citizen scientists in research. Eos Trans. AGU. 2011;92(38):313–315. [Google Scholar]
- Gregor I., Schönhuth A., McHardy A.C. Snowball: strain aware gene assembly of metagenomes. Bioinformatics (Oxford, England) 2016;32:i649–i657. doi: 10.1093/bioinformatics/btw426. [DOI] [PubMed] [Google Scholar]
- Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schäffer A.A., Brister J.R. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn M.R., Boutwell C.L., Charlebois P., Lennon N.J., Power K.A., Macalalad A.R., Berlin A.M., Malboeuf C.M., Ryan E.M., Gnerre S., Zody M.C., Erlich R.L., Green L.M., Berical A., Wang Y., Casali M., Streeck H., Bloom A.K., Dudek T., Tully D., Newman R., Axten K.L., Gladden A.D., Battis L., Kemper M., Zeng Q., Shea T.P., Gujja S., Zedlack C., Gasser O., Brander C., Hess C., Günthard H.F., Brumme Z.L., Brumme C.J., Bazner S., Rychert J., Tinsley J.P., Mayer K.H., Rosenberg E., Pereyra F., Levin J.Z., Young S.K., Jessen H., Altfeld M., Birren B.W., Walker B.D., Allen T.M. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. RNA consensus structure prediction with RNAalifold. Methods Mol. Biol. 2007;395:527–544. doi: 10.1007/978-1-59745-514-5_33. [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Otto C., Doose G., Tanzer A., Langenberger D., Christ S., Kunz M., Holdt L.M., Teupser D., Hackermüller J., Stadler P.F. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15(2):R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. Oxford University Press; New York: 2009. The Evolution and Emergence of RNA Viruses. [Google Scholar]
- Holmes E.C. What does virus evolution tell us about virus origins? J. Virol. 2011;85(11):5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzer M., Krähling V., Amman F., Barth E., Bernhart S.H., Carmelo V.A.O., Collatz M., Doose G., Eggenhofer F., Ewald J., Fallmann J., Feldhahn L.M., Fricke M., Gebauer J., Gruber A.J., Hufsky F., Indrischek H., Kanton S., Linde J., Mostajo N., Ochsenreiter R., Riege K., Rivarola-Duarte L., Sahyoun A.H., Saunders S.J., Seemann S.E., Tanzer A., Vogel B., Wehner S., Wolfinger M.T., Backofen R., Gorodkin J., Grosse I., Hofacker I., Hoffmann S., Kaleta C., Stadler P.F., Becker S., Marz M. Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells. Sci. Rep. 2016;6:34589. doi: 10.1038/srep34589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M., Tomonaga K. Non-retroviral fossils in vertebrate genomes. Viruses. 2011;3(10):1836–1848. doi: 10.3390/v3101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo C., de Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I., Le Mercier P. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 2011;39(Database issue):D576–D582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Rupp R., Scornavacca C. Cambridge University Press; New York, NY: 2011. Phylogenetic Networks: Concepts, Algorithms and Applications. ISBN 0521755964, 9780521755962. [Google Scholar]
- Jain M., Olsen H.E., Paten B., Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.M., Rambaut A., Pybus O.G., Holmes E.C. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 2002;54(2):156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen A., Shackelton L.A., Holmes E.C. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc. Natl. Acad. Sci. 2011;108(1):238–243. doi: 10.1073/pnas.1011090108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C., Yusim K., Boykin L., Richardson R. The Los Alamos hepatitis C sequence database. Bioinformatics (Oxford, England) 2005;21:379–384. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lartillot N., Lepage T., Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics (Oxford, England) 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lerat E. Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity. 2010;104(6):520–533. doi: 10.1038/hdy.2009.165. [DOI] [PubMed] [Google Scholar]
- Ling P., Wang M., Chen X., Campbell K.G. Construction and characterization of a full-length cDNA library for the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) BMC Genomics. 2007;8:145. doi: 10.1186/1471-2164-8-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wimmer E., Paul A.V. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys. Acta. 2009;1789(9):495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V. Hepatitis C virus RNA replication. Curr. Top. Microbiol. Immunol. 2013;369:167–198. doi: 10.1007/978-3-642-27340-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani F., Bull R.A., Lloyd A.R. Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol. 2012;30:443–452. doi: 10.1016/j.tibtech.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., Tang J., Wu G., Zhang H., Shi Y., Liu Y., Yu C., Wang B., Lu Y., Han C., Cheung D.W., Yiu S.M., Peng S., Xiaoqian Z., Liu G., Liao X., Li Y., Yang H., Wang J., Lam T.W., Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhugiri R., Fricke M., Marz M., Ziebuhr J. RNA structure analysis of alphacoronavirus terminal genome regions. Virus Res. 2014;194:76–89. doi: 10.1016/j.virusres.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E.R. DNA sequencing technologies: 2006–2016. Nat. Protoc. 2017;12:213–218. doi: 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- Marschall T., Marz M., Abeel T., Dijkstra L., Dutilh B.E., Ghaffaari A., Kersey P., Kloosterman W., Makinen V., Novak A. Computational pan-genomics: status, promises and challenges. BioRxiv. 2016:043430. doi: 10.1093/bib/bbw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M., Beerenwinkel N., Drosten C., Fricke M., Frishman D., Hofacker I.L., Hoffmann D., Middendorf M., Rattei T., Stadler P.F., Töpfer A. Challenges in RNA virus bioinformatics. Bioinformatics. 2014;30(13):1793–1799. doi: 10.1093/bioinformatics/btu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy E.M., McDonald J.F. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19(3):362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Peng Y., Leung H.C.M., Yiu S., Chin F.Y.L. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., Zhou L., Larson C.N., Dietrich J., Klem E.B., Scheuermann R.H. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(Database issue):D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Tamayo S., Sánchez-Rodríguez A., Swings T., Van den Bergh B., Dubey A., Steenackers H., Michiels J., Fostier J., Marchal K. Frequency-based haplotype reconstruction from deep sequencing data of bacterial populations. Nucleic Acids Res. 2015;43:e105. doi: 10.1093/nar/gkv478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., Liang S., Zhang W., Guan Y., Shen D. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A., Ouédraogo N., Afrough B., Bah A., Baum J.H.J., Becker-Ziaja B., Boettcher J.P., Cabeza-Cabrerizo M., Camino-Sánchez A., Carter L.L., Doerrbecker J., Enkirch T., García-Dorival I., Hetzelt N., Hinzmann J., Holm T., Kafetzopoulou L.E., Koropogui M., Kosgey A., Kuisma E., Logue C.H., Mazzarelli A., Meisel S., Mertens M., Michel J., Ngabo D., Nitzsche K., Pallasch E., Patrono L.V., Portmann J., Repits J.G., Rickett N.Y., Sachse A., Singethan K., Vitoriano I., Yemanaberhan R.L., Zekeng E.G., Racine T., Bello A., Sall A.A., Faye O., Faye O., Magassouba N., Williams C.V., Amburgey V., Winona L., Davis E., Gerlach J., Washington F., Monteil V., Jourdain M., Bererd M., Camara A., Somlare H., Camara A., Gerard M., Bado G., Baillet B., Delaune D., Nebie K.Y., Diarra A., Savane Y., Pallawo R.B., Gutierrez G.J., Milhano N., Roger I., Williams C.J., Yattara F., Lewandowski K., Taylor J., Rachwal P., Turner D.J., Pollakis G., Hiscox J.A., Matthews D.A., O’Shea M.K., Johnston A.M., Wilson D., Hutley E., Smit E., Di Caro A., Wölfel R., Stoecker K., Fleischmann E., Gabriel M., Weller S.A., Koivogui L., Diallo B., Keïta S., Rambaut A., Formenty P., Günther S., Carroll M.W. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16(4):395–399. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- Rhoads A., Au K.F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13(5):278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sanjuan R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J. Virol. 2010;84(19):9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuch M., Höper D., Beer M. RIEMS: a software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinf. 2015;16:69. doi: 10.1186/s12859-015-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Wheeler J., Grimwood J., Dickson M., Yang J., Caoile C., Bajorek E., Black S., Chan Y.M., Denys M. Quality assessment of the human genome sequence. Nature. 2004;429(6990):365–368. doi: 10.1038/nature02390. [DOI] [PubMed] [Google Scholar]
- Schulz M.H., Zerbino D.R., Vingron M., Birney E. Oases: robust de novo RNA-Seq assembly across the dynamic range of expression levels. Bioinformatics (Oxford, England) 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Simmonds P. Evaluating the evidence for virus/host co-evolution. Curr. Opin. Virol. 2011;1(5):436–441. doi: 10.1016/j.coviro.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J.M., Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A.F.A., Hubley, R., Green, P., n.d. Repeat masker open-3.0 http://www.repeatmasker.org. Accessed 2017.
- Sperber G., Lövgren A., Eriksson N.E., Benachenhou F., Blomberg J. Retrotector online, a rational tool for analysis of retroviral elements in small and medium size vertebrate genomic sequences. BMC Bioinf. 2009;10(6):S4. doi: 10.1186/1471-2105-10-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinf. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpfer A., Höper D., Blome S., Beer M., Beerenwinkel N., Ruggli N., Leifer I. Sequencing approach to analyze the role of quasispecies for classical swine fever. Virology. 2013;438:14–19. doi: 10.1016/j.virol.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Töpfer A., Marschall T., Bull R.A., Luciani F., Schönhuth A., Beerenwinkel N. Viral quasispecies assembly via maximal clique enumeration. PLoS Comput. Biol. 2014;10:e1003515. doi: 10.1371/journal.pcbi.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. Cross-species infections. In: Salomon D.R., Wilson C., editors. Springer; Berlin Heidelberg, Germany: 2003. pp. 47–71. (Xeno-Transplantation). [Google Scholar]
- Will S., Reiche K., Hofacker I.L., Stadler P.F., Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007;3(4):e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Charlebois P., Gnerre S., Coole M.G., Lennon N.J., Levin J.Z., Qu J., Ryan E.M., Zody M.C., Henn M.R. De novo assembly of highly diverse viral populations. BMC Genomics. 2012;13:475. doi: 10.1186/1471-2164-13-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagordi O., Däumer M., Beisel C., Beerenwinkel N. Read length versus depth of coverage for viral quasispecies reconstruction. PLoS One. 2012;7:e47046. doi: 10.1371/journal.pone.0047046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]