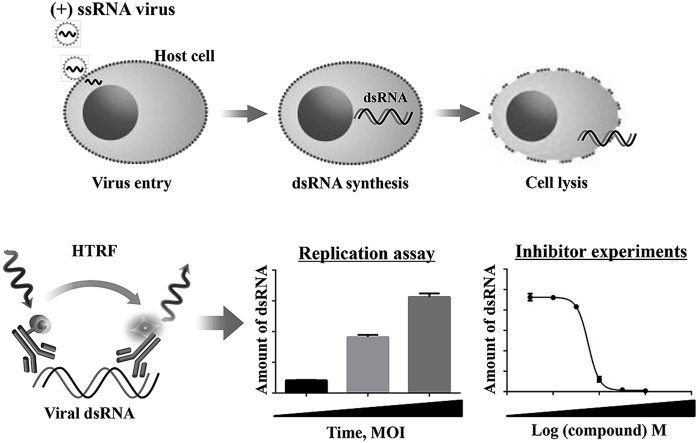
Abstract
The group of positive-sense single-stranded RNA ((+) ssRNA) viruses includes many important human pathogens. However, specific antiviral agents are not currently available for many RNA viruses. For screening of antiviral agents, methods that are simple, rapid, and compatible with high-throughput are required. Here, we describe a novel method for measurement of double-stranded RNA using a homogeneous time-resolved fluorescence assay. This method allowed detection of human rhinovirus (HRV), enterovirus, coxsackievirus, and murine norovirus. Furthermore, this method detected antiviral activity of a HRV 3C protease inhibitor. The assay may be useful for discovery of antiviral agents against (+) ssRNA viruses.
Keywords: RNA virus, Double-stranded RNA, Homogeneous time-resolved fluorescence assay, Viral replication, Antiviral agents
Abbreviations used: HTRF, homogeneous time-resolved fluorescence; dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; HRV, human rhinovirus; EV, enterovirus; CV, coxsackievirus; MNV, murine norovirus; CPE, cytopathic effect; qPCR, quantitative PCR; ELISA, enzyme-linked immunosorbent assay; MEM, Eagle's Minimum Essential Medium; FBS, fetal bovine serum; DMEM, Dulbecco's modified Eagle's medium; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; MOI, multiplicity of infection; EDTA, ethylenediaminetetraacetic acid; Eu, europium; poly(I:C), polyinosinic-polycytidylic acid; RT, room temperature; FRET, fluorescence resonance energy transfer; SDS, sodium dodecyl sulfate; S/B, signal-to-background; EC50, 50% effective concentrations; HTS, high-throughput screening
Graphical abstract
Highlights
-
•
We present a method to measure viral double-stranded RNA by HTRF assay.
-
•
This assay can detect several positive-sense RNA viruses.
-
•
This assay may be applicable for broad-spectrum detection of positive-sense RNA viruses.
-
•
This assay may be useful for discovery of antiviral agents.
1. Introduction
The group of positive-sense single-stranded RNA ((+) ssRNA) viruses (e.g., the Picornaviridae, Caliciviridae, Coronaviridae, Togaviridae, and Flaviviridae) includes many important human pathogens. However, specific antiviral agents are not currently available for many RNA viruses.
A double-stranded RNA (dsRNA) intermediate is commonly produced during the replication stage of (+) ssRNA viruses [1]. Viral RNA released into the cytoplasm of infected cells is translated and the negative-sense strand is subsequently produced, leading to the formation of dsRNA replicative intermediates during the replication stage. Positive-strand progeny viral RNA is generated using the negative-sense RNA as a template. Therefore, the detection of viral dsRNA may be useful for monitoring virus replication and proliferation.
To date, evaluation of virus infection has generally been performed by plaque assay to measure virus infectivity, cytopathic effect (CPE) assay to measure morphological changes or virus infectivity in infected cells, quantitative PCR (qPCR) assay to measure viral RNA, and enzyme-linked immunosorbent assay (ELISA) to measure viral proteins. Each assay has a number of advantages and disadvantages. CPE assay and plaque assay are widely used and are cost-effective, but relatively time-consuming and have low throughput. Although qPCR assay is a highly specific method, it is expensive and requires preparation of RNA and specific primers for viral factors. ELISA is also a highly specific method, but it requires multiple washing and incubation steps due to the heterogeneous assay conditions. Therefore, a simple, rapid, cost-effective, and high-throughput assay is required for basic research and screening of new antiviral agents.
Here, we describe a simple, rapid, cost-effective, high-throughput method using a homogeneous time-resolved fluorescence (HTRF) assay [2] focusing on viral dsRNA. The dsRNA-HTRF assay may be a useful method for screening of antiviral agents against (+) ssRNA viruses.
2. Materials and Methods
2.1. Reagents
Anti-dsRNA antibodies produced in rabbits immunized with rice dwarf virus-RNA-methylated bovine serum albumin complexes have been described previously [3]. Rupintrivir was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
2.2. Cells
MRC-5 (CCL-171) and H1-Hela (CRL-1958) were obtained from ATCC (Manassas, VA). These cells were maintained in Eagle's Minimum Essential Medium (MEM; Sigma-Aldrich, St. Louis, MO) with 10% FBS (Moregate Biotech, Queensland, Australia) at 37 °C in a humidified atmosphere containing 5% CO2.
2.3. Virus propagation
Human rhinovirus (HRV)-B14 was kindly provided by Dr. Shigehiro Sato (Department of Microbiology, School of Medicine, Iwate Medical University, Japan). HRV-B14 was propagated in MRC-5 cells cultured in MEM supplemented with 2% FBS (Moregate Biotech), 5% tryptose phosphate broth (Sigma-Aldrich) and 30 mM MgCl2 at 35 °C for 48 h. Cells and culture supernatants were collected and frozen at −80 °C. The cells and culture supernatant were defrosted at 35 °C and then centrifuged at 3000 rpm at 4 °C for 10 min. Supernatants were stored at −80 °C. HRV-B14 was titered by CPE assay using Viral ToxGlo Assay (Promega, Madison, WI).
2.4. Virus infection
Cells plated on 24-well plates (Falcon, Lincoln Park, NJ) at a density of 1.0 × 105 cells/well for dsRNA-HTRF assay or real-time PCR assay, or on 96-well plates (Iwaki, Tokyo, Japan) at a density of 2.5 × 104 cells/well for CPE reduction assay were infected at the indicated multiplicity of infection (MOI). For inhibitor testing, cells were infected at MOI of 0.1. Following 1 h incubation, cells were treated with rupintrivir at the indicated concentration. After 24 or 48 h, the amount of virus was determined by dsRNA-HTRF assay, real-time PCR assay, or CPE reduction assay.
2.5. DsRNA-HTRF assay
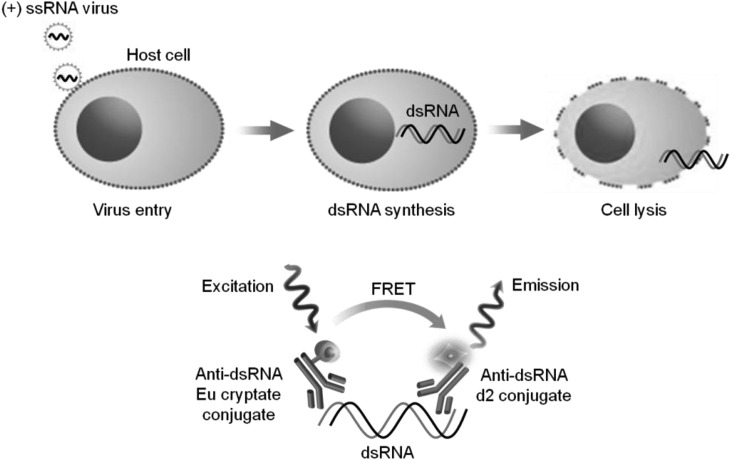
We used a prototype of the Viral double-stranded RNA detection kit (Cat# 64RNAPEG), recently launched by Cisbio K. K. (Chiba, Japan). Briefly, infected cells were lysed in ice-cold lysis buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.2% Triton-X) for 10 min on ice, collected, and then centrifuged at 15000 rpm for 10 min at 4 °C. Aliquots of 10 μL of lysate supernatants were transferred to 384-well plates (#2010-00110; Aurora Biotechnologies, Carlsbad, CA) followed by addition of 5 μL of anti-dsRNA-d2 and 5 μL of anti-dsRNA-europium (Eu) cryptate conjugate diluted in detection buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.2 M KF, 0.1% BSA). Polyinosinic-polycytidylic acid (poly(I:C)) was used as a dsRNA standard on the same plates. Poly(I:C) (1.56–100 ng/mL) was serially diluted at 1:2 dilution in lysis buffer. The plates were covered with ultraviolet optical plate seals (fluorescent permeable) and incubated at 4 °C or room temperature (RT). For each well, fluorescence was read on an Infinite® M1000 PRO (Tecan Japan, Tokyo, Japan) using the following filter settings: excitation 317 nm and emission 665 nm (for acceptor signals) and 620 nm (for donor signals), integration delay 60 μs, and integration time 500 μs. The HTRF signal ratio was calculated as follows: (signal at 665 nm/signal at 620 nm) × 10000. Fluorescence resonance energy transfer (FRET) signals represented by the Delta F were calculated as: [(Sample Ratio − Negative control Ratio)/(Negative control Ratio)] × 100 (%).
2.6. Data analysis
Calculation of EC50 (50% effective concentrations) and statistical analyses were performed using Graph Pad Prism Software version 5 (Graph Pad, San Diego, CA). Z′ values of the HTRF signal ratio were calculated as described previously [4]. Data are presented as the means ± SEM of three independent experiments.
3. Results
The assay scheme is depicted in Fig. 1 . To optimize this assay, we first determined the assay temperature using a synthetic analog of dsRNA, i.e., poly(I:C), and the reaction at 4 °C showed a higher HTRF signal level compared to the reaction at RT (Supplementary Fig. 1A). Moreover, we determined whether the dsRNA-HTRF assay could detect the concentration- and time-dependent signals. Increasing the concentration of poly(I:C) resulted in a significant increase in HTRF signal (Delta F%) (Supplementary Fig. 1B). Furthermore, these signals increased in a time-dependent manner (Supplementary Fig. 1C). In addition, these signals were abolished by dsRNA-specific nuclease RNase III (Supplementary Fig. 1D), indicating that these HTRF signals were specifically detected for dsRNA. Accordingly, in further experiments, the reaction conditions of 4 °C for 24 h were used, and serial dilution of poly(I:C) was used as a standard curve to determine the HTRF signal.
Fig. 1.
Schematic diagram of the double-stranded RNA (dsRNA) -homogeneous time-resolved fluorescence (HTRF)-based assay. Viral dsRNA is produced during replication of (+) ssRNA viruses in the cytosol of host cells. After cell lysis, dsRNA is detected in sandwich HTRF assay using anti-dsRNA antibody labeled with Eu cryptate (donor) and an anti-dsRNA antibody labeled with d2 (acceptor). When both antibodies simultaneously recognize dsRNA, induced proximity between the donor and acceptor units results in FRET under excitation at 317 nm and then leads to acceptor emission (665 nm).
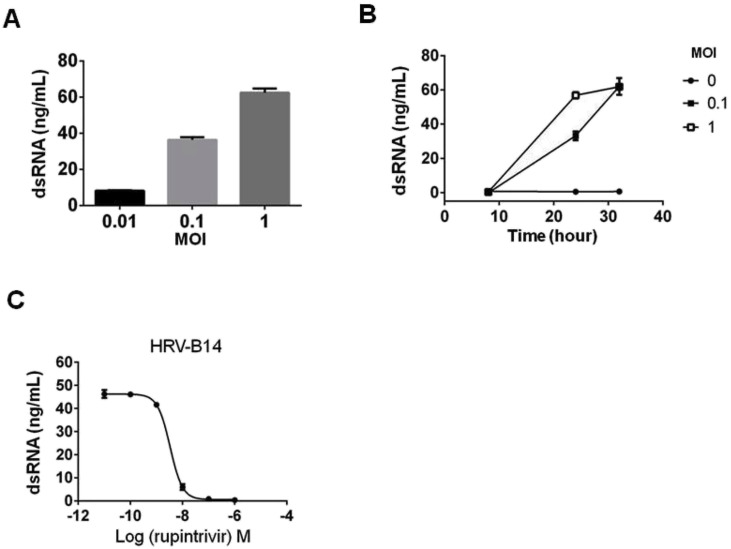
Picornaviruses are widely used for analysis of dsRNA intermediates [[5], [6], [7]]. Therefore, to evaluate whether the dsRNA-HTRF assay can detect (+) ssRNA viruses, we focused on infection by picornaviruses represented by HRV. When H1-HeLa cells were infected with HRV-B14, HTRF signals were clearly detected in a MOI-dependent manner (Fig. 2 A). Furthermore, these potentiated signals were also detected in a time-dependent manner, while non-infected cells hardly showed detectable HTRF signals (Fig. 2B). Cells infected with HRV-B14 at MOI of 1 for 24 h and MOI of 0.1 for 32 h showed cytotoxicity and cell detachment. The Z′ factor for this HTRF assay was 0.72, and the signal-to-background (S/B) ratio was 68, on comparison of mean values between MOI of 0 (non-infection) and MOI of 0.1 at 24 h post infection. In addition, we examined whether the dsRNA-HTRF assay could detect a single cycle of viral replication. Single-cycle growth of HRV is considered to take about 7 h [8]. H1-HeLa cells were infected with HRV-B14 at MOI of 10 to achieve single-cycle growth, and viral dsRNA was detected at 6 h post-infection using the dsRNA-HTRF assay (Supplementary Fig. 2), indicating that this assay could evaluate the single-cycle growth of HRV-B14. Cytotoxicity was observed at 24 h post-infection at MOI of 10. Moreover, significant amounts of dsRNA were detected for enterovirus (EV)-A71, coxsackievirus (CV)-A21, EV-D68, and CV-B3, which belong to the picornaviruses, in a manner dependent on the MOI by the dsRNA-HTRF assay (Supplementary Fig. 3A – D). The amounts of murine norovirus (MNV)-S7 dsRNA, which belongs to the genus Norovirus, were also measured by the dsRNA-HTRF assay (Supplementary Fig. 3E). No cytotoxicity was observed under these conditions. The RNase III abolished the infection-induced potentiated signals (Supplementary Fig. 4A – C). These results indicated that the dsRNA-HTRF assay can be used to evaluate the proliferation of (+) ssRNA viruses.
Fig. 2.
Detection of HRV-B14 proliferation using the dsRNA-HTRF assay. (A) H1-HeLa cells were infected with serial titers of HRV-B14 and cultured for 24 h. (B) H1-HeLa cells were infected with HRV-B14 for the indicated time periods. (C) Dose-response curve of rupintrivir on dsRNA production induced by HRV-B14. H1-HeLa cells were infected with HRV-B14 (MOI = 0.1). After 1 h, infected cells were incubated with rupintrivir at the indicated concentration and cultured for 24 h. Infected cells were subjected to the dsRNA-HTRF as described in Materials and Methods. Data are presented as the means ± SEM of three independent experiments.
Rupintrivir (AG7088) is an inhibitor of the HRV 3C protease and suppresses the proliferation of not only HRV but also EV [9]. We examined whether the dsRNA-HTRF assay could measure the inhibitory effect of rupintrivir on HRV and EV. In the dsRNA-HTRF assay, rupintrivir inhibited the proliferation of HRV-B14, CV-A21, and EV-D68 with EC50 of 3.3, 45, and 15, respectively (Fig. 2C, Supplementary Fig. 5A and B). The Z′ factor for this HTRF assay of inhibitor experiments was 0.71 ± 0.087. The observed EC50 values were comparable to those reported previously against HRV-B14 [9], CV-A21 [9], and EV-D68 [10].
We compared the dsRNA-HTRF assay with qPCR assay and CPE assay with regard to equivalency for detecting the effects of antiviral compounds. The anti-HRV-B14 activity detected by the dsRNA-HTRF assay was comparable to those determined by qPCR assay (Supplementary Fig. 6A) and CPE assay (Supplementary Fig. 6B) (rupintrivir EC50 = 5.1, 2.3 nM, respectively).
4. Discussion
There have been several reports showing the utility of HTRF assay for discovery of antiviral agents against human immunodeficiency virus [11], hepatitis C virus [12], and HRV [13]. However, there have been no reports of HTRF assay focused on viral replication. Here we report the first study regarding development of the HTRF assay to detect proliferation of (+) ssRNA viruses. Several anti-dsRNA antibodies are commercially available, which detect multiple (+) ssRNA viruses, e.g., picornaviruses [14], caliciviruses [15], coronaviruses [16], togaviruses [17], and flaviviruses [18,19] using immunofluorescence analysis and/or ELISA. In the present study, we showed that the dsRNA-HTRF assay could detect the proliferation of HRV, CV, EV, and MNV. As most positive-strand RNA viruses produce dsRNA during the replication stage, this assay may be applicable for broad-spectrum detection of other RNA viruses. Further investigations using the dsRNA-HTRF assay are needed to examine the possibility of broad-spectrum detection of RNA viruses, i.e., coronaviruses, togaviruses, and flaviviruses.
Normally, a Z′-factor value > 0.5 is considered acceptable for the high-throughput screening (HTS) platform [4]. In the present study, we showed that the dsRNA-HTRF assay had a Z′ factor of 0.71 ± 0.087 in inhibitor experiments. This assay had a good S/B ratio. Moreover, HTRF technology is simple, rapid, highly robust, and suitable for high-throughput analyses. Therefore, the dsRNA-HTRF assay may provide a new HTS-compatible strategy for the discovery of novel antiviral agents. Further experiments are needed to determine whether this assay can be optimized and miniaturized to meet HTS requirements for screening compound libraries.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of interest
Koji Adachi is an employee of Cisbio K. K., the company that supplied the reagents used in the homogenous time-resolved fluorescence assays.
Acknowledgments
The authors are grateful to Dr. Yunike Akasaka of Kyorin Pharmaceutical Co., Ltd. for helpful advice and comments on the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ab.2018.10.021.
Appendix. Supplementary data
The following is the supplementary data to this article:
References
- 1.Linden L., Wolthers K., van Kuppeveld F. Replication and inhibitors of enteroviruses and parechoviruses. Viruses. 2015;7:4529–4562. doi: 10.3390/v7082832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathis G. HTRF(R) technology. J. Biomol. Screen. 1999;4:309–314. doi: 10.1177/108705719900400605. [DOI] [PubMed] [Google Scholar]
- 3.Kitagawa Y., Okuhara E. Anti-poly(I)·poly(C) antibody bound to cellulose and its use in the specific separation of double-stranded RNAs. Anal. Biochem. 1981;115:102–108. doi: 10.1016/0003-2697(81)90531-5. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J.-H., Chung Oldenburg. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 5.Ahlquist P., Noueiry A.O., Lee W.-M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards A.L., Soares-Martins J.A.P., Riddell G.T., Jackson W.T. Generation of unique poliovirus RNA replication organelles. mBio. 2014;5 doi: 10.1128/mBio.00833-13. e00833-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Q., Hato S.V., Langereis M.A., Zoll J., Virgen-Slane R., Peisley A., Hur S., Semler B.L., van Rij R.P., van Kuppeveld F.J.M. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/J.CELREP.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haff R.F., Wohlsen B., Force E.E., Stewart R.C. Growth characteristics of two rhinovirus strains in WI-26 and monkey kidney cells. J. Bacteriol. 1966 doi: 10.1128/jb.91.6.2339-2342.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patick A.K., Binford S.L., Brothers M.A., Jackson R.L., Ford C.E., Diem M.D., Maldonado F., Dragovich P.S., Zhou R., Prins T.J., Fuhrman S.A., Meador J.W., Zalman L.S., Matthews D.A., Worland S.T. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 1999;43:2444–2450. doi: 10.1128/AAC.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhoden E., Zhang M., Nix W.A., Oberste M.S. In Vitro efficacy of antiviral compounds against enterovirus D68. Antimicrob. Agents Chemother. 2015;59:7779–7781. doi: 10.1128/AAC.00766-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings R.T., McGovern H.M., Zheng S., Park Y.W., Hermes J.D. Use of a phosphotyrosine-antibody pair as a general detection method in homogeneous time-resolved fluorescence: application to human immunodeficiency viral protease. Anal. Biochem. 1999;269:79–93. doi: 10.1006/abio.1999.4021. [DOI] [PubMed] [Google Scholar]
- 12.Kota S., Scampavia L., Spicer T., Beeler A.B., Takahashi V., Snyder J.K., Porco J.A., Hodder P., Strosberg A.D. A time-resolved fluorescence–resonance energy transfer assay for identifying inhibitors of hepatitis C virus core dimerization. Assay Drug Dev. Technol. 2010;8:96–105. doi: 10.1089/adt.2009.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton P., O'Shea D., Wells E., Moakes K., Dunmore R., Butler R.J., Wilkinson T., Ward A., Casson N., Strain M., Vousden K., Lowe D.C., Pattison D.V., Carruthers A.M., Sleeman M.A., Vaughan T.J., Harrison P. Development of a homogeneous high-throughput screening assay for biological inhibitors of human rhinovirus infection. J. Biomol. Screen. 2013;18:237–246. doi: 10.1177/1087057112469047. [DOI] [PubMed] [Google Scholar]
- 14.Jurgeit A., Moese S., Roulin P., Dorsch A., Lötzerich M., Lee W.-M., Greber U.F. An RNA replication-center assay for high content image-based quantifications of human rhinovirus and coxsackievirus infections. Virol. J. 2010;7:264. doi: 10.1186/1743-422X-7-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyde J.L., Sosnovtsev S.V., Green K.Y., Wobus C., Virgin H.W., Mackenzie J.M. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J. Virol. 2009;83:9709–9719. doi: 10.1128/JVI.00600-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canton J., Fehr A.R., Fernandez-Delgado R., Gutierrez-Alvarez F.J., Sanchez-Aparicio M.T., García-Sastre A., Perlman S., Enjuanes L., Sola I. MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jose J., Taylor A.B., Kuhn R.J. Spatial and temporal analysis of alphavirus replication and assembly in mammalian and mosquito cells. mBio. 2017;8 doi: 10.1128/mBio.02294-16. e02294-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Targett-Adams P., Boulant S., Mclauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication †. J. Virol. 2008;82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien C.A., Hobson-Peters J., Yam A.W.Y., Colmant A.M.G., McLean B.J., Prow N.A., Watterson D., Hall-Mendelin S., Warrilow D., Ng M.L., Khromykh A.A., Hall R.A. Viral RNA intermediates as targets for detection and discovery of novel and emerging mosquito-borne viruses. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.