Abstract
Background
Human Bocavirus (HBoV) is a newly discovered parvovirus whose role as a causative agent of respiratory disease remains unclear.
Study design
We investigated the presence of HBoV by quantitative PCR in the nasopharyngeal samples of 192 French children consecutively hospitalized for acute bronchiolitis. Other common respiratory viruses were detected using immunofluorescence assays, cell culture detection, or RT-PCR assays.
Results
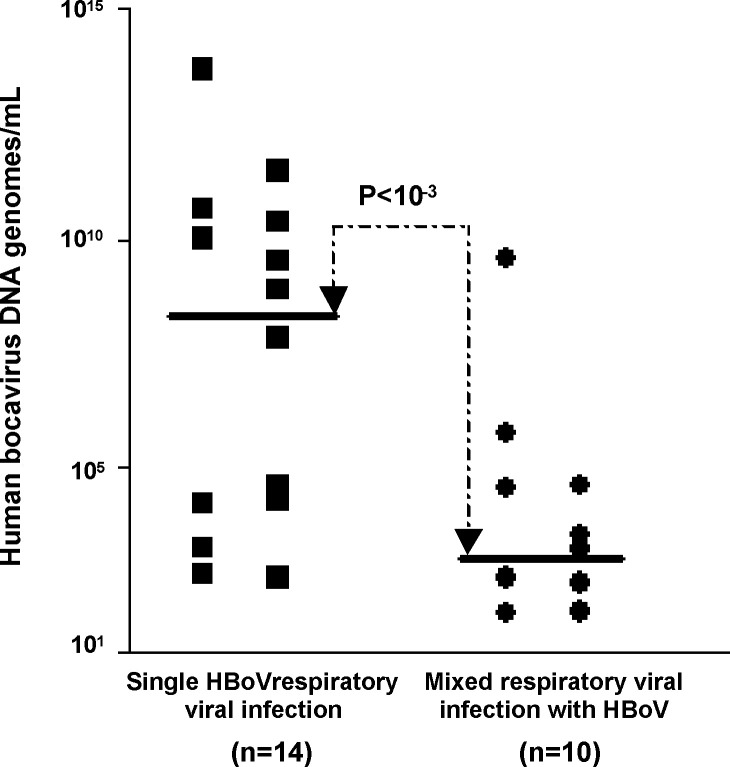
HBoV was detected in 24 (12.5%) of 192 study children. In 14/192 cases (7%) HBoV was the sole isolate and in 10/192 (5%) it was part of a mixed viral infection. HBoV was the third most common pathogen detected after respiratory syncytial virus (45/192; 23%) and rhinovirus (24/192; 12%). It occurred more often in infants aged 1–12 months (P = 0.002). Median levels of HBoV DNA genome in respiratory samples were significantly higher in patients with single HBoV infection than in patients with mixed respiratory viral infection with HBoV (4 × 108 copies/ml vs. 2 × 103 copies/ml, P < 0.001).
Conclusions
Our data suggest that HBoV at a high viral load could be an etiologic agent of respiratory tract disease, whereas the exact role of HBoV at a low viral load, as etiological cause or as pathophysiological co-factor of respiratory diseases, remains to be determined.
Keywords: Human Bocavirus, Bronchiolitis, Respiratory viral infections
1. Introduction
Human Bocavirus (HBoV) is a recently discovered pathogen identified as a cause of lower respiratory tract infections (LRTIs) in humans (Allander et al., 2005, Allander et al., 2007). This new virus, a member of the Parvoviridae family has been primarily identified in children with acute expiratory wheezing or bronchiolitis, bronchitis or bronchopneumonia (Allander et al., 2005, Kesebir et al., 2006). Recent studies have reported the presence of HBoV DNA sequences in 1.5–19% of respiratory tract samples of infants with various respiratory diseases hospitalized in North America, Europe, Asia, Africa and Australia suggesting that this virus is an ubiquitous respiratory pathogen (Allander et al., 2005, Allander et al., 2007, Arnold et al., 2006, Bastien et al., 2006, Choi et al., 2006, Foulongne et al., 2006, Ma et al., 2006, Sloots et al., 2006, Weissbrich et al., 2006). In these previous reports, HBoV was identified by PCR assays either as a unique viral pathogen or associated with respiratory syncytial virus (RSV) or other common respiratory viruses, suggesting that this virus could play a role as etiological agent or as cofactor of bronchiolitis. At present, the potential causal role of HBoV in the development of childhood bronchiolitis remains to be assessed in well-defined respiratory disease cohort of paediatric patients (Simon et al., 2007, Allander et al., 2007). In the present report, we retrospectively analyzed the epidemiological, virological and clinical features of HBoV respiratory infections in a cohort of French children hospitalized for acute bronchiolitis. To assess the causal role of HBoV in bronchiolitis, all the study respiratory samples were tested for the presence of other common respiratory viruses, including human metapneumovirus (HMPV) by classical immunofluorescence antigen detection, cell culture or molecular detection assays. Moreover, we determined and compared the viral genome loads and the sequences of VP1/VP2 gene of the HBoV strains detected in the nasopharyngeal samples of infants with single or multiple viral infections.
2. Patients and methods
2.1. Patients
Of 465 consecutive patients admitted to the pediatric department (University Medical Hospital Center of Reims, Champagne-Ardenne, France) from September 2001 to June 2002 for acute respiratory wheezing, we retrospectively selected 192 infants aged ≤36 months (mean age: 8.6 months; range: 12 days to 36 months) with clinical signs of bronchiolitis, hospitalized in the pediatric unit within 3 days of the onset of symptoms. Infants with cystic fibrosis, congenital heart disease, rhinitis, chronic allergic rhinitis and otitis or with genetic or acquired-immunodepression were excluded. Nasopharyngeal secretions were collected in sterile physiological saline fluid with a disposable mucus extractor at the time of hospital admission according to the recent European Respiratory Society Guideline (Anonymous, 2001). Nasopharyngeal aspirates were then rapidly transported to the virology laboratory where they were divided into two sterile tubes. Tube 1 was used to perform the immunofluorescence assays for viral antigens and viral cell-culture detection assays. Tube 2 was immediately frozen and stored at −80 °C prior to RT-PCR assays (Freymuth et al., 1997). The hospital's ethics committee approved the study and informed consent was obtained from parents.
2.2. Classical respiratory virus detection assays
Immunofluorescence assays for the detection of RSV, influenza viruses A and B, parainfluenza virus and adenovirus antigens were performed on tube 1 using monoclonal antibodies according to the manufacturer's recommendations (Argene Biosoft, Varhiles, France). This aliquot was diluted in 3 ml of standard viral transport medium as described previously (Freymuth et al., 1997). Moreover, 200 μl of this diluted nasopharyngeal secretion specimen was inoculated in duplicate onto 24-well plates covered with monolayers of continuous human diploid fibroblasts (MRC-5) or Rhesus monkey kidney (MA-104) cells as previously described (Freymuth et al., 1997). Virus isolates were typed by the standard method of virus neutralization on cell culture for enteroviruses, and by classical immunofluorescence antigen detection assays on infected cell monolayers for adenovirus, influenza A and B viruses, parainfluenza virus types 1–3 and RSV (Freymuth et al., 1997).
2.3. Molecular respiratory virus detection assays
Retrospectively, total nucleic acids were extracted from 200 μl of the nasal aspirate sample in tube 2 using a rapid extraction protocol according to the manufacturer's recommendations (High Pure Viral Nucleic Acid Kit, Roche Diagnostics, Mannheim, Germany). Nucleic acids were eluted in a final volume of 50 μl of diethyl-pyrocarbonate (DEPC) sterile water, as described by the manufacturer's recommendations and stored at −80 °C until used. Detection of human rhinovirus and enterovirus RNA sequences was carried out using a picornavirus RT-PCR detection assay followed by differential Southern blotting as previously described (Andreoletti et al., 2000). HMPV RNA genomic sequences were detected by real-time RT-PCR amplification of the nucleoprotein (N) gene as described previously (Bouscambert-Duchamp et al., 2005). HBoV DNA was detected by real-time PCR using two primers, HBoV-F (TATGGCCAAGGCAATCGTCCAAG) and HBoV-R (GCCGCCTGAACATGAGAAACAGA) previously designed by Sloots et al. (2006) and allowing the amplification of a 291-bp fragment of the putative NS-1 gene. This PCR screening assay was performed using an iCycler iQ Real-time PCR Detection System and the iQ™SYBR®Green Supermix 2X (Bio-Rad, Marnes-la-coquette, France) according to the manufacturer's recommendations and using 5 μl of extracted DNA solution per reaction tube. Real-time PCR cycling conditions were 95 °C for 15 min for the activation of the DNA polymerase, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, followed in turn by a melting curve. This quantitative PCR assay allowed a range of detection from 1.4 × 102 (C t value = 33) to 1.4 × 1014 (C t value = 10) copies of a reference plasmid containing the full length HBoV DNA genome per ml of nasopharyngeal aspirate sample (Allander et al., 2005). In order to check the quality of each DNA sample and to obtain a normalization of quantitative HBoV DNA values per ml of sample, a quantitative detection of GAPDH per ml of sample was also carried out using the “TaqMan GAPDH Control Reagents” according to the manufacturer's recommendations (Applied Biosystem, Courtaboeuf, France) (Bouscambert-Duchamp et al., 2005). The ratio of the number of HBoV genomes to the number of GAPDH DNA copies was calculated, which allowed us to take into account the inter-sample variability of the number of recovered respiratory epithelial cells (Gravitt et al., 2003). All the quantitative PCR DNA results were finally expressed as the number HBoV DNA genomes per ml of nasopharyngeal aspirate.
For each respiratory sample positive for HBoV by real-time PCR detection assay, a 404-bp fragment of the putative VP1 and VP2 genes was amplified in a second PCR assay using previously published primers and PCR amplification procedures (Bastien et al., 2007). The PCR products were sequenced on an ABI 3130 Sequencer using 150 ng of DNA and a fluorescent dye-terminator kit (Applied Biosystems). The DNA sequences were assembled and analyzed with the SeqScape softwareVersion 2.1 (Applied Biosystem, Courtaboeuf, France). Phylogenetic trees were generated by the neighbor-joining method using MEGA programs Version 3.1 (Nei and Kumar, 2000, Saitou and Nei, 1987). For bootstrap analysis, sequences were added randomly, and one tree was held at each step (1000 bootstrap replicates). All of the HBoV sequences reported in this publication have been submitted to EMBL (accession numbers AM849111–AM849120).
2.4. Statistical analyses
Fischer exact test, Student's t-test or Mann–Whitney U-tests were carried out when necessary, using the SAS software Version 8.2 (SAS Institute, Cary, NC, USA). P-values lower than 0.05 were considered as significant.
3. Results
3.1. Viral findings in infants with acute bronchiolitis
A potential viral pathogen was detected in 73% of the 192 study infants (Table 1 ). Twenty-four patients (12.5%) were positive for HBoV DNA sequences in the nasopharynx. HBoV was the only virus detected in 14 (7%) patients, whereas it was associated with another viral respiratory pathogen in 10 (5%) cases (7% vs. 5%, P > 0.5). The viruses most frequently associated with HBoV were picornaviruses (HRV and enteroviruses; 40% and 30%, respectively) and RSV (30%) (Table 2 ). An association of HBoV with both HMPV and rhinovirus was identified in one case of the 10 respiratory co-infections (Table 2). Interestingly, HBoV appeared to be more prevalent among patients with symptoms of unexplained viral etiology (14/51; 27%) than among those in which other viruses (10/117; 9%) were detected (9% vs. 27%, P < 0.001) (data not shown).
Table 1.
Viral etiology of respiratory infection in 192 children with bronchiolitis during a 10-month study period
| Virus | Number (%) of children infected with virus | Number (%) of children with virus as sole agent |
|---|---|---|
| Respiratory syncytial virus | 59 (31) | 45 (23) |
| Rhinovirus | 47 (24) | 24 (12) |
| Human Bocavirus | 24 (12) | 14 (7) |
| Enterovirus | 18 (9) | 4 (2) |
| Influenza A virus | 11 (6) | 10 (5) |
| Human metapneumovirus | 8 (4) | 6 (3) |
| Parainfluenza virus type 3 | 5 (2) | 4 (2) |
| Adenovirus | 3 (1.5) | 2 (1) |
| Total (%) | 141 (73) | 109 (57) |
Note. Human Bocavirus, rhinovirus and human metapneumovirus were studied only by PCR; enterovirus was studied by culture and PCR; and all other viruses were studied by culture, and antigen detection.
Table 2.
Clinical, virological and demographic features of children with Human Bocavirus DNA detected in nasopharyngeal aspirate samples
| Patient number | Month detected | Sex | Age (months) | Days in hospital | Oxygenotherapy on admission | Respiratory distress | Previous LRTIsa or bronchiolitis | Underlying diseases | Clinical complication | Associated respiratory viral infections | Nb of HBoV DNA genomes/mLb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 56 | November 2001 | M | 6 | 2 | Adenovirus | 3.6 × 104 | |||||
| 96 | December 2001 | M | 12 | 2 | RSV | RSV | 3.9 × 103 | ||||
| 109 | December 2001 | M | 14 | scc | + | + | RSV | 4.2 × 102 | |||
| 121 | January 2002 | F | 1 | 2 | HRV, HMPV | 8.5 × 101 | |||||
| 143 | January 2002 | F | 16 | 3 | + | Pneumonia | 9.5 × 109 | ||||
| 144 | January 2002 | F | 15 | 2 | + | Asthma | IVA | 6.0 × 105 | |||
| 164 | Febuary 2002 | M | 14 | 4 | Yes | + | + | 7.5 × 108 | |||
| 171 | March 2002 | M | 8 | 6 | + | + | Asthma, prematurity | EV, HRV | 1.7 × 103 | ||
| 173 | March 2002 | M | 14 | 2 | 4.5 × 1013 | ||||||
| 174 | March 2002 | M | 15 | 2 | Yes | + | 3.1 × 1011 | ||||
| 175 | March 2002 | M | 13 | 3 | Asthma, prematurity | EV, HRV | 3.6 × 109 | ||||
| 176 | March 2002 | F | 1 | 2 | 1.9 × 104 | ||||||
| 177 | March 2002 | F | 2 | 61 | Yes | + | Bronchodysplasia asthma | HRV | 4.4 × 104 | ||
| 178 | March 2002 | F | 4 | 2 | + | + | 4.3 × 104 | ||||
| 179 | March 2002 | F | 8 | 4 | 3.2 × 109 | ||||||
| 180 | March 2002 | M | 16 | 14 | + | + | 2.0 × 104 | ||||
| 184 | March 2002 | F | 4 | 5 | 7.1 × 107 | ||||||
| 185 | April 2002 | M | 4 | 2 | 1.9 × 103 | ||||||
| 186 | April 2002 | F | 11 | 3 | 2.3 × 1010 | ||||||
| 187 | April 2002 | M | 4 | 4 | 5.3 × 102 | ||||||
| 188 | April 2002 | M | 7 | 2 | RSV | 6.9 × 101 | |||||
| 190 | April 2002 | M | 9 | 2 | 4.6 × 102 | ||||||
| 191 | April 2002 | F | 8 | 4 | 4.1 × 102 | ||||||
| 203 | May 2002 | F | 11 | scc | + | EV | 3.3 × 102 |
Low respiratory tract infections.
ml of nasopharyngeal aspirate (HBoV DNA load results were normalized taking into account the GAPDH DNA load per ml of sample).
Simple consultation.
HBoV DNA genome loads in the nasopharyngeal aspirates ranged from 6.9 × 101 to 4.5 × 1013 copies/ml of sample material (Fig. 1 ). Interestingly the median level of the genome loads appeared to be significantly higher in patients with single HBoV infection (4 × 108 copies/ml; range: 4.1 × 102 to 4.5 × 1013 copies/ml) than in patients with a mixed respiratory viral infection with HBoV (2 × 103 copies/ml; range: 6.9 × 101 to 3.6 × 109 copies/ml) (P < 0.001) (Fig. 1).
Fig. 1.
Comparison of Human Bocavirus (HBoV) DNA genome loads in single infection or mixed infection among all 24 nasopharyngeal aspirates positive for HBoV. Each sample is represented by a single dot. The continuous line indicates the median level for each group.
3.2. Clinical features of Human Bocavirus respiratory infections
Comparison between age groups showed that HBoV occurred more frequently in infants aged 1–12 months (mean 9 months; S.D.: 4 months) (P = 0.002) (Table 2), whereas RSV occurred significantly more often infants aged 6–12 months than other stratified age groups of patients (P < 0.05). The sex ratio of HBoV-infected patients was 13 males and 11 females (P < 0.5). Of the 24 HBoV-positive patients, 20 had classical clinical bronchiolitis, associated with respiratory distress in five patients (Anonymous, 2001). Three of these required oxygen therapy on admission. Of the 24 HBoV-positive patients, 17 (70%) were previously healthy. In the rest, four had a history of asthma, three had had previous LRTI, two were ex-premature infants and one had bronchopulmonary dysplasia.
Human Bocavirus-infected cases were hospitalized for a mean of 6 days (median: 2; S.D.: 12). This mean duration of hospital stay was not statistically different from that observed in cases infected by other respiratory viruses. Moreover, the mean duration of hospital stay was not significantly different between in patients with a single viral infection and those with a dual virus infection (8 ± 15.5 days vs. 2.1 ± 1.6 days; P > 0.1).
3.3. Seasonality of virus infections
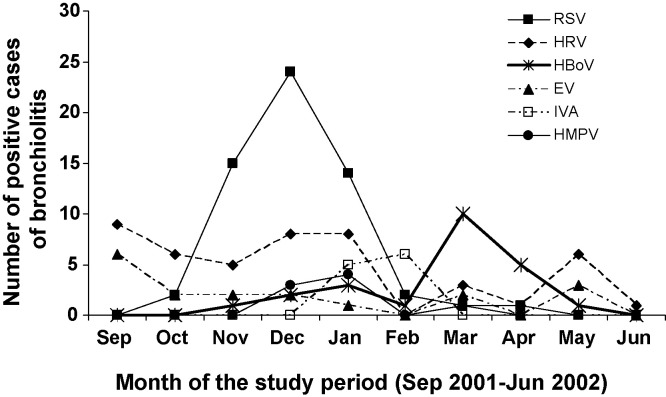
In the present study, HBoV circulated during two periods, from December 2001 to January 2002 and from March to April 2002 (Fig. 2 ). From December 2001 to January 2002, HBoV respiratory infections accounted for 6% of all hospitalized bronchiolitis cases at a time when RSV was epidemic, resulting in mixed RSV and rhinovirus or enterovirus respiratory tract infections. During March 2002, a second epidemic peak occurred in which HBoV accounted for 62% of all hospitalized bronchiolitis cases, suggesting that HBoV might be a common etiological cause of bronchiolitis during this period (Fig. 2).
Fig. 2.
Monthly distribution of bronchiolitis cases with acute viral respiratory infection during a 10-month period (September 2001–June 2002). RSV: respiratory syncytial virus, HRV: human rhinovirus, HBoV: Human Bocavirus, EV: enterovirus, IVA: influenza virus A, HMPV: human metapneumovirus.
3.4. Phylogenetic analysis of HBoV strains
Comparative phylogenetic analysis of VP1/VP2 gene sequences of the HBoV genome was performed for 10 (42%) of 24 isolates detected in nasopharyngeal samples during the cohort study. For the other cases, the PCR amplification of VP1/VP2 genes prior to PCR sequencing analysis was impossible, perhaps because VP1/VP2 gene PCR amplification appeared to be 10,000-fold less sensitive than real-time HBoV PCR detection assay (not shown). Sequence comparison with previously published North America, French, Spanish and African isolates shows that the VP1 and VP2 genes of the HBoV strains detected during the present study (fall 2001 and spring 2002), had high identity rates with other strains detected between 2002 and 2005 (not shown).
4. Discussion
We detected HBoV DNA genome in the nasopharynx of 12.5% of children with acute bronchiolitis. It was the third most commonly detected virus after RSV and rhinoviruses. These findings show that HBoV is a common virus in French children with respiratory infection, particularly infants aged 1–12 months. The only previous study from France reported nine cases of HBoV respiratory infection in a cohort of 262 infants (3%) hospitalized for various respiratory pathologies (Foulongne et al., 2006). Similarly, other recent studies outside France have reported HBoV in 1.5–19% of cohorts of children hospitalized for acute respiratory pathologies including acute wheezing illnesses or bronchiolitis and asthma (Allander et al., 2005, Allander et al., 2007, Arnold et al., 2006, Bastien et al., 2006, Choi et al., 2006, Foulongne et al., 2006, Ma et al., 2006, Sloots et al., 2006, Weissbrich et al., 2006). The higher prevalence of HBoV in our study may be the result of technical differences in study design and technique including sensitivity of PCR assays, technique of nasopharyngeal sampling, differences in patient groups and their epidemiology or the duration of study periods. Moreover, the present study included only clinically well-characterized patients with acute bronchiolitis, which may be the major manifestation of HBoV infection, whereas previous studies included a broader selection of clinical presentations.
Our findings support a role of HBoV as a cause of respiratory disease and indicate that HboV may be the third most important potential etiological agent of acute childhood bronchiolitis after RSV and human rhinovirus. Our data clearly support the hypothesis that HboV plays a significant role as etiological cause of bronchiolitis in children. However, it is possible that we underestimated the prevalence of infections by other common respiratory viruses because PCR was not applied to all possible respiratory pathogens, especially parainfluenza 4a and 4b, and coronaviruses OC43, 229E, NL64 and HKU1), which are considered minor causes of childhood bronchiolitis (Kaplan et al., 2008, Theamboonlers et al., 2007, Vabret et al., 2008). Moreover, we did not test respiratory samples from a control group of children matched for age, residence area and month of hospitalization and who were free of known respiratory symptoms or diseases. Therefore, the exact prevalence of respiratory HBoV infection as cause of bronchiolitis remains to be assessed in prospective studies which test by real-time PCR for the presence of all common viral pathogens in the respiratory tract of pediatric patients at the onset of respiratory symptoms and in appropriate, immunocompetent, asymptomatic controls (Lam et al., 2007).
In the present report, we identified HBoV as the unique viral pathogen in 14 (7%) of 192 French infants hospitalized for acute bronchiolitis (Table 1). HBoV appeared to be more prevalent among patients with symptoms of unexplained viral etiology than among those in whom other viruses were detected (8% vs. 27%, P < 0.001). The median level of the genome loads appeared to be significantly higher in patients with single HBoV infection (4 × 108 copies/ml) than in patients with mixed respiratory viral infections with HBoV (2 × 103 copies/ml) (P < 0.001) (Fig. 1). Taken together, these findings suggest an association between high HBoV load (high level of replicative viral infection) and acute bronchiolitis of otherwise unknown etiology. Other reports have assessed the HBoV DNA level in nasopharyngeal aspirates of infants hospitalized for bronchiolitis (Allander, 2008). However, none of these previous studies normalized their results according to the number of GAPDH DNA copies detected. Therefore, in the previously published studies, the differences observed in viral loads might be attributable to differences in the quality of nasopharyngeal specimens (Allander, 2008). Allander et al. previously defined two subgroups characterized by HBoV DNA levels lower or higher than 104 ml−1. According to this study, an HBoV DNA level higher than 104 ml−1 is a high viral load and one statistically associated with symptoms (Allander, 2008). Taking into account the previous published results on HBoV DNA levels, our quantitative data suggest that HBoV was a potential etiologic agent in 14 patients with HBoV infection alone, of whom 7 (50%) had a high viral load of ≥4 × 108 copies/ml (Fig. 1) (Allander, 2008). At the same time, shedding of HBoV (continuous or secondary to other infections) also appears to be common and was detected in mixed infections in 10 of our patients, of whom 1 (10%) had a high viral load, suggesting a potential etiological role of HboV in this case (Fig. 1). Of these 10 patients, 5 with a mixed viral infection had a medium HBoV DNA level ranging from 2 × 103 to 4 × 108 ml−1 suggesting a potential etiological role of this virus in the development of bronchiolitis. Four patients with mixed viral infection demonstrated an HBoV DNA level lower than 2 × 103 ml−1, suggesting an absence of etiological role of this virus in the development of bronchiolitis (Fig. 1). In fact, the diagnostic value of the viral load in individual patients remains to be determined in further prospective multicenter clinical studies (Allander, 2008). Several studies have also reported that dual viral infections are associated with more severe disease than infections with a single viral agent, therefore the clinical relevance of high or low levels of HBoV viral load will require further large additional studies (Greensill et al., 2003, Papadopoulos et al., 2002, Semple et al., 2005) (Table 2).
In the present study, HBoV-associated acute bronchiolitis did not differ clinically from that induced by rhinoviruses, enteroviruses, or RSV. Our results indicate that acute bronchiolitis is almost invariably associated with virus infection and often with simultaneous infection with two or three viruses (Table 1). Risk factors for severe disease with HBoV may be similar to those for RSV in the present study: prematurity and asthma (Table 2). Interestingly, the seasonal distribution of HBoV in our study was characterized by two peaks—winter and spring (Fig. 2), and therefore appeared to be different from those previously observed in Canada where no seasonal prevalence was reported (Bastien et al., 2006, Bastien et al., 2007). Close clustering of HBoV isolates recovered from Canada, France, and South Africa suggests that the evolutionary pattern of HBoV does not correlate with geographic or temporal variation during the last 4 years of HBoV circulation.
In conclusion, our findings suggest that HBoV is a common and frequent causative agent of bronchiolitis in French children and that it is the third most prevalent virus detected after RSV and rhinovirus. Our data suggest that HBoV at a high viral load could be an etiologic agent of respiratory tract disease, whereas the exact role of HBoV at a low viral load, as etiological cause or as pathophysiological co-factor, remains to be determined. Quantitative analysis normalized by the number of GAPDH DNA copies detected in nasal secretions, may be important to ensure comparability of future studies of HBoV infection. These further investigations would be of major interest for the development of future diagnostic, therapeutic and preventive strategies to fight against the viral causes of childhood bronchiolitis.
Conflict of interest
None of the authors of the present manuscript have a commercial or other association that might pose a conflict of interest.
Acknowledgement
This work was supported in part by grant for Clinical and Virological research (EA-3798: DAT/PPCIDH) from the Medical University and School of Medicine of Reims, France.
References
- Allander T. Human bocavirus. J Clin Virol. 2008;41:29–33. doi: 10.1016/j.jcv.2007.10.026. [DOI] [PubMed] [Google Scholar]
- Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T., Jartti T., Gupta S., Niesters H.G., Lehtinen P., Osterback R. Human bocavirus and acute wheezing in children. Clin Infect Dis. 2007;1(44):904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoletti L., Lesay M., Dewilde A., Lambert V., Wattre P. Differential detection of rhinoviruses and enteroviruses RNA sequences associated with classical immunofluorescence assay detection of respiratory virus antigens in nasopharyngeal swabs from infants with bronchiolitis. J Med Virol. 2000;61:341–346. doi: 10.1002/1096-9071(200007)61:3<341::AID-JMV10>3.0.CO;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous Proc Arch Pediatr; Paris, France, September 21; 2001. [PubMed] [Google Scholar]
- Arnold J.C., Singh K.K., Spector S.A., Sawyer M.H. Human bocavirus: prevalence and clinical spectrum at a children's hospital. Clin Infect Dis. 2006;43:283–288. doi: 10.1086/505399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N., Brandt K., Dust K., Ward D., Li Y. Human bocavirus infection. Can Emerg Infect Dis. 2006;12:848–850. doi: 10.3201/eid1205.051424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N., Chui N., Robinson J.L., Lee B.E., Dust K., Hart L. Detection of human bocavirus in canadian children in a 1-year study. J Clin Microbiol. 2007;45:610–613. doi: 10.1128/JCM.01044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouscambert-Duchamp M., Lina B., Moret H., Trompette A., Motte J., Andreoletti L. Detection of human metapneumovirus RNA sequences in nasopharyngeal aspirates of young French children with acute bronchiolitis by real-time reverse transcriptase PCR and phylogenetic analysis. J Clin Microbiol. 2005;43:1411–1414. doi: 10.1128/JCM.43.3.1411-1414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.H., Lee H.J., Kim S.J., Eun B.W., Kim N.H., Lee J.A. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children 2000–2005. Clin Infect Dis. 2006;43:585–592. doi: 10.1086/506350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V., Rodiere M., Segondy M. Human bocavirus in children. Emerg Infect Dis. 2006;12:862–863. doi: 10.3201/eid1205.051523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F., Vabret A., Galateau-Salle F., Ferey J., Eugene G., Petitjean J. Detection of respiratory syncytial virus, parainfluenzavirus 3, adenovirus and rhinovirus sequences in respiratory tract of infants by polymerase chain reaction and hybridization. Clin Diag Virol. 1997;8:31–40. doi: 10.1016/s0928-0197(97)00060-3. [DOI] [PubMed] [Google Scholar]
- Gravitt P.E., Peyton C., Wheeler C., Apple R., Higuchi R., Shah K.V. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J Virol Methods. 2003;112:23–33. doi: 10.1016/s0166-0934(03)00186-1. [DOI] [PubMed] [Google Scholar]
- Greensill J., McNamara P.S., Dove W., Flanagan B., Smyth R.L., Hart C.A. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg Infect Dis. 2003;9:372–375. doi: 10.3201/eid0903.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N.M., Dove W., Abd-Eldayem S.A., Abu-Zeid A.F., Shamoon H.E., Hart C.A. Molecular epidemiology and disease severity of respiratory syncytial virus in relation to other potential pathogens in children hospitalized with acute respiratory infection in Jordan. J Med Virol. 2008;80:168–174. doi: 10.1002/jmv.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesebir D., Vazquez M., Weibel C., Shapiro E.D., Ferguson D., Landry M.L. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis. 2006;194:1276–1282. doi: 10.1086/508213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W.Y., Yeung A.C., Tang J.W., Ip M., Chan E.W., Hui M. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol. 2007;45:3631–3640. doi: 10.1128/JCM.00280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Endo R., Ishiguro N., Ebihara T., Ishiko H., Ariga T. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006;44:1132–1134. doi: 10.1128/JCM.44.3.1132-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S. Oxford University Press; 2000. Molecular evolution and phylogenetics. [Google Scholar]
- Papadopoulos N.G., Moustaki M., Tsolia M., Bossios A., Astra E., Prezerakou A. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med. 2002;165:1285–1289. doi: 10.1164/rccm.200112-118BC. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Semple M.G., Cowell A., Dove W., Greensill J., McNamara P.S., Halfhide C. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis. 2005;191:382–386. doi: 10.1086/426457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A., Groneck P., Kupfer B., Kaiser R., Plum G., Tillmann R.L. Detection of bocavirus DNA in nasopharyngeal aspirates of a child with bronchiolitis. J Infect. 2007;54:125–127. doi: 10.1016/j.jinf.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots T.P., McErlean P., Speicher D.J., Arden K.E., Nissen M.D., Mackay I.M. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theamboonlers A., Samransamruajkit R., Thongme C., Amonsin A., Chongsrisawat V., Poovorawan Y. Human coronavirus infection among children with acute lower respiratory tract infection in Thailand. Intervirology. 2007;50:71–77. doi: 10.1159/000097392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Dina J., Gouarin S., Petitjean J., Tripey V., Brouard J. Human (non-severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J Paediatr Child Health. 2008;44:176–181. doi: 10.1111/j.1440-1754.2007.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbrich B., Neske F., Schubert J., Tollmann F., Blath K., Blessing K. Frequent detection of bocavirus DNA in German children with respiratory tract infections. BMC Infect Dis. 2006;6:109. doi: 10.1186/1471-2334-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]