Abstract
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed for detecting the structural glycoprotein gene of yellow head virus (YHV). The RT-LAMP assay is a novel method of gene amplification that amplifies nucleic acid with high specificity, sensitivity and rapidity under isothermal conditions with a set of four specially designed primers that recognize six distinct sequences of the target. The whole procedure is very simple and rapid, and reaction time and temperatures were optimized for 60 min at 65 °C, respectively. Detection of gene amplification could be accomplished by agarose gel electrophoresis. The standardized RT-LAMP procedure was used to detect YHV in the heart and gill from infected shrimp. Thus, the RT-LAMP assay is extremely rapid, cost-effective, sensitive and specific and has potential usefulness for rapid diagnosis for YHV detection in shrimp.
Keywords: LAMP, Yellow head disease virus, Shrimp
1. Introduction
Yellow head virus (YHV) is an enveloped, rod-shaped particle (approximately 40 nm × 170 nm) with prominent surface projections (approximately 11 nm) and an inner helical nucleocapsid (Chantanachookin et al., 1993, Wang and Chang, 2000, Loh et al., 1997). Based on the virion morphology and the presence of a single-stranded RNA genome (Wongteerasupaya et al., 1995), YHV was previously reported as a rhabdovirus (Nadala et al., 1997). However, it was subsequently demonstrated that the YHV genome is positive-sense RNA (Tang and Lightner, 1998).
Sequence identity, genome organization and gene expression have indicated that gill associated virus (GAV) and YHV are related to coronaviruses, toroviruses and arteriviruses and are classified in new taxa (family Roniviridae, genus Okavirus) within the order Nidovirales (Cowley et al., 1999, Cowley et al., 2000, Sittidilokratna et al., 2002, Cowley and Walker, 2002).
The development of a loop-mediated isothermal amplification (LAMP) assay for detection of white spot disease virus (WSDV) DNA was described by Kono et al. (2004). The LAMP assay is a novel approach to nucleic acid amplification that amplifies DNA with high specificity, selectivity and rapidity under isothermal conditions. Therefore, a thermal cycler is not needed. The LAMP assay originally described by Notomi et al. (2000) is based on the principle of the reaction performed by a DNA polymerase with strand displacement activity and a set of two specially designed inner primers and two outer primers. LAMP is highly specific for the target sequence because of the recognition of the target sequence by six independent sequences in the initial stage and by four independent sequences in the later stages of the LAMP reaction. The amplification efficiency of the LAMP method is extremely high because of its no time loss for thermal change, based on its isothermal reaction. Therefore, the LAMP assay has the advantage in specificity, selectivity and rapidity over other nucleic acid amplification methods (Mori et al., 2001).
The LAMP assay is also useful for RNA template detection upon the use of reverse transcriptase (RTase) together with DNA polymerase. In this paper, the RT-LAMP assay for detection of YHV RNA in shrimp is described.
2. Materials and methods
2.1. Shrimp
Black tiger shrimp (Penaeus monodon) with or without YHV clinical signs were collected from shrimp farms in Songkhla, Thailand. Shrimp samples were kept on ice for RNA extraction.
2.2. RNA extraction
RNA extraction from the gills was carried out using an RNA extraction kit (High Pure RNA Tissue Kit (Roche Diagnostics, Germany) according to the manufacturer's instructions. Briefly, gill tissues (20–25 mg) were homogenized with the lysis buffer. RNA was then eluted from spin columns in a final volume of 100 μl of elution buffer and stored at −80 °C until use.
2.3. Design of primers for RT-LAMP
YHV-specific RT-LAMP primers were designed according to published sequence of YHV structural glycoprotein gene (GenBank accession number: AF540644; Jitrapakdee et al., 2003) using Primer Explorer version 3 (https://primerexplorer.jp/lamp3.0.0/index.html). A set of four primers composed of two outer and two inner primers was designed. The two outer primers are known as the forward outer primer (F3) and the backward outer primer (B3), which helps in strand displacement. The inner primers are known as the forward inner primer (FIP) and the backward inner primer (BIP). Each primer has two distinct sequences corresponding to the sense and anti-sense sequences of the target, one for priming in the first stage and the other for self-priming in later stages. FIP contains F1c region (complementary to F1), a TTTT spacer and the F2 region. BIP contains the B1c region (complementary to B1), a TTTT spacer and the B2 region. FIP and BIP were HPLC-purified. The location of the primers within the RNA fragment is shown in Fig. 1 .
Fig. 1.
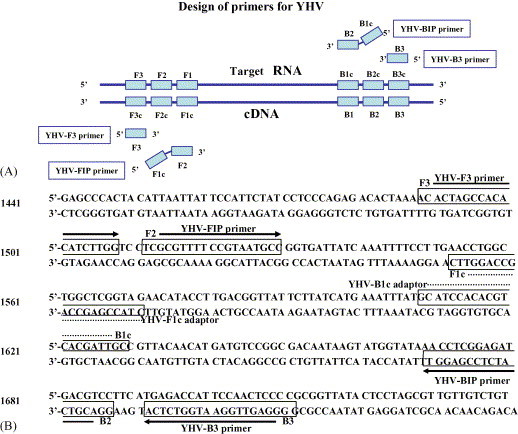
(A) Schematic diagram of two inner (FIP and BIP) and two outer (F3 and B3) primers for RT-LAMP. This diagram was partially quoted from Eiken Chemical Co. Ltd. (B) Nucleotide sequence of YHV structural glycoprotein gene (GenBank accession number: AF540644) used for the inner and outer primers. RNA sequences used for primer design are shown by boxes and arrows.
2.4. Optimization of reaction time and temperature
The RT-LAMP was carried out in a total volume of 25 μl reaction mixture with a Loopamp RNA Amplification Kit (Eiken Chemical Co. Ltd., Japan) according to the manufacturer's instructions. Briefly, 5 μl of target RNA was mixed with 2 μl (40 pmol) of each YHV-FIP and YHV-BIP, 1.0 μl (5 pmol) of YHV-F3 and YHV-B3, 12.5 μl of 2 × Reaction Mix, 0.5 μl of distilled water and 1.0 μl Enzyme Mix containing Bst DNA polymerase and AMV reverse transcriptase. After incubation at 65 °C for 15, 30, 45 or 60 min, the reaction was terminated by heating at 80 °C for 2 min. The reaction temperature (60, 63 and 65 °C) was also optimized. The RT-LAMP products were electrophoresed in a 2% agarose gel to determine the optimal condition.
2.5. Determination of RT-LAMP sensitivity
Ten-fold serial dilutions (10−1 to 10−8 diluted) of RNA extracted from YHV-infected shrimp was used as a template for RT-LAMP according to determined conditions. After the reaction, RT-LAMP products were electrophoresed on a 2% agarose gel and visualized using a gel document system (Ultra-Violet Products, Japan).
2.6. Nested RT-PCR for YHV detection
Nested RT-PCR was carried out using a commercial kit, IQ 2000 (Farming IntelliGene Technology Corporation, Taiwan) for detecting YHV and GAV. The first-step RT-PCR was carried out in 8 μl reaction volume containing 7.0 μl of RT-PCR PreMix (reaction buffer, dNTPs and YHV/GAV-specific primers), 0.5 μl of IQzyme DNA polymerase (2 U/μl), 0.5 μl of RT Enzyme Mix, 2.0 μl of RNA extracted from YHV. Ten-fold serial dilutions (10−1 to 10−8) of template RNA were used to determine the sensitivity of the detection. The amplification regime was 30 min at 42 °C, 2 min at 94 °C followed by 15 cycles of 94 °C for 20 s, 62 °C for 20 s and 72 °C for 30 s, then final elongation for 30 s at 72 °C and 30 s at 20 °C. After RT-PCR reaction was completed, 14.0 μl of nested PCR PreMix and 1.0 μl of IQzyme DNA polymerase were added. Two-step PCR reaction profile was followed by 30 cycles of 94 °C for 20 s, 62 °C for 20 s and 72 °C for 30 s, then final elongation for 30 s at 72 °C and 30 s at 20 °C. Two-step PCR products were electrophoresed in a 2% agarose gel to visualize the specific products.
2.7. Specificity of RT-LAMP detection
To determine specificity of RT-LAMP method, RT-LAMP was carried out with the different sources of RNA template, i.e. RNAs or DNAs of WSDV-infected shrimp, Taura syndrome virus (TSV)-infected shrimp or healthy shrimp using commercial RNA extraction kit, High Pure RNA Tissue Kit.
2.8. Quick extraction for RT-LAMP detection
Instead of using the commercial kit for extraction of RNA from shrimp, 0.5 M NaOH (Wang et al., 1993) was used. RNA was extracted from gill tissues of shrimp samples showing yellow head disease clinical signs. Twenty-five micrograms of sample was homogenized in 200 μl 0.5 M NaOH on ice. Then, 5 μl homogenization was diluted with 495 μl Tris–HCl buffer. The same weight of shrimp gill sample was used for the extraction of RNA using High Pure RNA Tissue Kit following the manufacturer's instruction. A series of 10-fold dilutions (10−1 to 10−5 diluted) of extracted RNA were used as template for RT-LAMP. RT-LAMP was performed using determined condition. RT-LAMP products were electrophoresed and analyzed in a 2% agarose gel.
3. Results
3.1. Optimization of RT-LAMP reaction condition for YHV detection
The RT-LAMP was carried out using RNA as template in order to determine the optimal temperature and reaction time. RT-LAMP products were detected at both 63 and 65 °C. However, the product at 65 °C showed a clearer reaction bands and this temperature was used as an optimal temperature. No amplification of template was found in the reaction time of 15 and 30 min. For the reaction time of 45 and 60 min at 65 °C, LAMP products were detected. However, for the complete amplification, the reaction time of 60 min was selected as an optimal reaction time. The results are shown in Fig. 2 .
Fig. 2.
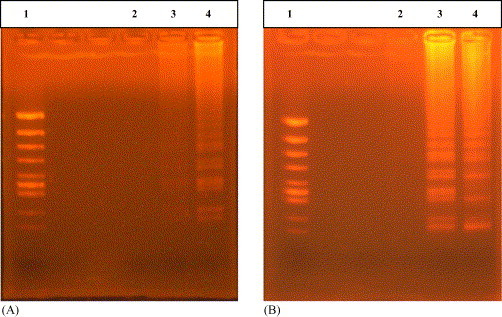
Results of optimization for RT-LAMP condition. (A) Varied temperature: lane 1, molecular size marker (φ/X174/Hinc II digest); lanes 2–4, RT-LAMP carried out at 60, 63 and 65 °C, respectively. (B) Varied reaction time: lane 1, molecular marker (φ/X174/Hinc II digest); lanes 2–5, LAMP carried out for 15, 30, 45 and 60 min, respectively. All products were electrophoresed on a 2% agarose gel and stained with ethidium bromide.
3.2. Comparison in sensitivity between RT-LAMP and nested RT-PCR
In order to determine the sensitivity of detection limit, RT-LAMP and nested RT-PCR were carried out using various concentrations (10−1 to 10−8 dilution) of RNA extracted from YHV-infected shrimp as template. RT-LAMP detected at a concentration of 10−6 dilution as a template, while the nested RT-PCR detected 10−7 diluted. The sensitivity of detection limit by RT-LAMP is 10 times lower than that of nested RT-PCR (Fig. 3 ).
Fig. 3.
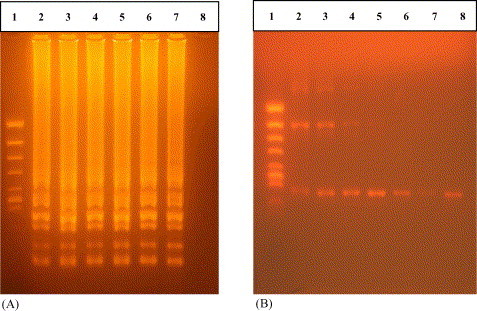
Sensitivity of RNA detection by RT-LAMP and nested RT-PCR: (A) RT-LAMP products and (B) nested RT-PCR products. Lane 1, molecular size marker (φ/X174/Hinc II digest); lanes 2–8, RT-LAMP and nested RT-PCR carried out using concentrations of RNA (10−1, 10−2, 10−3, 10−4, 10−5, 10−6 and 10−7), respectively. All products were electrophoresed on a 2% agarose gel and stained with ethidium bromide.
3.3. Specificity of RT-LAMP detection
The cross-reaction with other shrimp disease viruses, i.e. WSDV, TSV and healthy shrimp RNA was also carried out to determine the specificity of RT-LAMP method. Positive result for RT-LAMP was found none of them (Fig. 4 ). This indicates that this RT-LAMP method is a high specificity for YHV.
Fig. 4.
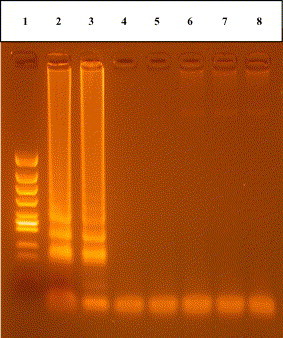
RT-LAMP products with different source of RNA/DNA template, i.e. from heart of YHV-infected shrimp (positive), TSV-infected shrimp (negative), WSDV-infected shrimp (negative) and healthy shrimp (negative). RT-LAMP products: lane 1, molecular size marker (φ/X174/Hinc II digest); lanes 2 and 3, YHV-infected shrimps; lanes 4 and 5, TSV-infected shrimp; lanes 6 and 7, WSDV-infected shrimp; lane 8, healthy shrimp. All the products were electrophoresed on a 2% agarose gel and stained with ethidium bromide.
3.4. Quick extraction for RT-LAMP detection
The template RNA extracted by the commercial RNA extraction kit (High Pure RNA Tissue Kit; Roche Diagnostics) provided higher sensitivity for RT-LAMP (10−5 dilution) than that by quick method (10−3 dilution) using 0.5 M NaOH (Fig. 5 ).
Fig. 5.
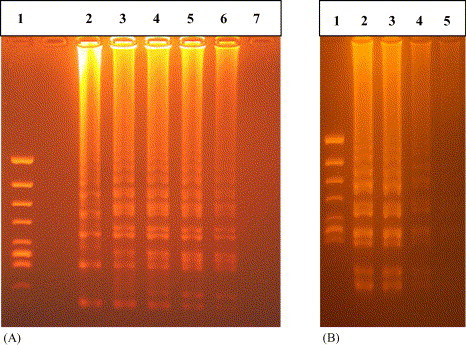
RT-LAMP products with various concentrations of template (10−1, 10−2, 10−3, 10−4, 10−5 and 10−6) extracted using commercial kit and quick method. (A) Commercial RNA extraction kit: lane 1, molecular size marker (φ/X174/Hinc II digest); lanes 2–7, 10−1, 10−2, 10−3, 10−4, 10−5 and 10−6, respectively. (B) 0.5 M NaOH method: lane 1, molecular size marker (φ/X174/Hinc II digest); lanes 2–5, 10−2, 10−3, 10−4 and 10−5. RT-LAMP was carried out using series of concentrations of RNA. All products were electrophoresed on 2% agarose gels and stained with ethidium bromide.
4. Discussion
In this study, the RT-LAMP diagnostic protocol was carried out for the detection of YHV in shrimp. Two sets of primer (outer and inner) used were able to amplify a 221 bp sequence of structural glycoprotein gene. The optimal condition of RT-LAMP reaction for the detection of YHV-RNA was shown as 65 °C and 60 min. However, it was also found that the reaction could be terminated within 45 min. This suggests RT-LAMP is a more rapid method for the detection of shrimp virus, compared to the RT-PCR method which takes 30–60 min for RT reaction and at least 2–3 h for conventional PCR method. In addition, it was shown that RT-LAMP method used for YHV detection specifically reacts only to YHV-infected shrimp. No cross-reaction with WSDV-infected shrimp was found.
The sensitivity of RT-LAMP was found to be 10 times lower than that of nested RT-PCR using an IQ2000 Kit for the detection of YHV/GAV. However, nested RT-PCR detection needs at least 2–3 h, comparing RT-LAMP within one hour. As an efficient diagnostic method, the rapidness for detection should be also considered.
The sensitivity of RT-LAMP also depends on the quality of RNA template, as the results showed quick method for RNA extraction using 0.5 M NaOH. This rapid technique resulted in lower sensitivity for the detection of virus, compared to using a commercial kit, although this method may take only 5 min for RNA extraction. Therefore, for the confirmatory diagnosis shrimp showing the typical signs of yellow head disease that are heavily infected with the virus can be diagnosed with this method in short period of time for RNA preparation.
This study proposed the first RT-LAMP protocol as an alternative method for the rapid detection of YHV with high sensitivity and specificity. This protocol is useful for the detection of low concentration of YHV from several tissues of cultured shrimp during early stages of infection. Additionally, this method could be used as both screening and confirmatory diagnosis for suspected shrimp, even though the virus titer is relatively low. This technique is recommended as an applied protocol for health management program and disease surveillance of shrimp in hatcheries as well as in grow-out pond, in order to prevent the disease outbreak.
Acknowledgement
This study was supported partly by a grant-in-aid for Science Research from the Ministry of Education, Science, Culture, Sports, Science and Technology of Japan.
References
- Chantanachookin C., Boonyaratpalin S., Kasornchandra J., Direkbusarakom S., Aekpanithanpong U., Supamattaya K., Sriuraitana S., Flegel T.W. Histology and ultrastructure reveal a new granulosis-like virus in Penaeus monodon affected by yellow-head disease. Dis. Aquat. Org. 1993;17:145–157. [Google Scholar]
- Cowley J.A., Dimmock C.M., Wongteerasupaya C., Boonsaeng V., Panyim S., Walker P.J. Yellow head virus from Thailand and gill-associated virus from Australia are closely related but distinct prawn viruses. Dis. Aquat. Org. 1999;36:153–157. doi: 10.3354/dao036153. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate nidovirus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. The complete sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S., Unajak S., Sittidilokratna N., Hodgson R.A.J., Cowley J.A., Walker P.J., Panyim S., Boonsaeng V. Identification and analysis of gp116 and gp64 structural glycoproteins of yellow head nidovirus of Penaeus monodon shrimp. J. Gen. Virol. 2003;84(Pt 4):863–873. doi: 10.1099/vir.0.18811-0. [DOI] [PubMed] [Google Scholar]
- Kono T., Savan R., Sakai M., Itami T. Detection of white spot syndrome virus in shrimp by loop-mediated isothermal amplification. J. Virol. Methods. 2004;115:59–65. doi: 10.1016/j.jviromet.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Loh P.C., Tapay L.M., Lu Y., Nadala E.C., Jr. Viral pathogens of the penaeid shrimp. Adv. Virus Res. 1997;48:263–312. doi: 10.1016/S0065-3527(08)60290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Nadala E.C.B., Tappy L.M., Loh P.C. Yellow-head virus: a rhabdovirus-like pathogen of penaeid shrimp. Dis. Aquat. Org. 1997;31:141–146. [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittidilokratna N., Hodgson R.A.J., Panyim S., Cowley J.A., Jitrapakdee S., Boonsaeng V., Walker P.J. The complete ORF1b-gene sequence indicates yellow head virus is an invertebrate nidovirus. Dis. Aquat. Org. 2002;50:87–93. doi: 10.3354/dao050087. [DOI] [PubMed] [Google Scholar]
- Tang K.F.J., Lightner D.V. A yellow head virus probe: application to in situ hybridization and determination of its nucleotide sequence. Dis. Aquat. Org. 1998;35:165–173. doi: 10.3354/dao035165. [DOI] [PubMed] [Google Scholar]
- Wang H., Qi M., Cutler A.J. A simple method of preparing plant samples for PCR. Nucleic Acids Res. 1993;21:4153–4154. doi: 10.1093/nar/21.17.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.C., Chang P.S. Yellow head virus infection in the giant tiger prawn Penaeus monodon cultured in Taiwan. Fish Pathol. 2000;35:1–10. [Google Scholar]
- Wongteerasupaya C., Sriurairatana S., Vicker J.E., Akrajamorn S., Boonsaeng V., Panyim S., Tassanakajon A., Withyachumnarnkul B., Flegel T.W. Yellow-head virus of Penaeus monodon is an RNA virus. Dis. Aquat. Org. 1995;22:45–50. [Google Scholar]


