Highlights
-
•
Effect of fulminant hepatic failure (FHF) associated mutations on helicase activities and virus replication were checked.
-
•
All the FHF mutants showed comparable unwinding activities with the wild type protein despite the differences in ATPase activities.
-
•
All the FHF mutant replicons showed marginal decrease in virus replication compared to the wild type replicon suggesting alternate function/s of the helicase protein.
-
•
Walker A motif and Walker B motif in the helicase domain are indispensable for HEV replication.
Keywords: Hepatitis E virus, Helicase, Mutations, Fulminant hepatic failure
Abstract
Fulminant hepatic failure (FHF) is the severe form of hepatitis E virus infection. Virus sequence analyses from severe cases have shown presence of unique and highly conserved mutations in the helicase domain of genotype 1, 3 and 4 viruses. We evaluated role of two amino acid replacements (L1110F) and (V1120I); found to be frequent in genotype 1 FHF-E viruses from India. Three mutant helicase proteins (two with single point mutations and one with dual mutations) were expressed in Escherichia coli and evaluated for their ATPase and RNA unwinding activities. Both L1110F and V1120I helicase mutants showed marginal decrease in ATPase activity, while L1110F/V1120I dual mutant showed normal ATPase activity. All three mutants proteins showed RNA unwinding activities comparable to wild type protein. Corresponding mutations were made in the helicase domain of HEV RLuc replicon and replication efficiencies were tested in the S10-3 (Huh 7) cells. The mutant replicon V1120I showed lower replication as compared to L1110F and L1110F/V1120I mutants. However, all three replicon mutants showed lower replication efficiencies as compared to the wild type replicon. Walker A and Walker B motif mutant HEV replicons were unable to replicate indicating essential role of the virus encoded helicase domain during HEV replication. FHF-E associated helicase mutations resulted in only marginal decrease in the virus replication suggesting alternate function/s of the helicase protein. Mutations in the helicase domain of FHF-E viruses may be responsible for changing virus or host-virus protein–protein interactions, causing alterations in the host responses, eventually leading to more severe disease manifestations.
1. Introduction
Hepatitis E virus (HEV) is a common cause of waterborne acute viral hepatitis in developing countries. Though hepatitis E is recognized as self limiting disease in general population, for unknown reasons pregnant women are more prone to infection and development of fulminant hepatic failure (FHF) with fatality rates approaching 15–20% (Khuroo et al., 1981). In industrialized countries zoonosis is the major cause of hepatitis (Miyamura, 2011).
HEV is non-enveloped virus, approximately 27–34 nm in diameter and belongs to genus Hepevirus in the family Hepeviridae. HEV genome is linear, single-stranded positive sense RNA of about 7.2 kb. The genome has short 5′- and 3′-untranslated regions, a 5′-methylguanine cap, a 3′ poly (A) stretch, and three open reading frames ORF1, ORF2 and ORF3. ORF1 codes for viral non-structural polyprotein and contains several conserved enzymatic domains, including putative methyltransferase, papain like cysteine protease, helicase and RNA-dependent RNA polymerase (Koonin et al., 1992). ORF2 codes for viral capsid protein and ORF3 for a small phosphoprotein that gets associated with cellular membranes and cytoskeleton fractions (Zafrullah et al., 1997). ORF3 also has pleiotropic effects on host cell pathways and recently reported to have an important role in viral egress from infected cell (Ahmad et al., 2011, Holla et al., 2013). HEV has been classified phylogenetically into four major genotypes 1, 2, 3 and 4. Genotypes 1 and 2 are restricted to humans and cause sporadic and epidemic hepatitis, mostly in developing countries. Genotypes 3 and 4 HEV are zoonotic and infect humans and wide range of animal species (International Committee for Taxonomy of Viruses; Ninth Report).
Helicases are molecular motor proteins that unwind nucleic acid strands using ATP derived energy and have important role during virus replication (Kadare and Haenni, 1997). Most positive sense RNA viruses encode their own RNA helicases with highly conserved domains. In addition to the viral genome replication, helicases are also involved in RNA translocation, genome packaging/unpackaging, protecting RNA at replication site, modulating RNA–protein interactions, capping etc. Helicases can also interact with host cell proteins and help in establishing virus infection.
Hepatitis E virus RNA helicase has been grouped into 5′–3′ class of superfamily 1 of helicases (SF1) containing seven signature motifs (I, Ia, II, III, IV, V and VI). These highly conserved motifs are also found in positive sense RNA viruses such as alphaviruses, arteriviruses, coronaviruses and rubiviruses. We have previously reported that HEV helicase has nucleotide triphosphatase (NTPase) activity and is able to hydrolyze all NTPs, although with differential efficiencies. The enzyme has ability to unwind RNA duplexes with 5′ overhangs and has sequence independent RNA-5′-triphosphatase (RTPase) activity suggestive of its role in forming viral cap structure.
Severity of HEV infection in pregnant women still remains a major question in HEV pathogenesis. Exact mechanism of liver damage in HEV infection is not yet known. Studies from our group have shown significantly higher levels of both Th1 (IFN-γ, IL-2 and TNF-α) and Th2 (IL-10) cytokines in FHF-E patients (Saravanabalaji et al., 2009) and association of TNF-α and IFN-γ cytokine gene polymorphism with the susceptibility and clinical outcome of hepatitis E (Mishra and Arankalle, 2011). Prabhu et al., 2011 have shown increased number of CD8+T cells in the liver biopsies of HEV infected patients suggesting their role in liver damage. Impairment of HEV specific T-cell responses was shown to be associated with chronic HEV infection in immunocompromised organ transplant patients (Suneetha et al., 2012). It is not yet clear whether severity of HEV infection is influenced alone by the host factors or there is involvement of viral factors also.
There are studies indicating association of point mutations in HEV genome with disease severity; however underlying mechanism still remains unknown. Silent substitutions of U at the nucleotide 3148 (in the helicase domain) and C at the nucleotide 5907 (in the capsid gene) in HEV strains of genotype 3 (comparison between 10 acute and 1 FHF case) and 4 (comparison between 23 acute and 5 FHF cases) have been documented to have association with fulminant hepatitis and disease severity in patients. C5907 was also associated with high viral loads in the infected individuals (Inoue et al., 2006, Inoue et al., 2009). Takahashi et al. (2009) have reported association of V239A mutation (in the helicase domain) (7/7 severe hepatitis cases) with increased virulence of genotype 3 virus. Similarly, comparative sequence analysis of genotype 1 sequences from acute viral hepatitis cases from India, Indian subcontinent and FHF cases from India showed sub clustering of 6/7 FHF sequences indicative of similarity in them. There were six unique amino acid substitutions in the ORF1 region of HEV sequences derived from FHF cases (F179S, A317T, T735I, L1110F, V1120I, and F1439Y) (Mishra et al., 2013). Two of these mutations L1110F (found in 5/7 FHF virus sequences) and V1120I (6/7 FHF virus sequences) were present in the HEV helicase region, between helicase motif IV and motif V. These reports suggested possible involvement of helicase domain in determining outcome of HEV infections. Present study addresses possible role of L1110F and V1120I amino acid replacements in the helicase domain of genotype 1 virus in altering enzymatic activities of the protein and replication efficiency of the virus.
2. Materials and methods
2.1. Cells
S10-3 cells (a subclone of the human hepatoma Huh7 cells) were a kind gift from Dr. S. Emerson (NIH, USA) and were maintained as described previously (Emerson et al., 2006).
2.2. Cloning, expression and purification of HEV NTPase/helicase domain
The HEV helicase domain (amino acids 960–1204 of HEV ORF1) clone (pET15b.HEV Hel) from genotype 1 virus was previously constructed in our laboratory (GenBank Accession No. DQ459342) (Karpe and Lole, 2010a). The Escherichia coli BL21 (DE3)/RIPL codon plus cells (Stratagene, La Jolla, CA) were transformed with recombinant plasmid for protein expression. Protein induction was done with 1.0 mM IPTG for 2 h at 37 °C. The N-terminal His tag fusion protein was purified from bacterial culture pellets using ProBond nickel chelating resin column (Invitrogen) in denatured condition. Briefly, cell pellet was lysed by using buffer containing 6 M guanidine hydrochloride, 50 mM HEPES buffer (pH 7.8) and 0.5 M NaCl. Cell lysate was centrifuged at 10,000 × g for 40 min and supernatant was loaded on to column pre-equilibrated with binding buffer (8 M urea, 0.5 M NaCl, 50 mM HEPES buffer, pH 7.8). Washing was done initially with wash buffer 1 (8 M urea, 0.5 M NaCl, 50 mM HEPES, pH 6.0) followed by wash buffer 2 (8 M Urea, 0.5 M NaCl, 50 mM HEPES, pH 5.3). Elution of the protein was performed using elution buffer (8 M urea, 0.5 M NaCl, 50 mM HEPES buffer, pH 4.0). Collected fractions were analyzed on 10% SDS-PAGE. Fractions containing protein of expected size were combined and concentrated using Amicon membrane columns (cutoff: 10 kDa) (Millipore).
Renaturation/refolding of the purified protein was done by incubating the protein overnight in buffer containing 50 mM HEPES (pH 7.2), 1 M PPS, 1 mM dithiothreitol (DTT) at 4 °C followed by quick dialysis in 50 mM HEPES buffer. Western blot analysis was done using anti-His monoclonal antibodies (Sigma Chemicals, St. Louis, MO). Protein concentration was determined by the Bradford's method. Glycerol, DTT, protease inhibitor (Roche) were added to the final concentrations of 20%, 2 mM and 1× respectively, and the protein was stored at −80 °C in aliquots until use.
2.3. Generation of helicase protein mutants by site directed mutagenesis
Construction of the Walker A motif (motif I) and Walker B motif (motif II) helicase mutants is previously reported (Karpe and Lole, 2010a). Mutants L1110F Hel, V1120I Hel and L1110F/V1120I Hel were generated with primers pairs as listed in the Table 1 using pET15b.HEV Hel clone as the template with QuickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Clones were confirmed by sequencing and host E. coli BL21 cells were transformed with these plasmids. Protein induction and purification were carried out as described above.
Table 1.
DNA oligonucleotides used in this study.
| Name | Sequence (5′–3′) | Mer |
|---|---|---|
| L1110F Hel For | GACCACTAGCCGGGTTTTCCGTTCGTTGTT | 30 |
| L1110F Hel Rev | AACAACGAACGGAAAACCCGGCTAGTGGTC | 30 |
| V1120I Hel For | CTGGGGTGAGCCTGCCATCGGGCAGAAACTAGTG | 34 |
| V1120I Hel Rev | CACTAGTTTCTGCCCGATGGCAGGCTCACCCCAG | 34 |
| Mut GDD For | GTGGCTGCCTTTAAAGGTGCTGCTTCGATAGTGCTTTGC | 39 |
| Mut GDD Rev | GCAAAGCACTATCGAAGCAGCACCTTTAAAGGCAGCCAC | 39 |
| Mut GKS For | GGTGTGCCTGGATCCGGCGCGTCCCGCTCTATTACC | 36 |
| Mut GKS Rev | GGTAATAGAGCGGGACGCGCCGGATCCAGGCACACC | 36 |
| Mut DE For | GGGCGCCGGGTTGTCATTGCTGCGGCCCCGTCCCTTC | 37 |
| Mut DE Rev | GAAGGGACGGGGCCGCAGCAATGACAACCCGGCGCCC | 37 |
| Mut L1110F For | ACCACTAGTCGGGTCTTCCGGTCGTTGTTCTGG | 33 |
| Mut L1110F Rev | CCAGAACAACGACCGGAAGACCCGACTAGTGGT | 33 |
| Mut V1120I For | GGTGAGCCCGCCATTGGGCAGAAGCTAGTGTTC | 33 |
| Mut V1120I Rev | GAACACTAGCTTCTGCCCAATGGCGGGCTCACC | 33 |
2.4. ATPase assay
ATPase assays were performed by sensitive and high throughput colorimetric method as described earlier (Karpe and Lole, 2010a). Briefly, reaction was performed in a 96-well plate using malachite green reagent. The 50 μl reaction contained, 300 pmol of enzyme, 50 mM HEPES (pH 7.2), 2 mM MgCl2, 10 mM KCl, 0.05 mg of BSA/ml, 2 mM DTT and 0.1 mM NTP. After incubation at 37 °C for 40 min color was developed by adding of an equal volume of malachite green-molybdate reagent and further incubating at room temperature for 30 min. The absorbance was measured at 630 nm by using μQuant 96-well plate reader (Bio-Tek). All reactions were carried out in triplicates, in two independent experiments and mean values were taken to calculate the NTPase activity in the form of amount of inorganic phosphate released through hydrolysis of NTP using the standard curve (made by using known concentrations of potassium dihydrogen phosphate). Comparative activities of mutant proteins were derived by assuming the activity of wild type helicase protein as 100%. The kinetic parameters (K m, V max) were calculated by using the double reciprocal plot. Briefly, each mutant protein (300 pmoles) was incubated with ATP at different concentrations from 2.5 μM to 100 μM. The reciprocals of substrate hydrolysis (1/V) were plotted against the reciprocals of substrate concentrations (1/[S]). Mean values from two independent experiments performed in triplicates were taken to calculate K m (μM) and V max (nmoles min−1 mg−1) values by fitting the data using MS excel 2007.
2.5. RNA unwinding assay
RNA unwinding assays were performed by non-radioactive ELISA based method as described previously (Mhaindarkar et al., 2014). Briefly, RNA duplexes were generated by annealing in vitro synthesized, biotin labeled 38 mer RNA (binding strand) and Digoxigenin (DIG) labeled 26 mer RNA strand (release strand). The biotin ylated strand binds to the streptavidin-coated wells while the helicase catalyzes removal of DIG labeled strand. DIG-labeled strand is detected using anti-DIG antibodies coupled to horseradish peroxidase which gives amount of intact RNA duplex after the enzymatic action. Taking absorbance thus allows indirect measurement of unwinding efficiency of the enzyme, i.e. decrease in absorbance is proportional to unwinding by the enzyme. All reactions were carried out in triplicates, in two independent experiments and mean values were taken to calculate percent unwinding activities of the helicase mutants.
2.6. Generation of mutants of HEV subgenomic replicon
HEV Rluc replicon encoding Renilla luciferase (Rluc) gene was a kind gift from Dr. X.J. Meng (Virginia Tech, Blacksburg, USA). This subgenomic clone was developed by them from pSKHEV-2, genotype 1 HEV infectious cDNA clone (GenBank Accession No. AF444002) (Cao et al., 2010). Following replicon mutants were developed using HEV-Rluc replicon as the template by site directed mutagenesis in the current study: Mut GKS (changing GKS to GAS in the Walker A motif), Mut DE (changing DE to AA in the Walker B motif), Mut L1110F, Mut V1120I, Mut L1110F/V1120I and Mut GDD (changing conserved RdRp GDD motif to GAA was previously shown to completely stop HEV replication by Graff et al., 2005, Cao et al., 2010). The primer sequences used for construction of the mutant clones are given in Table 1.
2.7. Generation of capped RNA transcripts and transfection
HEV replicon plasmids were linearized by utilizing unique BglII site located immediately downstream of the poly (A) tract of the HEV sequence and capped RNA transcripts were synthesized by in vitro transcription using mMessage mMachine T7 ultra kit (Ambion). Following transcription, DNA template was removed by DNase I treatment, transcribed RNA was purified by lithium chloride precipitation method as per the manufacturer's instructions and quantified on Nanodrop spectrophotometer (ND-1000, Nanodrop technologies). Integrity of the transcripts was checked by doing denaturing agarose gel electrophoresis. For each experiment, S10-3 cells were grown up to 60–70% confluence in 24-well cell culture plates and washed with serum free medium, OptiMEM (Invitrogen, Life technologies) prior to transfection. Capped RNA transcripts, diluted appropriately in the OptiMEM were transfected in cells (2 μg/well of the 24 well plate) using 1,2-dimyristyl Rosenthal inhibitor ether (DMRIE-C) reagent (Invitrogen) as per the manufacturer's instructions. Firefly luciferase plasmid DNA was co-transfected (PGL-3 promoter vector, 100 ng/well) with HEV-Rluc RNA to normalize cell transfection efficiency and the Renilla luciferase signal. After 4 h of incubation at 34.5 °C, transfection mixture was replaced with DMEM containing 10%FBS. All transfections were carried out in triplicates and each set of experiments was repeated twice.
2.7.1. Reporter gene assay
Monolayer of the RNA transfected cells was washed two times with phosphate buffered saline, cells were lysed in 100 μl of 1× Passive Lysis Buffer (Promega) and the lysates were immediately frozen at −80 °C until use. For the assay, samples were thawed, centrifuged at 5000 RPM for 2 min and 20 μl cell extracts were used for measuring the dual luciferase activities (Renilla luciferase: RLuc and firefly luciferase: Luc) using Dual luciferase assay system (Promega) and readings were taken on the Perkin Elmer 2030 Reader (Victor X3). RLuc values were divided with Luc values to normalize transfection efficiencies.
2.8. Detection of HEV replicative intermediate (negative strand RNA) by strand specific RT-PCR
S10-3 cells were transfected with capped RNA transcripts as described above, incubated at 34.5 °C for 4 h, the transfection mixture was removed and the cell monolayer was washed 10 times with 1× PBS. RNA transfected cells were harvested at different time intervals and cell pellets were processed for the total cellular RNA extraction using Ribopure kit (Ambion Life technologies, USA) as per the manufacturer's instructions. Detection of negative sense RNA (nsRNA) (replicative intermediate) was done using strand specific tagged primer-based reverse-transcription PCR as described previously (Chatterjee et al., 2012).
3. Results
3.1. Construction of HEV helicase mutants, protein expression and purification
We have previously reported cloning and expression of helicase domain protein (Karpe and Lole, 2010a). On comparison of Indian FHF-E derived HEV helicase domain sequences with this expression construct, except for the two amino acid changes (L1110F and V1120I) remaining sequences were 100% identical. Hence this helicase domain clone was used as the template to develop FHF helicase mutant clones in view of checking their enzymatic activities. Three mutant proteins were generated by site directed mutagenesis (L1110F Hel, V1120I Hel and L1110F/V1120I Hel) while previously developed Walker A (GKS to GAS) and Walker B (DE to AA) mutant proteins (Karpe and Lole, 2010a) were used for comparison in the present study. Locations of point mutations in the HEV helicase encoding region are shown in the schematic diagram of the helicase in Fig. 1 . All proteins were expressed along with N-terminal fusion histidine tag and purified using nickel affinity chromatography in denaturing buffer system followed by refolding in renaturing buffer (Mhaindarkar et al., 2014). Renaturation of the proteins was monitored by doing ATPase assay of the wild type protein, purified in parallel with mutants in identical buffer conditions and at similar protein concentrations. Overall, protein purity was more than 90% for all proteins. All mutants including wild type protein showed identical band size (26 kDa) when analyzed by 10% SDS-PAGE (Fig. 2A) and western blot (Fig. 2B).
Fig. 1.
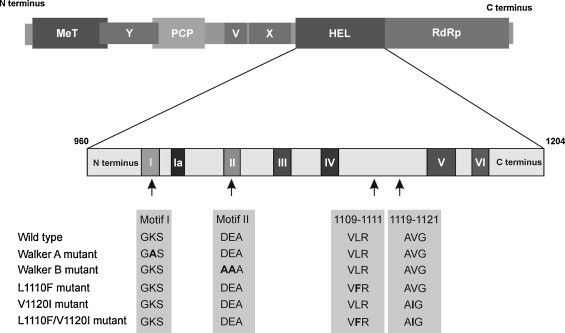
Schematic representation of hepatitis E virus ORF1: HEV ORF1 region encodes nonstructural polyprotein that contains methyltransferase (MeT), Y, papain-like cysteine protease (PCP), variable (V), X (macro), RNA helicase (Hel), and RNA-dependent RNA polymerase (RdRp) domains. The conserved motifs, typical of the helicase superfamily I (designated as I–VI) are shown in the enlarged view of HEV helicase (960–1204 aa). The amino acid changes done in the helicase region of pET15b.HEV Hel expression clone and HEV Rluc replicon are indicated in ‘Bold’ text.
Fig. 2.
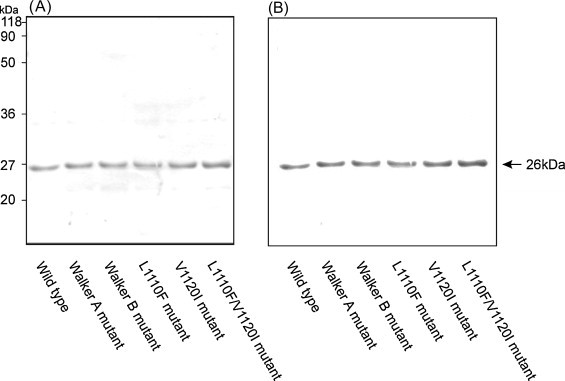
SDS-PAGE and Western blot analysis: (A) SDS-PAGE (10%) analysis of wild-type and mutant helicase proteins (Walker A motif, Walker B motif, L1110F Hel, V1120I Hel and L1110F/V1120I Hel), (B) Western blot analysis was carried out using anti-histidine monoclonal antibodies.
3.2. ATP hydrolysis activity of helicase mutants
Comparative analysis of ATPase activities of the mutant helicase proteins was done using equimolar amounts of purified proteins (300 pmoles/assay) as reported previously (Mhaindarkar et al., 2014). Reactions were carried out in parallel for all mutant proteins including wild type helicase protein as a positive control and Walker A motif (GKS to GAS) and Walker B motif (DE to AA) mutants as negative controls in a 96-well plate. The assay measured amount of released phosphate during ATP hydrolysis. Activity of the wild type protein (5081 ± 167.67 picomoles of released PO4) was taken as 100% and comparative activities of the mutants were calculated as described in the methods. L1110F Hel (78 ± 4.3%) and V1120I Hel (69 ± 7.4%) mutant proteins showed significant decrease in the ATPase activities as compared to wild type protein (p = 0.02 and p = 0.005 respectively), while the double mutant L1110F/V1120I Hel showed activity (98 ± 2.3%) comparable to the wild-type helicase protein (Table 2 ).
Table 2.
Comparative NTPase and unwinding activities of helicase mutants.
| Helicase protein | % NTPase activity | % unwinding activity |
|---|---|---|
| L1110F Hel | 80.3 ± 4.3 | 112.2 ± 10.2 |
| V1120I Hel | 70.4 ± 4.4* | 103.3 ± 9.7 |
| L1110F/V1120I Hel | 98.2 ± 2.3 | 110.6 ± 7.5 |
| Walker A | 25.1 ± 3.9* | 18.6 ± 7.3* |
| Walker B | 4.43 ± 6.9* | 0 ± 0* |
| WILD | 100 ± 3.3 | 100 ± 8.3 |
Comparative activities were calculated by assuming the activity of wild type helicase as 100%. ±: standard error of the mean.
p ≤ 0.05: significant difference as compared to wild type protein.
Similar results were obtained when these mutants were tested for their enzyme kinetics. L1110F Hel and V1120I Hel both exhibited higher K m and lower V max values as compared to wild type enzyme indicating lowered enzymatic efficiencies of both these single mutants. While the dual mutant L1110F/V1120I Hel exhibited K m and V max values comparable to wild type enzyme indicating normal level of catalytic efficiency (Table 3 ).
Table 3.
Enzyme kinetics (ATPase) of helicase mutants.
| Protein | Km (μM) | Vmax (nmol min−1 mg−1) |
|---|---|---|
| L1110F Hel | 43.75 ± 1.7 | 2.5 ± 1.9 |
| V1120I Hel | 52.43 ± 3.6 | 2.6 ± 2.1 |
| L1110F/V1120I Hel | 36.01 ± 3.9 | 3.1 ± 2.1 |
| Walker A | – | 1.1 ± 1.6 |
| Walker B | – | – |
| WILD | 33.62 ± 4.7 | 3.1 ± 1.2 |
Walker A and B mutant proteins showed variable Km values. ±: standard deviation of the mean.
Walker A motif and Walker B motif mutants showed negligible ATPase activities as expected (16 ± 3.9 and 0 respectively). Critical role of Walker A and B mutations in the NTPase activity of HEV helicase has been previously reported by us (Karpe and Lole, 2010a). We used these mutants as negative controls in the study to rule out carryover contamination of host cell NTPases during protein purification.
3.3. RNA duplex unwinding activities of helicase mutants
Knowing that the helicase unwinding activity requires energy generated from ATP hydrolysis, FHF helicase mutants were further assessed for RNA unwinding activity using non-radioactive quantitative assay described in the methods. This assay also included both positive and negative controls similar to ATPase assay. L1110F Hel and V1120I Hel mutants showed comparable duplex unwinding activities (106 ± 10.2 and 107 ± 9.7% respectively). While, the double mutant L1110F/V1120I Hel showed slight increase in the unwinding activity (116 ± 7.5%) (Table 2), though not significantly higher compared to wild type protein. Overall, all three FHF mutants showed similar unwinding activities comparable to wild type protein. Walker A mutant showed 18 ± 7.3% unwinding while, Walker B motif mutant showed complete loss of RNA duplex unwinding (Table 2). In our previous study we have shown that HEV helicase retains its ability to unwind RNA even with 40% ATPase activity as compared to wild type protein (Mhaindarkar et al., 2014). Hence, despite some loss in the ATPase activity, L1110F Hel and V1120I Hel both mutants displayed unwinding activities comparable to wild type protein.
3.4. Replication of HEV replicon mutants
We used HEV RLuc replicon to see the effect of helicase domain mutations on HEV replication. Construction of HEV RLuc subgenomic replicon by replacement of nt 5148–5816 in the ORF2 region of the genotype 1 infectious clone (pSK-HEV-2, Genbank Accession No. AF444002) with Renilla luciferase gene is reported by Cao et al., 2010. Complete genome sequence analysis of 7 FHF-E derived viral sequences with the pSK-HEV-2 showed overall 93–92% nucleotide identity; however, helicase domain amino acid sequences (245 aa) were completely identical in all the sequences except for the two above mentioned unique changes in FHF-E virus strains. Viral RNA dependent RNA polymerases have two conserved catalytic aspartate residues in their catalytic sites which are known to be essential for coordination of the catalytic Mg2+ ions during RNA polymerization. The first aspartate residue in GDD motif of HEV RdRp is essential for virus replication (Graff et al., 2005). We developed RdRp mutant (Mut GDD) from HEV Rluc replicon by site directed mutagenesis (GDD to GAA) and used as a negative control. Capped RNA transcripts were made from wild type and Mut GDD HEV Rluc replicons and transfected into S10-3 cells. Virus replication was evaluated in terms of cell associated Renilla luciferase activities, at different time points from 1 to 6 days post transfection. At 24 h, though the luciferase activity of the wild type HEV Rluc replicon was negligible (data not shown) it increased significantly 2 days onwards. The luciferase activity of Mut GDD remained negligible till 6 days without any increase from the base level values (Fig. 3A). These results confirmed replication competence of wild type HEV Rluc replicon and negligible (no) replication of the GDD mutant in S10-3 cells.
Fig. 3.
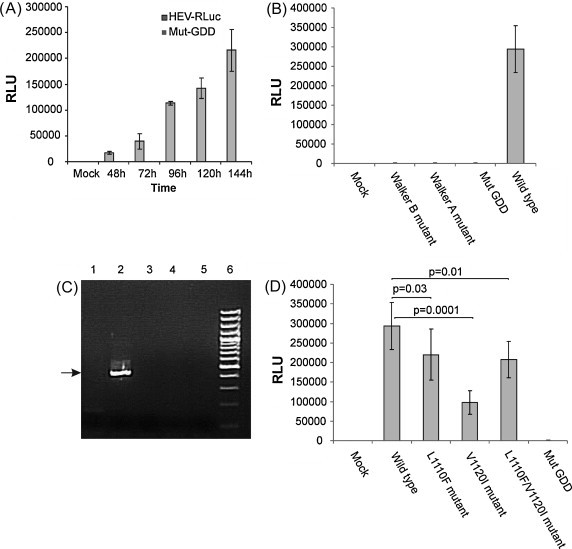
Replication of HEV Rluc replicon in S10-3 (Huh7) cells. (A) Replication kinetics: capped RNA transcripts of wild type and GDD mutant replicons were transfected in to the S10-3 cells, harvested at different time intervals as indicated and assayed for the dual luciferase activity. Renilla luciferase values were normalization with Firefly luciferase values of the respective time point. Mock sample represents cells without transfection. Bars indicate the SD of three independent transfection experiments. (B) Walker A motif and Walker B motif in the helicase domain are indispensable for HEV replication: capped RNA transcripts of wild type, Walker A mutant, Walker B mutant and GDD mutant (Mut GDD) replicons were transfected in S10-3 cells. Cells were assayed for luciferase activity to monitor the virus replication, 5 days post-transfection. Bars indicate the SD of three independent transfection experiments. (C) Negative strand RNA detection: Total RNA isolated from mock transfected, HEV Rluc, Mut GDD, Mut GKS and Mut DE RNA transfected S10-3 cells were processed for negative strand specific tag primer- based reverse transcription PCR at 48 h post-transfection. A representative 2% agarose gel of two independent experiments shows PCR product (415b p) and the lanes are – Mock cells (1), HEV Rluc (2), Mut GDD (3), Mut GKS (4), Mut DE (5) and 100 bp DNA ladder (6). (D) Effect of FHF-E associated helicase mutations on HEV replication: S10-3 cells were transfected with wild type, Mut L1110F, Mut V1120I and Mut L1110F/V1120I replicon RNA transcripts as indicated. Transfection of Mut GDD RNA served as a negative control. Cells were assayed for renilla luciferase activity at 5 days post-transfection as described in (A). The differences between signals produced by HEV Rluc mutants and the wild-type Rluc replicon were compared by one-way analysis of variance (ANOVA).
3.4.1. Effect of Walker A motif and Walker B motif mutations on HEV replication
Though we have previously shown that Walker A and Walker B helicase mutant proteins have negligible ATPase, RTPase and unwinding activities (Karpe and Lole, 2010a, Karpe and Lole, 2010b), it was not seen how such low level enzymatic activities affect the HEV replication. For that, we developed Walker A (Mut GKS) and Walker B (Mut DE) mutants from HEV Rluc replicon by site directed mutagenesis and checked their replication efficiencies in S10-3 cells. These experiments included wild type HEV replicon as the positive control and Mut GDD as the negative control. Replication efficiency was evaluated by measuring RLuc activity in the RNA transfected cells over the period of 5 days. Mutations in both Walker A and Walker B motifs affected RLuc synthesis and there was no RLuc activity (Fig. 3B).
RNA replication occurs through negative strand RNA intermediate, which then is used as the template for synthesis of positive sense subgenomic RNA and genomic RNA by RdRp during HEV replication. Five prime end of HEV genome encodes for the non-structural polyprotein while the capsid protein by the ORF2 region at 3′ end. As HEV-RLuc replicon was made by inserting RLuc gene into ORF2 encoding region, RLuc expression requires subgenomic RNA synthesis. Since both Walker A and Walker B mutants showed no RLuc activity there were two possibilities (1) helicase is essential during the transcription of subgenomic RNA, (2) helicase is required at the preceding step of virus replication, i.e. ns RNA (negative sense RNA) intermediate synthesis. If second possibility is true, then it will affect synthesis of both subgenomic RNA and new genomic RNA. To see whether Mut GKS and Mut DE were able to synthesize nsRNA intermediate, RNA transfected cells were harvested at 24 and 48 h post transfection and processed for strand specific RT-PCR. There was no nsRNA synthesis at both 24 and 48 h time points in both Mut GKS and Mut DE transfected cells (Fig. 3C, lanes 4 and 5) while cells transfected with wild type HEV-RLuc were positive (Fig. 3C, lane 2). As expected the Mut GDD also did not show any nsRNA due to lack of RdRp activity (Fig. 3C, lane 3). These results confirmed that helicase plays a major role during nsRNA synthesis during HEV replication.
3.4.2. Effect of L1110F and V1120I helicase domain mutations on HEV replication
To evaluate the role of FHF-E associated helicase domain mutations in HEV replication, three subgenomic replicon mutants were generated by site directed mutagenesis (Mut L1110F, Mut V1120I and Mut L1110F/V1120I). Capped RNA transcripts generated from the mutant clones were transfected into S10-3 cells and Renilla luciferase activity was measured at 5 days post-transfection. Mut V1120I replication was significantly lowered as compared to wild-type HEV Rluc replicon (p = 0.0001). Mut L1110F and Mut L1110F/V1120I replicons showed comparatively better replication as compared to Mut V1120I, however, it was lower as compared to wild type replicon (Fig. 3D). This showed that amino acid change L1110F was complementing V1120I change, since the negative effect of V1120I was at least partially rescued in the dual mutant (Fig. 3D). Overall, luciferase activity levels of all the three mutants remained significantly lower as compared to wild type HEV replicon indicating that both FHF-E mutations in the helicase domain of the virus have negative effect on the virus replication.
4. Discussion
HEV entry occurs primarily via oral route however, it is not yet known how the virus reaches liver. Main target of the virus is hepatocyte, where it replicates and gets released in to bile and gastrointestinal tract. There are no cytopathic changes in the infected cells and hence causes of liver pathology and severe clinical disease are still unknown. Hepatitis E is the major cause of FHF in adults in India (Arankalle et al., 1995, Madan et al., 1998). In addition to high mortality among pregnant women, the virus can also cause fulminant hepatitis in men and non-pregnant women (Arankalle et al., 1995).
Majority of positive sense RNA virus families encode their own RNA helicases with highly conserved sequences across different virus families. There are several studies suggesting possible involvement of viral helicases in activities other than replication. Mutations in viral helicases have been shown to alter virus replication and disease severity. A single amino acid substitution at position 249 (Thr to Pro) of the NS3 helicase region in West Nile virus (WNV) has been reported to confer a highly virulent phenotype on the viral strains otherwise weakly virulent in American crows (Brault et al., 2007). RNA helicase was suggested to be an attenuation or fitness determinant of WNV in different hosts (Ebel et al., 2011). Paired mutations in virus envelope protein and NS3 helicase domain of Dengue virus were shown to synergistically enhance RNA synthesis and viral load in human and mosquito derived cell lines (de Borba et al., 2012). Reversion of live attenuated porcine reproductive and respiratory syndrome virus vaccine strain to virulent virus by point mutations in papain-like cysteine protease and helicase regions has been reported (Nielsen et al., 2001). There are reports showing adaptive mutations in the helicase region of HCV replicons leading to better replication efficiencies in cells (Blight et al., 2000, Krieger et al., 2001, Grobler et al., 2003).
Multiple studies have attempted to identify hepatitis E virus associated virulence factors. Characterization of viral full genomes from FHF-E cases have shown occurrence of unique point mutations in the helicase domain of genotype 1, 3 and 4 FHF-E virus strains (Mishra et al., 2013, Inoue et al., 2006, Inoue et al., 2009, Takahashi et al., 2009). Majority of human HEV infections are caused by genotype-1 virus in developing countries. This genotype is predominantly transmitted by enteric route causing epidemics in contrast to sporadic cases caused by zoonotic transmissions of genotype-3 and genotype-4 viruses in developed countries. Associations of Th1 bias and higher HEV viral load have been documented in genotype-1 hepatitis E infections leading to FHF during pregnancy (Bose et al., 2011). The present study addresses effect of two point mutations (L1110F and V1120I) in the helicase domain of viral genomes from FHF-E cases from India, on the enzymatic activities of the helicase protein and virus replication. Sequence analysis of 121 HEV isolates (genotype 1–4) done by Mishra et al. (2013) showed that mutation L1110F was genotype-1 FHF specific (except for one type-4 sequence from Japan). Mutation V1120I was absent in genotype-2, predominant in genotype-3, and rare in genotype-4 HEV sequences. These dual mutations were present in 5/7 HEV genomes analyzed from FHF-E cases from India. In view of knowing critical role of helicases during positive sense RNA virus replication and recent report of Bose et al. (2011), showing higher viral loads in FHF cases as compared to self limiting hepatitis E cases we hypothesized that these highly conserved mutations in helicase domain could be responsible for enhancing helicase enzymatic activities, which plausibly would change the magnitude of virus replication. To check this we developed three helicase protein mutants, two with individual point mutations and the third one with dual mutations and tested their signature biochemical activities.
Comparable ATPase activities were shown by L1110F and V1120I single mutants however, they were significantly less than the corresponding wild type protein activity. The dual mutant L1110F/V1120I showed ATPase activity similar to wild type protein. This suggested that these amino acid changes are complementary to each other. Despite differences in ATPase activities of the mutants, all three mutant proteins showed normal duplex unwinding activities comparable to wild-type protein. These mutations in helicase domain did not alter enzymatic activities of these proteins possibly because mutations were present in the flanking region between the conserved functional motifs IV and V of the enzyme.
We further evaluated effect of these mutations on the HEV replication using subgenomic replicon with RLuc reporter gene in hepatoma cells. Mutant V1120I showed significantly lower replication as compared to L1110F replicon. However, this negative effect of Mut V1120I was partially rescued by complementary L1110F mutation. Replication of L1110F was comparable to the dual mutant. Overall, all three FHF mutant replicons showed lower replication levels as compared to wild type replicon. This suggested that mutations in helicase domain were not increasing virus replication efficiency and there could be alternative host/viral interactions resulting in to high viral loads in FHF-E pregnant women as noted by Bose et al. (2011). Reduced replication efficiencies of all three mutant replicons, despite having normal levels of ATPase and unwinding activities, possibly resulted due to altered interaction/s of the helicase protein with other either viral proteins or host cell proteins. It is known that host cell factors are required in most, if not all, steps of positive-strand RNA virus infections, including entry, viral gene expression, virion assembly, and release.
Walker A motif and Walker B motifs are the most common and highly conserved motifs in helicases and play crucial role during ATP hydrolysis. We have previously reported that mutations in the Walker A and Walker B motifs significantly reduce ATPase and RNA unwinding activities of HEV helicase (Karpe and Lole, 2010a). In the present study we confirmed indispensable role of HEV encoded helicase during virus replication. Replacement of critical lysine residue in the Walker A motif (GKS to GAS) and replacement of DE to AA in the Walker B motif in the HEV-Rluc replicon resulted in complete loss of viral RNA replication of both mutants as seen by negligible levels of RLuc activities. These mutants were also unable to synthesize replicative intermediate, which is a preceding step to subgenomic RNA transcription confirming that HEV encoded helicase is essential in the initial step of viral genome replication.
In conclusion, we report importance of HEV encoded helicase domain in virus replication as Walker A and Walker B motif mutations in the helicase domain completely abolished virus replication. FHF-E associated mutations in the helicase domain did not alter helicase protein associated enzymatic activities significantly. However, corresponding mutations in the HEV subgenomic replicon significantly reduced virus replication. There is possibility of involvement of these mutations in changing viral l/host-virus protein–protein interactions, causing alterations in host responses, eventually leading to more severe disease manifestations. In absence of small animal model for HEV studies, delineating role of viral helicase in disease severity would be a big challenge. However, it will be worthwhile to see early gene responses of hepatocytes on infection with these mutant viruses.
Acknowledgements
The work described here was financially supported by Department of Biotechnology (DBT), Ministry of Science and Technology ( BT/PR13535/Med/29/170/2010) and National Institute of Virology, India. We also thank Dr. S. Emerson (NIH, USA) and Dr. X.J. Meng (Virginia Tech, Blacksburg, USA) for providing S10-3 cell line and HEV-luciferase subgenomic replicon respectively.
References
- Ahmad I., Holla R.P., Jameel S. Molecular virology of hepatitis E virus. Virus Res. 2011;161:47–58. doi: 10.1016/j.virusres.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle V.A., Jha J., Favorov M.O., Chaudhari A., Fields H.A., Banerjee K. Contribution of HEV and HCV in causing fulminant non-A, non-B hepatitis in western India. J. Viral Hepat. 1995;2:189–193. doi: 10.1111/j.1365-2893.1995.tb00028.x. [DOI] [PubMed] [Google Scholar]
- Blight K.J., Kolykhalov A.A., Rice C.M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1975. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Bose P.D., Das B.C., Kumar A., Gondal R., Kumar D., Kar P. High viral load and deregulation of the progesterone receptor signaling pathway: association with hepatitis E-related poor pregnancy outcome. J. Hepatol. 2011;54:1107–1113. doi: 10.1016/j.jhep.2010.08.037. [DOI] [PubMed] [Google Scholar]
- Brault A.C., Huang C.Y., Langevin S.A., Kinney R.M., Bowen R.A., Ramey W.N., Panella N.A., Holmes E.C., Powers A.M., Miller B.R. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat. Genet. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Huang Y.W., Meng X.J. The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J. Virol. 2010;84:13040–13044. doi: 10.1128/JVI.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S.N., Devhare P.B., Lole K.S. Detection of negative sense RNA in packaged hepatitis E virions using improved strand specific RT-PCR method. J Clin. Microbiol. 2012;50:1467–1470. doi: 10.1128/JCM.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strottmann D.M., de Noronha L., Mason P.W., Duarte dos N., Santos C. Synergistic interactions between the NS3hel and E proteins contribute to the virulence of Dengue virus Type 1. PLoS Negl. Trop. Dis. 2012;6(4):e1624. doi: 10.1371/journal.pntd.0001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel G.D., Fitzpatrick K.A., Lim P.Y., Bennett C.J., Deardorff E.R., Jerzak G.V., Kramer L.D., Zhou Y., Shi P.Y., Bernard K.A. Nonconsensus West Nile virus genomes arising during mosquito infection suppress pathogenesis and modulate virus fitness in vivo. J. Virol. 2011;85:12605–12613. doi: 10.1128/JVI.05637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S.U., Nguyen H., Torian U., Purcell R.H. ORF3 Protein of Hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 2006;80:10457–10464. doi: 10.1128/JVI.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Nguyen H., Kasorndorkbua C., Halbur P.G., St Claire M., Purcell R.H., Emerson S.U. In vitro and in vivo mutational analysis of the 3′-terminal regions of hepatitis E virus genomes and replicons. J. Virol. 2005;79:1017–1026. doi: 10.1128/JVI.79.2.1017-1026.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler J.A., Markel E.J., Fay J.F., Graham D.J., Simcoe A.L., Ludmerer S.W., Murray E.M., Migliaccio G., Flores O.A. Identification of a key determinant of hepatitis C virus cell culture adaptation in domain II of NS3 helicase. J. Biol. Chem. 2003;278:16741–16746. doi: 10.1074/jbc.M212602200. [DOI] [PubMed] [Google Scholar]
- Holla R.P., Ahmad I., Ahmad Z., Jameel S. Molecular virology of hepatitis E virus. Semin. Liver Dis. 2013;33:3–14. doi: 10.1055/s-0033-1338110. [DOI] [PubMed] [Google Scholar]
- Inoue J., Nishizawa T., Takahashi M., Aikawa T., Mizuo H., Suzuki K., Shimosegawa T., Okamoto H. Analysis of the full-length genome of genotype 4 hepatitis E virus isolates from patients with fulminant or acute self-limited hepatitis E. J. Med. Virol. 2006;78:476–484. doi: 10.1002/jmv.20565. [DOI] [PubMed] [Google Scholar]
- Inoue J., Takahashi M., Mizuo H., Suzuki K., Aikawa T., Shimosegawa T., Okamoto H. Nucleotide substitutions of hepatitis E virus genomes associated with fulminant hepatitis and disease severity. Tohoku J. Exp. Med. 2009;218:279–284. doi: 10.1620/tjem.218.279. [DOI] [PubMed] [Google Scholar]
- Kadare G., Haenni A.L. Virus-encoded RNA helicases. J. Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y.A., Lole K.S. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 2010;84:3595–3602. doi: 10.1128/JVI.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y.A., Lole K.S. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J. Virol. 2010;84:9637–9641. doi: 10.1128/JVI.00492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuroo M.S., Teli M.R., Skidmore S., Sofi M.A., Khuroo M.I. Incidence and severity of viral hepatitis in pregnancy. Am. J. Med. 1981;70:252–255. doi: 10.1016/0002-9343(81)90758-0. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Gorbalenya A.E., Purdy M.A., Rozanov M.N., Reyes G.R., Bradley D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger N., Lohmann V., Bartenschlager R. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 2001;75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan K., Gopalkrishna V., Kar P., Sharma J.K., Das U.P., Das B.C. Detection of hepatitis C and E virus genomes in sera of patients with acute viral hepatitis and fulminant hepatitis by their simultaneous amplification in PCR. J. Gastroenterol. Hepatol. 1998;13:125–130. doi: 10.1111/j.1440-1746.1998.tb00626.x. [DOI] [PubMed] [Google Scholar]
- Mhaindarkar V., Sharma K., Lole K.S. Mutagenesis of hepatitis E virus helicase motifs: effects on enzyme activity. Virus Res. 2014;179:26–33. doi: 10.1016/j.virusres.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Mishra N., Arankalle V.A. Association of polymorphisms in the promoter regions of TNF-a (−308) with susceptibility to hepatitis E virus and TNF-a (−1031) and IFN-γ (+874) genes with clinical outcome of hepatitis E infection in India. J. Hepatol. 2011;55:1227–1234. doi: 10.1016/j.jhep.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Mishra N., Walimbe A.M., Arankalle V.A. Hepatitis E virus from India exhibits significant amino acid mutations in fulminant hepatic failure patients. Virus Genes. 2013;46:47–53. doi: 10.1007/s11262-012-0833-7. [DOI] [PubMed] [Google Scholar]
- Miyamura T. Hepatitis E virus infection in developed countries. Virus Res. 2011;161:40–46. doi: 10.1016/j.virusres.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Nielsen H.S., Oleksiewicz M.B., Forsberg R., Stadejek T., Bøtner A., Storgaard T. Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations. J. Gen. Virol. 2001;82:1263–1272. doi: 10.1099/0022-1317-82-6-1263. [DOI] [PubMed] [Google Scholar]
- Prabhu S.B., Gupta P., Durgapal H., Rath S., Gupta S.D., Acharya S.K., Panda S.K. Study of cellular immune response against Hepatitis E Virus (HEV) J. Viral Hepat. 2011;18:587–594. doi: 10.1111/j.1365-2893.2010.01338.x. [DOI] [PubMed] [Google Scholar]
- Saravanabalaji S., Tripathy A.S., Dhoot R.R., Chadha M.S., Kakrani A.L., Arankalle V.A. Viral load, antibody titers and recombinant open reading frame 2 protein-induced TH1/TH2 cytokines and cellular immune responses in self-limiting and fulminant hepatitis E. Intervirology. 2009;52:78–85. doi: 10.1159/000214862. [DOI] [PubMed] [Google Scholar]
- Suneetha P.V., Pischke S., Schlaphoff V., Grabowski J., Fytili P., Gronert A., Bremer B., Markova A., Jaroszewicz J. Hepatitis E virus (HEV)-specific T-cell responses are associated with control of HEV infection. Hepatology. 2012;55:695–708. doi: 10.1002/hep.24738. and other authors. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Okamoto H., Abe N., Kawakami M., Matsuda H., Mochida S., Sakugawa H., Suginoshita Y., Watanabe S., Yamamoto K. Virulent strain of hepatitis E virus genotype 3, Japan. Emerg. Infect. Dis. 2009;15:704–709. doi: 10.3201/eid1505.081100. and other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M., Ozdener M.H., Panda S.K., Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 1997;71:9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


