Abstract
WU and KI polyomaviruses represent novel viruses discovered in respiratory secretions from human patients with acute respiratory tract infection. However, the association between WU/KI polyomaviruses and human disease has remained unclear. In this study, the prevalence of these two novel viruses and occurrence of co-infection with other respiratory viruses were determined in Thai pediatric patients with respiratory disease. Previously described PCR assays were applied to detect WU/KI polyomaviruses as well as other respiratory viruses in 302 nasopharyngeal suction specimens collected from February 2006 through February 2007. The results revealed the anneal prevalence of WU and KI polyomaviruses in the Thai population was 6.29% and 1.99%, respectively. The frequency of co-detection of WU and KI polyomaviruses with other respiratory viral pathogens was 42.11% and 33.33%, respectively. Moreover, each of the two complete genome sequences of WU (CU_295 and CU_302) and KI (CU_255 and CU_258) polyomaviruses were genetically and phylogenetically characterized. Sequence analysis showed that they contained features common to those found in previous studies. However, there were several nucleotide variations within the non-coding regulatory regions and various non-synonymous mutations within the coding regions which may influence virulence and pathogenesis of these viruses. Nevertheless, it is still possible that these viruses are not the causative agents of clinical respiratory disease. Therefore, judging the association of WU/KI polyomavirus infections with a particular disease will be challenging and require more comprehensive case control investigations.
Keywords: Prevalence; Characterization; WU, KI polyomaviruses; Respiratory disease
1. Introduction
Globally, respiratory virus infections are responsible for significant morbidity and mortality, especially in children (Mulholland, 2003). The viruses associated with respiratory disease that are well recognized in human populations include influenza viruses, parainfluenza viruses, rhinoviruses, respiratory syncytial viruses, adenoviruses, human coronaviruses, human metapneumoviruses and human bocaviruses. Recently, KI polyomavirus (Allander et al., 2007) and WU polyomavirus (Gaynor et al., 2007) were identified in respiratory secretions from human patients with acute respiratory tract infection.
Polyomaviruses are small (40–50 nanometers in diameter) DNA viruses belonging to the Polyomaviridae family. They are non-enveloped icosahedral particles containing a circular double stranded DNA genome of approximately 5000 base pairs in length. They can infect a number of hosts of various species such as birds, rodents and primates. Four distinct polyomaviruses – JC (John Cunningham) virus, BK virus, KI (Karolinska Institute) virus and WU (Washington University) virus – have been reported to infect humans. Both JC virus and BK virus were discovered in 1971 (Gardner et al., 1971 and Padgett et al., 1971) and are genetically closely related to each other. They usually cause asymptomatic infections; yet, they can become oncogenic or induce disease in immunosuppressed or immunocompromised patients (Khalili et al., 2006, Hirsch et al., 2006, Fioriti et al., 2005).
Two novel, closely related polyomaviruses (KI and WU) were recently identified in respiratory secretions from children with acute respiratory tract infections (Allander et al., 2007 and Gaynor et al., 2007). Thus, KI and WU polyomaviruses were reported as novel viruses potentially causing respiratory disease in humans. However, both KI and WU polyomaviruses were frequently co-detected with other respiratory viruses. Moreover, a previous study reported that no evidence for an aetiological link between KI or WU polyomavirus infections and respiratory disease (Norja et al., 2007). Therefore, the clinical significance of KI and WU polyomaviruses requires further investigation. This study was aimed at establishing prevalence and clinical association of KI and WU polyomavirus infections in children with respiratory disease in Thailand. Moreover, KI and WU polyomaviruses isolated in Thailand were subjected to genetic and phylogenetic analysis.
2. Materials and methods
2.1. Clinical samples
At King Chulalongkorn Memorial Hospital, Thailand, nasopharyngeal suction specimens were collected from 302 hospitalized pediatric patients (age range 5 days to 14 years) with respiratory illness from February 2006 through February 2007. NP suction samples were collected in 1 ml of transport medium with antibiotics (Penicillin G 2 × 106 U/l and Streptomycin 200 mg/l) and stored at −70 °C until tested. The study was approved by the ethics committee of Chulalongkorn University and informed consent was obtained from the patient's parents.
2.2. Nucleic acid extraction and cDNA synthesis
DNA and RNA were extracted using TRI REAGENT® LS (Molecular Research Center, Inc., Cincinnati, OH) and the resulting pellets were dissolved in 20 μl of 8 mM NaOH solution or DEPC-treated water for DNA or RNA, respectively. Reverse transcription (RT) was performed at 37 °C for 2 h using the M-MLV reverse-transcription system (Promega, Madison, WI) consisting of 200 units of M-MLV reverse transcriptase, 5 μl of 5× M-MLV reaction buffer, 5 μl of 10 mM dNTP, 25 units of RNasin® ribonuclease inhibitor, 0.5 μg/μl of random hexamer, 0.5 μg of RNA and nuclease-free water to a final volume of 25 μl.
2.3. Detection of respiratory viruses by PCR amplification
PCR amplifications of KI and WU polyomaviruses were performed by using specific primer sets described in previous publications (Allander et al., 2007 and Gaynor et al., 2007). In addition, PCR amplifications of other respiratory viruses including human metapneumovirus, human bocavirus, influenza A virus, influenza B virus, parainfluenza virus, adenovirus and respiratory syncytial virus were performed by using specific primers described in previous reports (Samransamruajkit et al., 2006, Allander et al., 2005, Chieochansin et al., 2008, Chutinimitkul et al., 2008, Chi et al., 2005, Krafft et al., 2005, Samransamruajkit et al., 2002). Amplification of the human GAPDH gene served as an internal control of specimen quality and integrity. The limits of detection were approximately 103 copies/μl and 10 copies/μl for conventional PCR and nested PCR, respectively. All specific primers used in this study are summarized in Table 1 .
Table 1.
Primers used for PCR amplification of specific viruses
| Target | Oligo name | Sequence (5′ → 3′) | Position | Gene | Product size (bp) | References |
|---|---|---|---|---|---|---|
| WU polyomavirus | WU_925F | 5′-GCAGGATACAATCCCCAAGGA-3′ | 925–948 | VP2 | 656 | Modified from Gaynor et al. (2007) |
| WU_1580R | 5′-GCTGCATAATGGGGAGTACC-3′ | 1580–1561 | ||||
| WU_1331F | 5′-TGTTACAAATAGCTGCAGGTCAA-3′ | 1331-1353 | VP2 | 250 | Gaynor et al. (2007) | |
| WU_1580R | 5′-GCTGCATAATGGGGAGTACC-3′ | 1580–1561 | ||||
| KI polyomavirus | KI_1534F | 5′-ACAAGGCCAAGAAGTCAAGTTC-3′ | 1534–1555 | VP1 | 329 | Allander et al. (2007) |
| KI_1862R | 5′-TCACACTCACTAACTTGATTTGG-3′ | 1862–1840 | ||||
| KI_1613F | 5′-CAGTACCACTGTCAGAAGAAAC-3′ | 1613–1634 | VP1 | 208 | ||
| KI_1821R | 5′-TTCTGCCAGGCTGTAACATAC-3′ | 1821–1801 | ||||
| Human Metapneumovirus | MPVP_F | 5′-ACGGGGTAGAGAAGAGCTGG-3′ | 389–408 | N | 616 | Samransamruajkit et al. (2006) |
| MPVP_R | 5′-GCAAAGTTGGGACAGTTGGC-3′ | 1004–985 | ||||
| MPVN_F | 5′-GCATCAACCATAGAAGTGGGAC-3′ | 556–577 | N | 259 | ||
| MPVN_R | 5′-GCATTGTTTGACCGGCCCCA-3′ | 814–795 | ||||
| Human Bocavirus | HBoV_188F | 5′-GACCTCTGTAAGTACTATTAC-3′ | 2351–2371 | NP1 | 354 | Allander et al. (2005) |
| HBoV_542R | 5′-CTCTGTGTTGACTGAATACAG-3′ | 2704–2684 | ||||
| HBoV_VPF2 | 5′-TTCAGAATGGTCACCTCTACA-3′ | 3639–3659 | VP1 | 648 | Chieochansin et al. (2008) | |
| HBoV_VPR2 | 5′-CTGTGCTTCCGTTTTGTCTTA-3′ | 4286–4266 | ||||
| Influenza A virus | FluA_M_F | 5′-RGGCCCCCTCAAAGCCGA-3′ | 76–93 | M1 | 160 | Chutinimitkul et al. (2008) |
| FluA_M_R | 5′-ACTGGGCACGGTGAGYGT-3′ | 235–218 | ||||
| Influenza B virus | FluB_M_F | 5′-ATGTCGCTGTTTGGAGACACAAT-3′ | 25–47 | M1 | 295 | Chi et al. (2005) |
| FluB_M_R | 5′-TCAGCTAGAATCAGRCCYTTCTT-3′ | 320–301 | ||||
| Parainfluenza virus | Para_F | 5′-GCTAAATACTGTCTTMAHTGGAGAT-3′ | 11254–11278 | L | 139 | Chieochansin et al. (2008) |
| Para_R | 5′-GTAAGGATCACCWACATADAWTGTA-3′ | 11392–11370 | ||||
| Adenovirus | Adeno_F | 5′-GATGGCCACCCCATCGATGMTGC-3′ | 18227–18249 | L3 | 96 | Krafft et al. (2005) |
| Adeno_R | 5′-GCGAACTGCACCAGACCCGGAC-3′ | 18324–18303 | ||||
| Respiratory syncytial virus (type A and B) | RSV_AB-F | 5′-GTCTTACAGCCGTGATTAGG-3′ | 1673–1692 | N | 838 | Samransamruajkit et al. (2002) |
| RSV_AB_R | 5′-GGGCTTTCTTTGGTTACTTC-3′ | 2510–2491 | ||||
| Respiratory syncytial virus (type A) | RSV_A_F | 5′-GATGTTACGGTGGGGAGTCT-3′ | 1908–1927 | N | 343 | |
| RSV_A_R | 5′-GTACACTGTAGTTAATCACA-3′ | 2242–2223 | ||||
| Respiratory syncytial virus (type B) | RSV_B_F | 5′-AATGCTAAGATGGGGAGTTC-3′ | 1905–1923 | N | 183 | |
| RSV_B_R | 5′-GAAATTGAGTTAATGACAGC-3′ | 2098–2070 | ||||
| Internal control | GAPDH_F | 5′-GTGAAGGTCGGAGTCAACGG-3′ | 112–131 | Human GAPDH | 492 | Chutinimitkul et al. (2008) |
| GAPDH_R | 5′-GTTGTCATGGATGACCTTGGC-3′ | 603–583 | ||||
The conventional PCR reaction mixture comprised 2 μl of DNA or cDNA, 0.5 μM of each primer, 10 μl of 2.5× Eppendorf MasterMix (Eppendorf, Hamburg, Germany), and nuclease-free water to a final volume of 25 μl. The amplification reactions were performed in a thermal cycler (Eppendorf, Hamburg, Germany) under the following conditions: initial denaturation at 94 °C for 3 min, followed by 40 amplification cycles consisting of 94 °C for 30 s, 50–60 °C (depending on the specific primer pair) for 30 s, and 72 °C for 1 min, and concluded by a final extension at 72 °C for 7 min. After 2% agarose gel electropholysis, the expected amplified products (Table 1) were stained with ethidium bromide and then visualized under UV transillumination.
2.4. Amplification of complete genome sequences of WU/KI polyomaviruses
In order to study the genome characteristics of WU and KI polyomaviruses isolated in Thailand, whole genome sequencing was performed on two specimens of WU polyomaviruses (CU_295 and CU_302) and two specimens of KI polyomaviruses (CU_255 and CU_258). The complete genomes of WU/KI polyomaviruses were amplified directly from clinical samples using four primer pairs to generate overlapping fragments of the circular genome. All primers used for whole genome sequencing were designed from regions conserved among WU and KI polyomaviruses as summarized in Table 2 . The reaction mixture was prepared using the Eppendorf MasterMix (Eppendorf, Hamburg, Germany) as described above. The amplification reactions were performed in a thermal cycler (Eppendorf, Hamburg, Germany) under the following conditions: initial denaturation at 94 °C for 3 min, followed by 40 amplification cycles consisting of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min, and concluded by a final extension at 72 °C for 7 min. After 2% agarose gel electropholysis, the expected amplified products were purified and subsequently sequenced using 12 primers as summarized in Table 2.
Table 2.
Conserved primers for whole genome amplification and sequencing of WU/KI polyomaviruses
| Oligo name | Sequence (5′ → 3′) | Positiona | Product (bp) |
|---|---|---|---|
| WU/KI_4969F | TTGCAGTTTTAAATATAAAGAATTAAG | 4969–4995 | 1395 |
| WU/KI_761R | ACAGTAGYAGCTGCTTCAGATAT | 761–739 | |
| WU/KI_1134R | GGCAATTCTATTGAACACGTCCT | 1134–1112 | |
| WU/KI_989F | CCCCCTACYCAGGAATGGCA | 989–1008 | 1560 |
| WU/KI_1990R | ATTWGGWATATCAGGGGGAGCAA | 1990–1968 | |
| WU/KI_2548R | CCTTCTTTGTCTAAARTGTAGCCTA | 2548–2524 | |
| WU/KI_2407F | ACTRTTGGATGAAAATGGCATTGG | 2407–2430 | 1695 |
| WU/KI_2870F | GCAAAYTCAGTAAGGCCTATATA | 2870–2892 | |
| WU/KI_4101R | CATAGAGTTAGTGCTRTTAATAACTT | 4101–4076 | |
| WU/KI_3780F | TACTAYTGCATTTTTCACACTCTTC | 3780–3804 | 1389 |
| WU/KI_4703R | TGTGGATAGAGTGYTACTGCTA | 4703–4682 | |
| WU/KI_5168R | AAGAAGGCAAAATGGATAAAACTTT | 5168–5144 |
Position based on nucleotide sequence of WU polyomavirus (NC_009539).
2.5. Nucleotide sequencing
For identification of nucleotide sequences, the PCR amplified products were purified using the Perfectprep Gel Cleanup kit (Eppendorf, Hamburg, Germany) according to the manufacturer's specifications. The resulting purified DNAs served as templates for DNA sequencing in the ABI PRISM® 310 automated DNA sequencer with the Big Dye Terminator V.3.0 Cycle Sequencing Ready Reaction kit (ABI, Foster City, CA). Determination of the nucleotide sequences were performed in duplicate and analyzed by both direction using forward and reverse primers to ensure that variations of nucleotide sequences were not due to sequencing errors. When a difference was observed, triplicate sequences were determined in order to confirm the consistency of the sequencing result.
2.6. Sequencing analysis and phylogenetic tree construction
Nucleotide sequences were analyzed and assembled using the Lasergene 6 Package® (DNASTAR, Inc., Madison, WI, USA) and BLAST analysis tool (http://www.ncbi.nlm.gov/BLAST). Complete genome sequences were prepared and aligned using Clustal W implemented in the BioEdit program (version 7.0.4.1). Phylogenetic trees were constructed by neighbor-joining analysis with the Tamura-Nei model implemented in the MEGA3© program (Kumar et al., 2004). All nucleotide sequences of WU/KI polyomavirus obtained from this study were submitted to the GenBank database and assigned accession numbers EU358745–EU358769 (Table 3 ).
Table 3.
Clinical characteristics of WU/KI polyomavirus positive patients
| Patient No. | Sex | Age | Datea (d/m/y) | Hospitalized days | Result | Co-infected viruses | Clinical signs and symptoms | Accession number |
|---|---|---|---|---|---|---|---|---|
| CU_4 | M | 7 months | 16/02/06 | 15 | WU | Adenovirus | Severe bronchopneumonia | EU358745 |
| CU_5 | F | 3 years | 22/02/06 | 4 | WU | – | Pneumonia | EU358746 |
| CU_12 | M | 1 year | 08/03/06 | 2 | WU | – | Pneumonia | EU358747 |
| CU_32 | F | 3 years | 13/04/06 | 6 | WU | Flu A | Pneumonia | EU358748 |
| CU_38 | F | 1 year | 28/04/06 | 3 | WU | Adenovirus | Pneumonia | EU358749 |
| CU_50 | M | 1 year | 29/05/06 | 14 | WU | – | Pneumonia | EU358750 |
| CU_62 | M | 1 year | 16/06/06 | 3 | WU | – | Pneumonia | EU358751 |
| CU_76 | M | 2 years | 10/07/06 | 4 | WU | – | Pneumonia (congenital musculoskeletal deformities) | EU358752 |
| CU_84 | M | 3 months | 14/07/06 | 4 | KI | – | Pneumonia | EU358762 |
| CU_93 | M | 5 months | 31/07/06 | 4 | WU | RSV | Pneumonia | EU358753 |
| CU_104 | F | 9 years | 27/07/06 | 45 (dead) | KI | MPV | Pneumonia, Germinoma of brain | EU358763 |
| CU_141 | M | 1 year | 29/08/06 | 12 | WU | Adenovirus | Pneumonia (gastro-esophageal reflex) | EU358754 |
| CU_182 | F | 2 years | 04/10/06 | 4 | WU | – | Pneumonia, Growth retardation | EU358755 |
| CU_187 | F | 9 months | 04/10/06 | 5 | WU | Adenovirus | Croup | EU358756 |
| CU_199 | M | 3 years | 17/10/06 | 10 | WU | – | Pneumonia, Down's syndrome | EU358757 |
| CU_205 | M | 1 year | 17/10/06 | 2 | WU | Bocavirus | Acute brochiolitis | EU358758 |
| CU_214 | M | 2 years | 27/10/06 | 6 | WU | – | Asthmatric bronchitis | EU358759 |
| CU_255 | M | 1 year | 17/12/06 | 2 | KI | – | Acute bronchiolitis | EU358766 |
| CU_257 | F | 1 year | 22/12/06 | 14 | KI | – | Pneumonia | EU358764 |
| CU_258 | M | 1 year | 15/12/06 | 79 | KI | – | Pneumonia (cirrhosis) | EU358767 |
| CU_262 | M | 3 years | 03/01/07 | 3 | WU | Flu A | Pneumonia | EU358760 |
| CU_265 | M | 1 year | 10/01/07 | 5 | KI | Bocavirus | Neuroblastoma pneumonia | EU358765 |
| CU_276 | M | 1 year | 18/01/07 | 8 | WU | – | Bronchopneumonia | EU358761 |
| CU_295 | F | 1 year | 17/02/07 | 3 | WU | – | Acute bronchiolitis | EU358768 |
| CU_302 | M | 1 year | 28/02/07 | 5 | WU | – | Pneumonia | EU358769 |
Date of specimen collection.
2.7. Statistical analysis
Statistical data were analyzed using SPSS for Windows version 11.5 software package. Statistical analyses were performed using Pearson χ 2 for determination of seasonal distribution of WU and KI polyomaviruses. The results were considered statistically significant at P < 0.05.
3. Results
3.1. Prevalence of WU polyomavirus in Thailand
PCR amplification and subsequent nucleotide sequencing demonstrated that 19 of the 302 nasopharyngeal suction specimens were positive for WU polyomavirus, amounting to a prevalence of 6.29% in this sample population. Single infection with WU polyomavirus was detected in 11 of the 19 positive cases (57.89%), whereas 8 cases (42.11%) showed evidence of co-infection with other respiratory viruses. Four cases were co-infected with adenovirus, two with influenza A virus, one with respiratory syncytial virus and one with human bocavirus (Table 3, Table 4 ).
Table 4.
Co-infection of WU/KI polyomavirus with other respiratory viruses
| Positive cases (% of positive) |
||
|---|---|---|
| WU polyomavirus | KI polyomavirus | |
| Single infection | 11 (57.81%) | 4 (66.67%) |
| Co-infection with | ||
| Adenovirus | 4 (21.05%) | 0 |
| Influenza A virus | 2 (10.53%) | 0 |
| Influenza B virus | 0 | 0 |
| Parainfluenza virus | 0 | 0 |
| Human Metapneumovirus | 0 | 1 (16.67%) |
| Human Bocavirus | 1 (5.26%) | 1 (16.67%) |
| Respiratory syncytial virus | 1 (5.26%) | 0 |
| Total | 19 | 6 |
3.2. Prevalence of KI polyomavirus in Thailand
KI polyomavirus was detected in 6 of the 302 respiratory specimens, indicating a prevalence of 1.99% in the Thai population. Single infection with KI polyomavirus was detected in four of six positive specimens (66.67%), whereas the remaining two cases (33.33%) were co-infected with human metapneumovirus and human bocavirus, respectively (Table 3, Table 4).
3.3. Clinical presentation in WU/KI polyomavirus positive patients
Patient positive for WU or KI polyomavirus suffered from a wide range of respiratory symptoms including bronchopneumonia, croup, bronchiolitis, bronchitis and pneumonia (Table 3). Most of the patients (11 of 15) only infected with WU or KI polyomaviruses showed clinical signs associated with pneumonia. However, associations between the presence of WU/KI polyomaviruses and respiratory diseases remain highly speculative due to the lack of comprehensive case control studies. Although WU/KI polyomavirus positive specimens were detected in subjects ranging in age from 3 months to 9 years, the majority were found in approximately 1-year-old babies (Table 3). There was no significant seasonal variation for WU polyomavirus infection (P = 0.11), but a predominance of infection was detected during the winter months (November–January in Thailand) for KI polyomavirus (P = 0.02), respectively (Fig. 1 ).
Fig. 1.
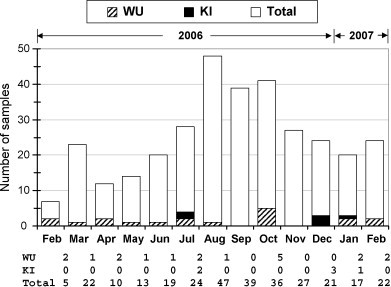
Seasonal variation and WU/KI polyomavirus infection. Bar graph shows number of WU or KI positive samples as well as total number of clinical specimens collected from 302 pediatric patients with respiratory disease in each month from February 2006 through February 2007.
3.4. Complete genome analysis of WU polyomavirus in Thailand
Whole genome sequences of WU polyomaviruses found in Thai patients (isolates CU_295 [EU358768] and CU_302 [EU358769]) were aligned with Australian isolates [EF444549–EF444554] (Gaynor et al., 2007). Analysis of WU polyomavirus nucleotide sequences showed 99.6–99.9% similarity among the Thai and Australian isolates. Phylogenetic analysis revealed that WU polyomaviruses isolated in Thailand (CU_295 and CU_302) were closely related to the reference strain of WU polyomavirus [NC_009539 or EF444549] (Fig. 2A). Additionally, there were numerous variations of the nucleotide sequences within non-coding regions including nucleotide positions 4, 32, 286, 2797, 4522 and 4559 (Table 5 ). Furthermore, analysis of deduced amino acids within the coding sequences revealed that several variations had occurred within coding sequences including the VP2 protein (amino acid positions 239, 250, 284 and 356), VP1 protein (amino acid position 79), and Large T Antigen (amino acid positions 360, 378, 440, 520 and 594) as shown in Table 5.
Fig. 2.
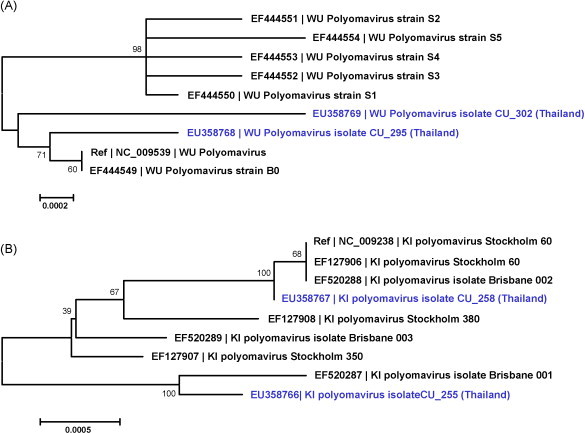
Phylogenetic tree constructed by neighbor-joining analysis with the Tamura-Nei model implemented in the MEGA3© program (version 3.1). Nodal confidence values indicate the results of bootstrap re-sampling (n = 1000). (A) Phylogenetic tree of whole genome sequences of WU polyomaviruses. (B) Phylogenetic tree of whole genome sequences of KI polyomaviruses.
Table 5.
Variations of nucleotide sequences within non-coding regions and amino acid sequences within coding regions of WU polyomaviruses
| Accession no./isolate name | Variations of nucleotide sequences in non-coding regions |
Variation of deduced amino acid in coding sequences |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 32 | 286 | 2797 | 4522 | 4559 | VP2 |
VP1 | LTAg |
||||||||
| 239 | 250 | 284 | 256 | 79 | 360 | 378 | 440 | 520 | 594 | |||||||
| NC_009539/WU Ref strain | t | g | c | a | a | t | E | E | D | T | T | D | D | V | G | I |
| EF444549/WU strain B0 | t | g | c | a | a | t | E | E | D | T | T | D | D | V | G | I |
| EF444550/WU strain S1 | t | g | c | a | g | t | E | Q | D | T | T | D | D | V | G | L |
| EF444551/WU strain S2 | t | g | c | a | g | t | E | Q | D | T | T | D | D | V | E | L |
| EF444552/WU strain S3 | t | g | c | t | g | t | E | Q | D | T | T | G | D | V | G | L |
| EF444553/WU strain S4 | t | c | c | a | g | t | E | Q | D | T | T | D | D | V | G | L |
| EF444554/WU strain S5 | c | g | c | a | g | t | E | Q | H | T | T | D | G | I | G | L |
| EU358768/WU Thailand CU_295 | t | g | c | a | g | t | E | E | D | T | T | D | D | V | G | I |
| EU358769/WU Thailand CU_302 | t | g | g | a | g | a | Q | E | D | I | G | D | D | V | G | I |
3.5. Complete genome analysis of KI polyomavirus in Thailand
The complete genome sequences of KI polyomaviruses isolated from Thai patients (isolates CU_255 [EU358766] and CU_258 [EU358767]) were aligned with Swedish sequences [EF127906–EF127908] (Allander et al., 2007) and Queensland sequences [EF520287–EF520289] (Bialasiewicz et al., 2007). Nucleotide sequence analysis of KI polyomaviruses revealed 99.4–99.9% identity among the Thai, Queensland and Swedish isolates. Phylogenetic analysis showed that one of the KI polyomavirus isolated from Thailand (CU_258) was closely related to the reference strain of KI polyomavirus [NC_009238 or EF127906] whereas the second one (CU_255) was more closely associated with KI polyomavirus isolated from Queensland (isolate Brisbane 001 [EF520287]) (Fig. 2B). The latter similarity was largely due to a 10 nucleotide insertion (AGGCGCTGCG) at positions 60–69 within the non-coding region which the Thai isolate CU_255 and Queensland isolate Brisbane 001 shared (Table 6 ). Moreover, there were several variations of the nucleotide sequences within non-coding regions as for example, at nucleotide positions 76, 107, 110, 233 and 4656 (Table 6). Analysis of deduced amino acids within the coding sequences showed a variety of variations within coding sequences for the VP2 protein (amino acid positions 246 and 316), the VP1 protein (amino acid positions 181, 267 and 368), the Large T Antigen (amino acid positions 124, 199, 337, 365 and 494) and the small T Antigen (STAg) (amino acid position 104) as shown in Table 6.
Table 6.
Variations of nucleotide sequences within non-coding regions and amino acid sequences within coding regions of KI polyomaviruses
| Accession no./isolate name | Variations of nucleotide sequences in non-coding regions |
Variation of deduced amino acid in coding sequences |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 60 | 76 | 107 | 110 | 233 | 4656 | VP2 |
VP1 |
LTAg |
STAg | ||||||||
| 246 | 316 | 181 | 267 | 368 | 124 | 199 | 337 | 365 | 494 | 104 | |||||||
| NC_009238/KI Ref strain | – | t | t | t | c | a | T | P | K | Q | A | E | Y | V | L | Q | D |
| EF127906/KI Stockholm 60 | – | t | t | t | c | a | T | P | K | Q | A | E | Y | V | L | Q | D |
| EF127907/KI Stockholm 350 | – | c | c | g | a | t | S | P | K | Q | T | E | Y | V | V | Q | E |
| EF127908/KI Stockholm 380 | – | a | c | g | a | a | T | P | K | Q | A | E | Y | V | V | H | D |
| EF520287/KI isolate Brisbane 001 | aggcgctgcg | t | c | a | a | t | S | S | R | Q | A | E | Y | L | V | Q | E |
| EF520288/KI isolate Brisbane 002 | – | t | t | t | c | a | T | P | K | Q | A | E | Y | V | L | Q | D |
| EF520289/KI isolate Brisbane 003 | – | t | a | g | a | t | S | P | K | H | A | E | Y | V | V | Q | E |
| EU358766/KI Thailand CU_255 | aggcgctgcg | t | c | a | a | t | S | S | R | Q | A | K | H | L | V | Q | E |
| EU358767/KI Thailand CU_258 | – | t | t | t | c | a | T | P | K | Q | A | E | Y | V | L | Q | D |
3.6. Characterization of the non-coding regulatory region
Alignment of the non-coding regulatory regions of WU and KI polyomaviruses revealed AT-rich sequences on the downstream of the putative replication origin. Both Thai WU polyomavirus isolates (CU_295 and CU_302) contained an A/T rich region (TTTTTTTTATTATAATAA) common to the Australian isolates [EF444549–EF444554] whereas both Thai KI polyomavirus isolates (CU_255 and CU_258) contained an A/T rich region (TTTTTTTCTATATTACA) similar to the Swedish sequences [EF127906–EF127908] and the Queensland sequences [EF520287–EF520289]. The origin of replication of WU/KI polyomaviruses contains three potential pentanucleotide Large T Antigen (LTAg) binding sites, as opposed to the four found in most polyomaviruses. Two of the LTAg binding sites have the typical sequence “GAGGC” found in all WU/KI polymaviruses analyzed, except for the Australian WU polyomavirus strain S5 [EF444554] that contains “GGGGC” at the second LTAg binding site. The third consensus LTAg binding site found in all isolates of WU polyomaviruses has the sequence “GCCTC” whereas all isolates of KI polyomaviruses display the sequence “GCCCC”.
3.7. Characterization of the Large T Antigen
Analysis of the deduced amino acid sequences of the Large T Antigen revealed domains common to both WU and KI polyomaviruses, including the DnaJ domain, Rb binding domain, zinc finger motif and ATPase-p53 binding domain. All WU/KI polyomaviruses analyzed contained the DnaJ domain at the N terminus with the highly conserved hexapeptide motif “HPDKGG” (amino acid positions 42–47) and the Rb binding domain “LRCNE” at amino acid positions 108–113. The zinc finger motif was found in WU polyomaviruses at amino acid residues C321, C324, H336 and H339 whereas KI polyomaviruses displayed it at amino acid residues C312, C315, H327 and H331 of the LTAg protein. Conserved motifs “GPINSGKT” (amino acid positions 443–450) and “GSVKVNLE” (amino acid positions 520–527) were detected in all WU polyomaviruses analyzed whereas the motifs “GPINSGKT” (amino acid positions 436–443) and “GCVEVNLE” (amino acid positions 513–520) were found in all KI polyomaviruses analyzed in this study (Pipas, 1992).
4. Discussion
Resulting from this research, we present here preliminary data on the role of WU/KI polyomaviruses as potential respiratory pathogens in Thai pediatric patients. The prevalence of WU polyomavirus in Thailand (6.29%) was higher than that reported in Australia (4.5%) (Bialasiewicz et al., 2008), the United States (1.2%) (Gaynor et al., 2007) and United Kingdom (1%) (Norja et al., 2007) but comparable to the prevalence reported in a previous study from South Korea (7%) (Han et al., 2007). This may be due to particular racial predispositions or different geographical distributions of the virus. In contrast, the incidence of KI polyomavirus infection in Thailand (1.99%) was comparable to that established in Sweden (1%) (Allander et al., 2007), Australia (2.6%) (Bialasiewicz et al., 2008) and the United Kingdom (1.4%) (Norja et al., 2007).
WU polyomavirus infection does not appear subjected to significant seasonal variation; yet, a predominance of infection was detected during the winter months for KI polyomavirus. These findings were not in perfect agreement with previous studies conducted in Australia (Bialasiewicz et al., 2007, Bialasiewicz et al., 2008) which reported no apparent seasonal variation for KI polyomavirus, but a predominance of infection with WU polyomavirus during late winter to early summer. This discrepancy may be due to seasonal differences in each country.
The genome of WU (5229 bp in length) and KI (5040 bp in length) polyomaviruses comprises an early coding region on one strand for the Small T Antigen and the Large T Antigen, and a late coding region on the opposite strand for the 3 capsid proteins VP1, VP2, and VP3. These two regions are separated by a regulatory non-coding region that contains typical polyomavirus characteristics (Allander et al., 2007 and Gaynor et al., 2007). Based on complete genome analysis, the WU/KI polyomaviruses isolated from Thai patients were closely related to the isolates described in previous reports (Gaynor et al., 2007, Allander et al., 2007 and Bialasiewicz et al., 2007). This may be due to the fact that both WU and KI polyomaviruses are DNA viruses which are less error prone in the DNA replication process. However, there were numerous nucleotide variations within the non-coding regulatory regions as well as several non-synonymous mutations within coding regions.
No other respiratory virus was detected in 57.81% of the WU-positive specimens and 66.67% of the KI-positive specimens in our study, hinting at an association between WU/KI polyomaviruses and respiratory disease. However, these viruses may not be the causative agents of clinical respiratory disease and their presence in the respiratory tract may simply reflect their mode of transmission similar to other polyomaviruses in humans such as JCV and BKV. Nevertheless, most known human viruses are pathogenic in certain conditions and thus, novel viruses must be considered as potential pathogens. Various degrees of clinical symptoms ranging from asymptomatic to mild and severe illness depend on several factors, such as the host's immune status and age, the virus strain, viral load and mode of transmission. The 10-nucleotide insertion and several nucleotide sequence variations within non-coding regulatory regions as well as various non-synonymous mutations within the coding regions should be taken into consideration as factors potentially involved in the pathogenesis of these novel viruses. The difficulty with persisting viruses is that they are often discovered out their symptomatic context, so that judging the association of WU/KI polyomavirus infections with a particular disease will be challenging and require further extensive case control investigations.
Acknowledgements
This study was kindly supported by funding from Chulalongkorn University (Ratchadapiseksompotch Fund) and Commission on Higher Education, ministry of Education. We would like to express our gratitude to the entire staffs from the Center of Excellence in Clinical Virology, Faculty of Medicine, Chulalongkorn University for generous supports. We also would like to thank Ms. Petra Hirsch for reviewing the manuscript and Mr. Wasan Punyasang for statistical analysis.
References
- Allander T., Andreasson K., Gupta S., Bjerkner A., Gordana B., Perrson M.A., Dalianis T., Ramqvist T., Andersson B. Identification of a third human polyomavirus. J. Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T., Tammi M.T., Eriksson M., Bjekner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1289–1296. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialasiewicz S., Whiley D.M., Lambert S.B., Jacob K., Bletchly C., Wang D., Nissen M.D., Sloots T.P. Presence of the newly discovered human polyomaviruses KI and WU in Australian patients with acute respiratory tract infection. J. Clin. Virol. 2008;41:63–68. doi: 10.1016/j.jcv.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialasiewicz S., Whiley D.M., Lambert S.B., Wang D., Nissen M.D., Sloots T.P. A newly reported human polyomavirus, KI virus, is present in the respiratory tract of Australian children. J. Clin. Virol. 2007;40:15–18. doi: 10.1016/j.jcv.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X.S., Hu A., Bolar T.V., Al-Rimawi W., Zhao P., Tam J.S., Rappaport R., Cheng S.M. Detection and characterization of new influenza B virus variants in 2002. J. Clin. Microbiol. 2005;43:2345–2349. doi: 10.1128/JCM.43.5.2345-2349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieochansin T., Samransamruajkit R., Chutinimitkul S., Payungporn S., Hiranras T., Theamboonlers A., Poovorawan Y. Human Bocavirus (HBoV) in Thailand; clinical manifestations in a hospitalized pediatric patient and molecular virus characterization. J. Infect. 2008;56:137–142. doi: 10.1016/j.jinf.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutinimitkul S., Chieochansin T., Payungporn S., Samransamruajkit R., Hiranras T., Theamboonlers A., Poovorawan Y. Molecular characterization and phylogenetic analysis of H1N1 and H3N2 human influenza A viruses among infants and children in Thailand. Virus Res. 2008;132:122–131. doi: 10.1016/j.virusres.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Fioriti D., Degener A.M., Mischitelli M., Videtta M., Arancio A., Sica S., Sora F., Pietropaolo V. BKV infection and hemorrhagic cystitis after allogenic bone marrow transplant. Int. J. Immunopathol. Pharmacol. 2005;18:309–316. doi: 10.1177/039463200501800213. [DOI] [PubMed] [Google Scholar]
- Gardner S.D., Field A.M., Coleman D.V., Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- Gaynor A.M., Nissen M.D., Whilley D.M., MacKay I.M., Lambert S.B., Wu G., Brennan D.C., Storch G.A., Sloots T.P., Wang D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:595–604. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.H., Chung J.Y., Koo J.W., Kim S.W., Hwang E.S. WU polyomavirus in children with acute lower respiratory tract infections, South Korea. Emerg. Infect. Dis. 2007;13:1766–1768. doi: 10.3201/eid1311.070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch H.H., Drachenberg C.B., Steiger J., Ramos E. Polyomavirus associated nephropathy in renal transplantation: critical issues of screening and management. Adv. Exp. Med. Biol. 2006;577:160–173. doi: 10.1007/0-387-32957-9_11. [DOI] [PubMed] [Google Scholar]
- Khalili K., Gordon J., White M.K. The polyoma virus, JCV and its involvement in human disease. Adv. Exp. Med. Biol. 2006;577:274–287. doi: 10.1007/0-387-32957-9_20. [DOI] [PubMed] [Google Scholar]
- Krafft A.E., Russell K.L., Hawksworth A.W., McCall S., Irvine M., Daum L.T., Connoly J.L., Reid A.H., Gaydos J.C., Taubenberger J.K. Evaluation of PCR testing of ethanol-fixed nasal swab specimens as an augmented surveillance strategy for influenza virus and adenovirus identification. J. Clin. Microbiol. 2005;43:1768–1775. doi: 10.1128/JCM.43.4.1768-1775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Mulholland K. Global burden of acute respiratory infections in children: implications for interventions. Pediatr. Pulmonol. 2003;36:469–474. doi: 10.1002/ppul.10344. [DOI] [PubMed] [Google Scholar]
- Norja P., Ubillos I., Templeton K., Simmonds P. No evidence for an association between infections with WU and KI polyomaviruses and respiratory disease. J. Clin. Virol. 2007;40:307–311. doi: 10.1016/j.jcv.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett B.L., Walker D.L., ZuRhein G.M., Eckroade R.J., Dessel B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- Pipas J.M. Common and unique features of T antigens encoded by the polyomavirus group. J. Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samransamruajkit R., Moonviriyakit K., Vanapongtipagorn P., Prapphal N., Deerojanawong J., Poovorawan Y. Plasma endothelin-1 in infants and young children with acute bronchiolitis and viral pneumonia. Asian Pac. J. Allergy Immunol. 2002;20:229–234. [PubMed] [Google Scholar]
- Samransamruajkit R., Thanasugarn W., Prapphal N., Theamboonlers A., Poovorawan Y. Human metapneumovirus in infants and young children in Thailand with lower respiratory tract infections; molecular characteristics and clinical presentations. J. Infect. 2006;52:254–263. doi: 10.1016/j.jinf.2005.07.001. [DOI] [PubMed] [Google Scholar]


