Highlights
-
•
PI3V strains can be typed using molecular tests into types A, B, and C.
-
•
Commercial MLV vaccines with PI3V strains are type A.
-
•
PI3V type C is antigenic different from type A.
-
•
PI3V strains from cattle with BRD signs and recently vaccinated with MLV type A strains may have type B or C strains.
-
•
Cattle with BRD signs may have type A, B, or C strains in tissues or swabs.
Keywords: Parainfluenza-3 viruses, MLV vaccines, Genomics, Antigenic variation
Abstract
This study investigated the genetic and antigenic characterization of parainfluenza-3 virus (PI3V) of cattle. Using molecular tests including real time PCR and viral genome sequencing, PI3V strains could be separated into PI3V types, including PI3V A, PI3V B, and PI3V C. Isolates from cattle with bovine respiratory disease clinical signs and commercial vaccines in the U.S. with MLV PI3V were typed using these molecular tests. All the MLV vaccine strains tested were PI3V A. In most cases PI3V field strains from calves receiving MLV vaccines were types heterologous to the vaccine type A. Also antigenic differences were noted as PI3V C strains had lower antibody levels than PI3V A in serums from cattle receiving MLV PI3V A vaccines. This study further demonstrates there is genetic variability of U.S. PI3V strains and also antigenic variability. In addition, isolates from cattle with BRD signs and receiving MLV vaccines may have heterologous types to the vaccines, and molecular tests should be performed to differentiate field from vaccine strains. Potentially the efficacy of current PI3V A vaccines should be evaluated with other types such a PI3V B and PI3V C.
1. Introduction
Bovine respiratory diseases (BRD) have etiologies of viruses, bacteria, and Mycoplasma spp. (Fulton,2008, 2009) Viruses included in BRD, either singly or in combination include bovine herpesvirus-1 (BoHV-1), parainfluenza −3 virus (PI3V), bovine viral diarrhea viruses (BVDV), bovine respiratory syncytial virus (BRSV), and bovine coronaviruses (BoCV)(Fulton, 2008, Fulton, 2009, Fulton et al., 2011, Fulton et al., 2013, Fulton et al., 2016). The bovine PI3V is a RNA virus in the genus Respirovirus in the family Paramyxoviridae (Chanock et al., 2001). In North America both killed/inactivated and modified live virus (MLV) vaccines are used for control of these viruses (Compendium of Veterinary Products, 2010). Clinicians seeking information on potential causative agents/etiologies often submit samples from cattle such as nasal swabs or blood samples from affected cattle and tissues from necropsy samples to the diagnostic laboratories. Samples may be tested by “traditional tests” including virus isolation for infectious virus or more often “modern tests”, molecular tests including polymerase chain reaction (PCR) and in some cases, sequencing of the agent’s genome (Fulton et al., 2016, Fulton and Confer, 2012). Interpretation of such results is important for clinicians and the diagnostic laboratory. Viruses may appear to be similar based on the test used such as infectious viruses from cultures or these from the molecular tests that are homologous based on the product produced using primers. Also there is the dilemma of whether the virus identified is a field virus or a vaccine strain. A recent study utilized samples from cattle with BRD signs after processing with MLV vaccines administered (Fulton et al., 2016). In that study there were isolates of BoHV-1, BVDV, and PI3V that utilized sequencing of the viral genome and found either field strains or vaccine strains. Thus it is important to be able to differentiate viral isolates of field or vaccine origin. Several PI3V strains in the U.S. have been examined using viral sequencing with three types identified, PI3VA, PI3VB, and PI3V C. (Neill et al., 2015)
The study purposes were three-fold: (1) characterize PI3V strains in available commercial vaccines by molecular tests, (2) compare PI3V strains from field cases of BRD by molecular tests and to determine if the strain was related to MLV strains, and (3) using serums from calves given MLV vaccines containing PI3VA strains to determine antigenic variation when serums were tested in viral neutralization test with PI3VA and two PI3V C strains.
2. Materials and methods
2.1. Cattle groups
Viruses from cattle in this study included 22 viruses from 4 different studies (Table 1 ) dated from 1999 until 2014. Study 1 were cattle from the Southeast (SE) region of the U.S. shipped to an experimental feed yard in New Mexico. (Fulton et al., 2001) These cattle had not received PI3V vaccination prior to shipment. Study 2 included cattle from the SE U.S shipped to the Willard Sparks Experimental Feedyard at Oklahoma State University in 2009. Samples included both nasal swabs from cattle with BRD signs and samples collected from lungs at necropsy. These cattle had received a MLV vaccine containing PI3V at entry to the feed yard. Study 3 samples were from an Oklahoma feed yard from cattle with BRD signs and they had received a MLV vaccine at entry to the feed yard. The Study 4 included isolates from cattle using nasal swabs from cattle with BRD signs (Fulton et al., 2016). For the Studies 2–4 the vaccine used in each particular study is listed in Table 1.
Table 1.
PI3V Isolates from BRD clinical cases.
| Study | Year | Identifcation | Collection site | Days after processing | MLV vaccine at procesing | Type by PCR |
|---|---|---|---|---|---|---|
| Study 1 | 1999 | OK P 100 | NSa | 28 | No PI3V MLV | PI3VA |
| OK P 141 | NS | 28 | No PI3V MLV | PI3VB | ||
| OK P 147 | NS | 28 | No PI3V MLV | PI3VA | ||
| OK P 242 | NS | 33 | No PI3V MLV | PI3VA | ||
| Study 2 | 2009 | OK 261 SP | NS | 11 | Vax 1 | PI3VC |
| OK 284 SP RLU | Right lung | 30 | Vax 1 | PI3VC | ||
| OK 309 SP LLU | Left lung | 18 | Vax 1 | PI3VC | ||
| OK 453 SP RLU | Right lung | 24 | Vax 1 | PI3VC | ||
| OK 467 SP RLU | Right lung | 17 | Vax 1 | PI3VC | ||
| OK 643 SP LLU | Left lung | 35 | Vax 1 | PI3VC | ||
| OK 643 SP RLU | Right lung | 35 | Vax 1 | PI3VC | ||
| Study 3 | 2012 | OK PR 2 NS | NS | 2 | Vax 2 | PI3VB |
| OK PR 7 NS | NS | 2 | Vax 2 | PI3VC | ||
| Study 4 | 2014 | OK P 5797 | NS | 12 | Vax 3 | PI3VC |
| OK P 778 | NS | 20 | Vax 4 | PI3VC | ||
| OK P 825 | NS | 20 | Vax 3 | PI3VC | ||
| OK P 438 | NS | 19 | Vax 3 | PI3VC | ||
| OK P 831 | NS | 20 | Vax 3 | PI3VC | ||
| OK P 9054 | NS | NA | Vax 3 | PI3VC | ||
| OK P NM 64 | NS | 18 | Vax 5 | PI3VA | ||
| OK 95806 | NS | 13 | Vax 6 | PI3VB | ||
| OK 95524 | NS | 15 | Vax 6 | PI3VB | ||
NS-nasal swab.
2.2. Viruses from field studies
The isolates from Table 1 from Studies 1 and 4 were grown in cell culture using MDBK or bovine turbinate monolayers and confirmed by direct fluorescent antibody tests and/or a gel based PCR test. (Fulton et al., 2016, Fulton et al., 2001) Isolates from Studies 2 and 3 were grown in MDBK monolayers and confirmed as PI3V using gel based PCR test (Fulton et al., 2016)
2.3. Vaccines used in PCR and genomic characterization
There were 7 different MLV vaccines available in the U.S. used for molecular testing and which are listed in Table 2 . (Compendium of Veterinary Products, 2010) These included Vax 1, 2, 4,5,7,8, and 9. Vax 3 and Vax 6 represented vaccines among several different ones given to calves in Study 4, and did not contain PI3V.
Table 2.
MLV vaccines with PI3VA.
| Vaccine no. | Vaccine |
|---|---|
| Vax 1 | BRD Shield ™ |
| Vax 2 | Pyramid ® 5 MLV |
| Vax 3 | Pyramid ® 3 MLV |
| Vax 4 | Titanium ™ 5 MLV |
| Vax 5 | BoviShield Gold 5 ® MLV |
| Vax 6 | BoviShield IBR-BVD ® MLV |
| Vax 7 | Vista ® 5 SQ MLV |
| Vax 8 | Express ® 5 MLV |
| Vax 9 | Arsenal ® 4.1 MLV |
2.4. Molecular tests
2.4.1. Genomic sequencing of PI3V strains
Sequence analysis of BPI3V genomic RNAs was done as previously described (Neill et al., 2014) and as modified for use on the Illumina MiSeq (Hause et al., 2015). Sequences were assembled using the LaserGene NextGen suite (DNASTAR, Madison, WI). Comparison of sequences and generation of phylogenetic trees were done using MEGA 7.0 (Kumar et al., 2016) .The viral sequence was a region of the viral nucleocapsid and had 1548 bases.
2.4.2. PI3V real time PCR
PI3V isolates were genotyped by differential real-time PCR. Three primer/probe sets were used. The PI3V sets are illustrated in Table 3 . Because of the amount of genetic diversity in PI3V A viruses, a PI3V universal set was designed that detected all isolates. PCR primer and probe sets for type B and C were designed that were specific for the respective genotypes (Table 3). The real time PCR primers were designed within the phosphoprotein (P) gene. PCR reactions were assembled using 0.4 μM of each primer and 0.2 μM probe in a 25 μl reaction using the QuantiFast Probe RT-PCR Kit (Qiagen, Inc., Valencia, CA). One microliter of cell culture fluid containing the virus was used as template. Cycling conditions were: 50 °C for 20 min, 95 °C for 5 min, then 40 cycles of 95 °C for 15 s and 62 °C for 30 s. Fluorescence was read following every cycle. The isolate was considered positive for a specific genotype if the Ct values were between 15 and 35. Above 35, the samples were considered negative.
Table 3.
Real time PCR primers for determination of PI3V types.
| Primer | Sequence |
|---|---|
| PI3V U Forwarda | AGA GCA CTC RAT TTA CAG ARA GGb |
| PI3V U Reverse | GTA TCY GCA TTG TTN AGG ACA TTc |
| PI3V U Probe | AGA CAA GAG AGT TGT RTG TGT GG |
| PI3V B Forward | TTG TGT GTG TGG CAA ATG TC |
| PI3V B Reverse | TTC TGT GTG ATT CGT CCA TTT |
| PI3V B Probe | ACT CCG ATC ATC AGG CCC GC |
| PI3V C Forward | AAT AGG TGA CCC ACC CAA AC |
| PI3V C Reverse | CAG ATC ATC GAC ACT CCC ATA TC |
| PI3V Probe | TTG AGA TGG AGC GGT CAA AGG |
U- Universal primer for detection of all PI3V isolates.
R-purine.
N-any base.
2.5. Serology
The antigenic properties of two PI3V C strains were compared to PI3V A using each virus in a microtitration virus neutralization test (VNT) in 96-well MDBK monolayer assay. (Fulton et al., 2001) The end point titer was the highest dilution of serum that inhibited viral CPE in both wells. The PI3V A strain was the SF4 strain obtained from the USDA National Veterinary Services Laboratory, Ames, IA. The virus titers of the PI3V B strains were insufficient for the VNT. The PI3V C viruses included OK P 9054 as PI3V C1 and OK P 438 as PI3V C2. Positive and negative control serums were included in each assay. The 97 serums were from post weaned calves bled prior to entering the feedlot and had been vaccinated with different MLV vaccines 4–6 weeks prior to collection. The differences in antibody levels to each virus in the VNT was analyzed with Fishers exact two sided test (Fulton et al., 2001). The test was performed at the 0.05 level of significance.
3. Results
3.1. PI3V types in field isolates from BRD cases
There were 22 isolates from cattle with BRD signs (Table 1). These were typed using the real time PCR test. There were 4 viruses that were PI3V A type, 4 were PI3V B type, and 14 were PI3V C. The 18 PI3V isolates (PI3V B and PI3V C) were different from the PI3V A in vaccine strains. As these isolates were tested only by PCR, the PI3V A isolates could not be differentiated from the PI3V A strains in MLV vaccines. There were 4 isolates in Study 1, three PI3V A and one PI3V B, and there were no PI3V MLV vaccines administered to these calves at entry to the study. The 1 PI3V B is definitely a field strain as the current MLV vaccines contain PI3V A.
In Study 2 there were 7 PI3V isolates and were all PI3V C type strains. The cattle had received a MLV vaccine containing the PI3V A type. Thus these isolates were field strains. Study 3 had 2 strains, one each of PI3V B and PI3V C. These cattle had received a MLV containing PI3V A prior at processing. Thus the two PI3V strains in this study were field strains.
Study 4 had 9 PI3V strains with 6 PI3V C, 2 PI3V B, and 1 PI3V A. These cattle had all received MLV vaccines with PI3V A at processing. The 8 isolates of PI3V C and PI3V B are field strains. The 1 PI3V A could not be determined as field or vaccine strain as sequencing was not performed.
In summary, the field strains of PI3V predominated (18/22, 81.8%) compared to those potentially with PI3V A in the MLV vaccines. However, if the 4 PI3V A had been sequenced they may have been different from PI3V A vaccine strains. This would be expected as the cattle had not received PI3V A vaccine at processing.
3.2. PI3V types in MLV vaccines and field strains detected by sequencing
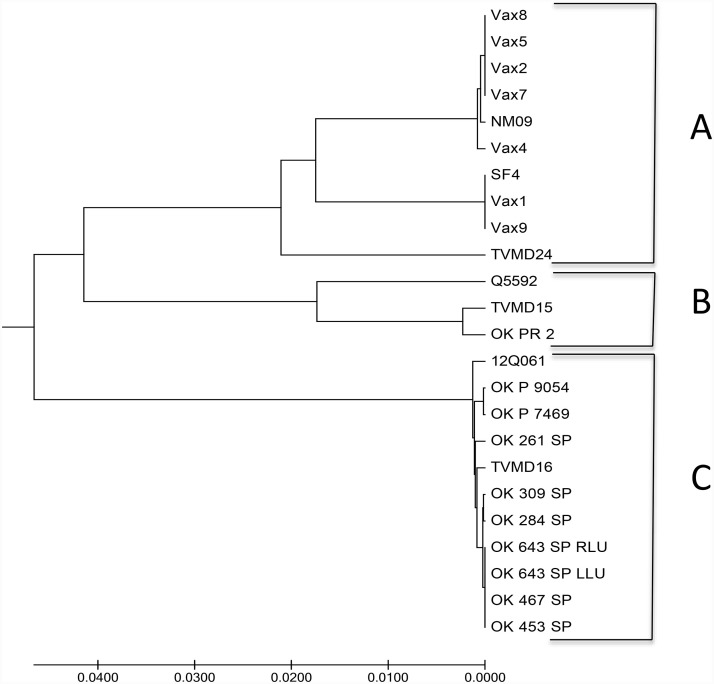
The MLV vaccine strains of PI3V were subjected to sequencing of the viral genome region for the nucleocapsid using whole genome sequencing with 24 viruses sequenced. These included 7 MLV vaccine strains, (Vax 1,2,4,5,7,8, and 9), 7 reference strains from a prior study, (NM09,SF4,TVMD24,Q5592,TVMD15,12Q06, and TVMD16) (Neill et al., 2015), 8 strains that were typed by real time PCR, (OK PR 2, OK 261 SP, OK 309 SP, OK 284 SP, OK 643 SP RLU, OK 643 SP LLU, OK 467 SP, and OK 453 SP), and 2 without real time PCR testing, (OK P 9054 and OK P 7469). The results of the sequencing and types are included in Fig. 1 . The MLV vaccine strains were all PI3V A. In the dendogram, the Vax 8, 5, 2, and 7 appear identical as PI3V A type, although they share some, but not complete identity with Vax 4 and the NM09 strain. Also in the PI3V A group are Vax 1 and 9 which are identical to the SF4 strain. There is a reference strain, TVMD24 which is a PI3V A, however it is different from the other PI3V A strains.
Fig. 1.
Dendogram of phylogenetic tree of PI3V types based on sequencing of nucleocapsid region of viral genome.
There was one of the field strains typed by PCR, OK PR 2 (PI3V B) which was identified as PI3V B as well. Reference strains Q5592 and TVMD15 were also identified as PI3V B by sequencing. There were 9 PI3V strains sequenced as PI3V C, and also included were reference strains 12Q061 and TVMD16.
3.3. PI3V types in cattle with BRD signs and MLV vaccinated
There were 20 PI3V isolates in this study from cattle with prior MLV PI3 vaccination with PI3V A strains, eighteen from Studies 2, 3, and 4 listed in Table 1, and two from Fig. 1 (OK P 9054 and OK P 7469). In this study, 19/20 (95.0%) were PI3V B or PI3V C strains. There was one strain OK NM P 64 as PI3V A strain detected by real time PCR, but not sequenced. Without sequencing, the field versus vaccine strain status could not be determined.
3.4. Serologic testing
There were 97 serums from calves receiving vaccines with PI3V A strains. The geometric mean titers and standard error of the mean (SE) are shown in Table 4 . The mean titer to the PI3V A SF 4 strain was 57.33, whereas the mean titers to PI3V C1 were 45.96 and to PI3V C2, 42.19. The titers in the serums of PI3V A vaccinated calves were higher to PI3V A than to both PI3V C1 and PI3V C2 with significance (<0.05)
Table 4.
Virus neutralizing antibody levels to PI3VA and PI3VC strains in serums from calves receiving PI3VA MLV vaccines.
| Virus | Mean and SE mean |
| PI3VA | 57.33a SE 11.35 |
| SF 4 strain | |
| PI3VC 1 | 45.96b SE 7.65 |
| OK P 9054 | |
| PI3VC 2 | 42.19b SE 6.64 |
| OK P 438 | |
Those with different letters for the mean were significant different (<0.05).
SE = standard error of the mean.
4. Discussion
This study provided additional information on the presence of PI3V types in both MLV vaccines and selected PI3V field strains as well as antigenic difference between PI3V A and other PI3V strains using serology. Using molecular tests including real time PCR and genomic sequencing of a region of the viral genome, nucleocapsid, and the PI3V strains could be differentiated into three types, PI3V A, PI3V B, and PI3V C. In this study the predominant PI3V type was other than PI3V A, and these included both PI3V B and C. Also with the vaccination history provided with the MLV vaccines given to the cattle, most of the isolates from cattle with BRD signs had been vaccinated with MLV PI3V A prior. It should be noted that cattle with the non MLV PI3V strains may have been exposed prior to transportation and commingling prior to vaccination. All of the commercial MLV available in the U.S, that were tested all were PI3V type A. using the viral sequencing with at least two groups each with identity to each other.
This study expands the knowledge of the characterization of viruses isolated from cattle with BRD signs with history of MLV vaccination. Cattle entering feedlots often have clinical signs of BRD within days of entering the feedlot. With samples submitted to the diagnostic laboratory and tested for viruses, resulting reports may indicate PI3V isolation from cultures or detection by molecular tests. Interpretation of those results is problematic as cell culture isolation or molecular tests will not differentiate field strains from the MLV vaccine strains. An earlier study (Fulton et al., 2016) found that when BoHV-1, PI3V, and BVDV isolates from cattle with BRD signs/lesions were tested using viral genome sequencing, the strains could be separated into field versus vaccine strains. This study adds more specific information on the PI3V strains compared to the prior study (Fulton et al., 2016) as the specific PI3V was identified (OK P 9054 and OK P 7469). This current study follows in line with a prior study where PI3V isolates in the U.S. include PI3V A strains along with PI3V B and PI3V C. (Neill et al., 2015) Several of those strains were included in reference strains in this study (Fig. 1).
The current MLV vaccines in the U.S. with PI3V contain the PI3V A type. And it was noted that there are at least two groups of PI3V A vaccines with unique genomic patterns. The origins of the seed stock strain for each vaccine was not available to the authors in this study. With the finding of PI3V B and PI3V C types in the cattle population, the questions becomes, does the genomic variation affect the antigenic properties? In this study, there was evidence of antigenic variation with antibody titers from calves receiving PI3V A vaccines had lower antibody titers when PI3V C viruses were used in the VNT. These results are similar to a prior study in the U.S. (Neill et al., 2015) these findings are similar to another bovine virus, BVDV with antigenic properties varied among the genotypes of BVDV, BVDV1a, BVDV1b, and BVDV2a. (Fulton, 2015) The efficacy of current MLV PI3V A vaccines should be evaluated using heterologous types such as PI3V B and PI3V C.
5. Conclusions
Points from this study include: (1) PI3V strains can be identified by molecular tests including real time PCR and viral sequencing with types A, B, and C identified, (2) commercial vaccines with MLV PI3V contain the PI3V A type, (3) PI3V strains from cattle receiving MLV vaccines at processing and later identified with BRD may have strains in the respiratory tract different from the MLV vaccine, and (4) the PI3V genomic differences may be manifested as antigenic differences.
Conflict of interest
The authors have no consulting or financial interest relationship with funding agency or companies involved with products in this study.
Acknowledgements
This research was supported by the Center for Veterinary Health Sciences, Oklahoma State University, Stillwater, OK via the research support from the Endowed Chair, McCasland Foundation for Food Animal Research support of Dr. R. Fulton. The Wentz Scholars program and the Niblack Research program supported the undergraduate research of Ms. Casey Landis. The research was supported by projects supported by the USDA ARS NADC, Ames, IA.
References
- Chanock R.M., Murphy B.M., Collins P.I. Parainfluenza viruses. In: Knipe M.K., Howley P.M., editors. Fields Virology. Lippincott, Williams, and Wilkins; Philadelphia, PA: 2001. pp. 1341–1379. [Google Scholar]
- 12th edition. North American Compendiums; Port Huron, MI: 2010. Compendium of Veterinary Products; pp. 1–1848. [Google Scholar]
- Fulton R.W., Confer A.W. Laboratory test descriptions for bovine respiratory disease diagnosis and their strengths and weaknesses: gold standards for diagnosis, do they exist. Can. Vet. J. 2012;53:754–761. [PMC free article] [PubMed] [Google Scholar]
- Fulton R.W., Ridpath J.F., Saliki J.T., Briggs R.E., Confer A.W., Burge L.J., Purdy C.W., Loan R.W., Duff G.C., Payton M.E. Bovine viral diarrhea virus (BVDV)1b: predominant subtype in calves with respiratory disease. Can. J. Vet. Res. 2001;66:181–190. [PMC free article] [PubMed] [Google Scholar]
- Fulton R.W., Step D.L., Wahrmund J., Burge L.J., Payton M.E., Cook B.J., Burken Richards C.J., Confer A.W. Bovine coronavirus (BCV) infections in transported and commingled beef cattle and sole-source calves. Can. J. Vet. Res. 2011;75:191–199. [PMC free article] [PubMed] [Google Scholar]
- Fulton R.W., Ridpath J.F., Burge L.J. Bovine coronaviruses from the respiratory tract: antigenic and genetic diversity. Vaccine. 2013;31:886–892. doi: 10.1016/j.vaccine.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton R.W., d’Offay J.M., Landis C., Miles D.G., Smith R.A., Saliki J.T., Ridpath J.F., Confer A.W., Neill J.D., Eberle R., Clement T.J., Chase C.C.L., Burge L.J., Payton M.E. Detection and characterization of viruses as field and vaccine strains in feedlot cattle with bovine respiratory disease. Vaccine. 2016;34:3478–3492. doi: 10.1016/j.vaccine.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton R.W. Viral diseases of bovine respiratory tract: bovine herpesvirus-1, parainfluenza −3 virus, bovine respiratory syncytial virus, bovine adenoviruses, bovine Coronavirus, and bovine viral diarrhea viruses. In: Anderson D.F., Rings D.M., editors. Current Veterinary Therapy-Food Animal Practice. Iowa State University Press; Ames, IA: 2008. pp. 171–191. [Google Scholar]
- Fulton R.W. Bovine respiratory disease research: 1983–2009. Anim. Health Res. Rev. 2009;10:131–140. doi: 10.1017/S146625230999017X. [DOI] [PubMed] [Google Scholar]
- Fulton R.W. Impact of species and subgenotypes of bovine viral diarrhea virus on control by vaccination. Anim. Health Res. Rev. 2015;16:40–54. doi: 10.1017/S1466252315000079. [DOI] [PubMed] [Google Scholar]
- Hause B.M., Collin E.A., Anderson J., Hesse R.A., Anderson G.A. Bovine rhinitis viruses are common in U.S. cattle with bovine respiratory disease. PLoS One. 2015;10:e0121998. doi: 10.1371/journal.pone.0121998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis (Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill J.D., Bayles D.O., Ridpath J.F. Simultaneous rapid sequencing of multiple RNA virus genomes. J. Virol. Methods. 2014;201:68–72. doi: 10.1016/j.jviromet.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill J.D., Ridpath J.F., Valayudhan B.T. Identification and genome characterization of genotype B and genotype C bovine parainfluenza type 3 viruses isolated in the United States. BMC Vet. Res. 2015;11(112):1–6. doi: 10.1186/s12917-015-0431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]