Abstract
Human adenovirus (Ad) serotype 3 causes respiratory infections. It is considered highly virulent, accounting for about 13% of all Ad isolates. We report here the complete Ad3 DNA sequence of 35,343 base pairs (GenBank accession DQ086466). Ad3 shares 96.43% nucleotide identity with Ad7, another virulent subspecies B1 serotype, and 82.56 and 62.75% identity with the less virulent species B2 Ad11 and species C Ad5, respectively. The genomic organization of Ad3 is similar to the other human Ads comprising five early transcription units, E1A, E1B, E2, E3, and E4, two delayed early units IX and IVa2, and the major late unit, in total 39 putative and 7 hypothetical open reading frames. A recombinant E1-deleted Ad3 was generated on a bacterial artificial chromosome. This prototypic virus efficiently transduced CD46-positive rodent and human cells. Our results will help in clarifying the biology and pathology of adenoviruses and enhance therapeutic applications of viral vectors in clinical settings.
Abbreviations: Ad, adenovirus; BAC, bacterial artificial chromosome; CAR, Coxsackie virus B and Ad receptor; ITR, inverted terminal repeat; kbp, kilo base pairs; MLP, major late promoter; MOI, multiplicity of infection; mu, map units; ORF, open reading frame
Keywords: Human adenovirus type 3, Species B1 adenovirus, Gene transfer vector, Bacterial artificial chromosome
Introduction
Today's major Ad vectors used in clinical applications are derived from the species C serotypes Ad2 and Ad5 which efficiently infect a variety of post-mitotic cells, including specialized tissues of the upper respiratory epithelium and the gut, in addition to many tumor cells. Their biology is very well characterized (for reviews, see, Meier and Greber, 2003, Russell, 2000, Shenk, 2001). Species C entry into epithelial cells occurs after virus binding to the Coxsackie virus B Ad receptor (CAR) (Bergelson et al., 1997) followed by engagement of αv containing heterodimeric integrins as secondary receptors (Wickham et al., 1993), which facilitate viral endocytosis and signaling into target cells (Meier et al., 2002, Suomalainen et al., 1999). Although species C Ads efficiently infect a number of cells and tissues, the lack of CAR or integrin expression may limit their general usefulness for gene therapy.
Recently, recombinant species B Ads or fiber swapped Ad vectors in which the fiber protein of the commonly used Ad5 is swapped with species B Ad fibers have gained interest for gene therapy and vaccination approaches. Species B Ads are divided into B1 and B2 subspecies. The B1 group comprises of Ad3, Ad7, Ad16, Ad21, and Ad50 and predominantly infects the upper respiratory tract, whereas the B2 group serotypes Ad11, Ad14, Ad34, and Ad35 are associated with kidney and urinary tract infections (Schmitz et al., 1983, Wadell, 2000). Infections with B1 Ads are a major cause of acute febrile and severe respiratory illness among military recruits (Dudding et al., 1972). In particular, the widespread Ad3 and Ad7 account for 13% and 19.7% of all Ad isolates typed and reported to WHO (Wadell, 2000). They are considered highly virulent and have been associated with acute clinical manifestations of considerable severity, residual lung damage, and fatal outcomes in children and military recruits in the US (for recent report, see Ryan et al., 2002). Seventeen genome types of Ad3 have been identified (Li and Wadell, 1988). These variants have been noticed to segregate in different geographic areas, time periods, and clinical conditions.
A major difference between the species C and the species B Ads is the receptor usage, which might help the species B Ads to overcome host restrictions, as suggested in early studies (Defer et al., 1990, Di Guilmi et al., 1995, Gall et al., 1996, Roelvink et al., 1998, Stevenson et al., 1995). Using different approaches, several groups have recently identified the membrane cofactor CD46 as an attachment receptor for species B serotypes, including Ad11 (Segerman et al., 2003), Ad35 (Gaggar et al., 2003), and Ad3 (Sirena et al., 2004). Most of the downstream steps of productive virus infection are, however, unknown. It is for example unclear if integrins or other proteins act as secondary receptors for virus uptake (Mathias et al., 1994, Shayakhmetov et al., 2000) or if virus attachment to CD46 is sufficient for uptake as CD46 is apparently internalized by multiple endocytic pathways in noninfected cells, including macropinocytosis and clathrin-mediated endocytosis (Crimeen-Irwin et al., 2003). Despite this shortage of knowledge, recombinant species B-based replication defective vectors have been developed for Ad35 (Gao et al., 2003, Sakurai et al., 2003, Seshidhar Reddy et al., 2003, Vogels et al., 2003) and Ad11 (Holterman et al., 2004, Stone et al., 2005). Interestingly, these vectors show an extended tropism compared to species C vectors and infect hematopoietic and dendritic cells. Similarly, replication-competent or replication-defective viral vectors derived from Ad7 have been used for vaccine strategies (Abrahamsen et al., 1997, Lubeck et al., 1989, Nan et al., 2003). An extended tropism was also demonstrated for fiber swapped Ad vectors containing fibers of species B viruses such as Ad3 (Kanerva et al., 2002, Stevenson et al., 1997, Von Seggern et al., 2000), Ad11 (Stecher et al., 2001), and Ad35 (Havenga et al., 2001, Havenga et al., 2002, Knaan-Shanzer et al., 2001, Rea et al., 2001, Shayakhmetov et al., 2000, Yotnda et al., 2001). To generate a tool for characterization of host cell–Ad3 interactions, we constructed a recombinant E1-deleted Ad3 vector expressing eGFP using the bacterial artificial chromosome (BAC) strategy, and we report here the complete nucleotide sequence of serotype Ad3 (GB) (GenBank accession no. DQ086466). This information will serve as a reference for further characterization of this prototype species B virus strain and facilitate the development of genome-based molecular diagnostic tools of Ad infections.
Results and discussion
Nucleotide sequence and genome organization
Based on primers deduced from partial Ad3 sequences (Table 1 ), the full genomic sequence of Ad3 prototype GB with an overall size of 35,343 base pairs was determined by sequencing both strands using the progressive specific primer method (GenBank accession no. DQ086466). Sequence identity between our sequence and the previously determined partial sequences varies between 95.8 and 100% (Table 1). Some of these differences may reflect genuine variations among strains or may have resulted from cloning artifacts or sequencing errors. This is also illustrated by a comparison of three recently determined prototype Ad11p sequences, where sequence discrepancies were reported in four or eleven of the 34,794 nucleotide positions (Holterman et al., 2004). Furthermore, contaminations of standard virus stocks have been noticed (Purkayastha et al., 2005a).
Table 1.
Previously published human adenovirus 3 sequences used for primer design
| Sequence origin | GenBank Accession No. | Reference | % Homology compared to the discussed sequence (number of differences) | Region (bp) |
|---|---|---|---|---|
| Ad3 complete sequence | DQ086466 | This publication | 1–35,342 | |
| Ad3 left end fragment containing ITR, E1A | (Cogan et al., 1992) | 99.24 (12/1572) | 1–1569 | |
| Ad3 ITR, left end | J01960 | (Tolun et al., 1979) | 99.4 (1/158) | 3–160 |
| Ad3 ITR, left end | J01963 | (Kosturko et al., 1982) | 95.8 (32/770) | 12–762 |
| Ad3 E1A 9S protein, E1A 13S protein, and E1A 12S protein genes, complete cds | AF492352 | (Avvakumov et al., 2002) | 100 | 576–1455 |
| Ad3 E1A protein gene, partial cds | AY380316 | Lin et al., 2003, unpublished | 99.3 (3/430) | 704–1134 |
| Ad3 polypeptide IX gene, complete cds | J01962 | (Engler, 1981) | 100 | 3413–3965 |
| Ad3 DNA polymerase gene, partial cds | AY780207 | Chmielewicz et al., 2004, unpublished | 100 | 5398–5646 |
| Ad3 virus-associated RNA, pre-terminal protein and 52/55-kDa protein genes, partial cds | U52534 | Ma et al., 1996, unpublished | 98.3 (10/582) | 10,305–10,890 |
| Ad3 virus-associated RNA I and RNA II genes | U10680 | (Kidd et al., 1995) | 99.7 (1/450) | 10,399–10,849 |
| Ad3 gene for pIIIa, pVII and penton base protein | Z29487 | (Cuzange et al., 1994) | 99.8 (3/1986) (penton 100% identical) | 13,686–15,668 (13,905–15,540) |
| Ad3 hexon gene | X76549 | (Pring-Akerblom et al., 1995) | 99.6 (10/2835) (hexon 3 aa differences) | 18,417–21,251 |
| Ad3 hexon gene, partial cds (nonfunctional) | AY380317 | Lin et al., 2003, unpublished | 97.7 (18/778) | 18,524–19,295 |
| Ad3 hexon gene, partial cds | AY684873 | Ju et al., 2004, unpublished | 98.7 (5/397) | 19,010–19,406 |
| Ad3 L3–23-kDa gene for chymotrypsin-like endoprotease | X13271 | (Houde and Weber, 1988) | 99.8 (2/1273) | 20,917–22,190 |
| Ad3 E3 region | M15952 | (Signas et al., 1986) | 99.9 (3/4379) | 26,993–31,372 |
| Ad3 fiber polypeptide gene | X01998 | (Signas et al., 1985) | 100 | 31,118–32,447 |
| Ad3 fiber protein gene, partial cds | AY380318 | Lin et al., 2003, unpublished | 98.7 (9/673) | 31,406–32,079 |
| Ad3 ITR, right end | J01961 | (Tolun et al., 1979) | 99.4 (1/158) | 35,183–35,340 |
Cds, coding sequence; ITR, inverted terminal repeat; aa, amino acid residue.
The Ad3 plus strand has a base composition of 25.29% A, 25.72% C, 25.33% G, and 23.66% T (Table 2 ). The overall GC content of Ad3 is 51.05%, which is well within the reported range of 50–52% for species B serotypes (Shenk, 2001) and very similar to the 51.03% GC content of Ad7 (Purkayastha et al., 2005b). In contrast, the GC content of B2 serotypes was reported to be 48.88% for both Ad11 and Ad35. The 51.22% GC content of Ad40 is closest to Ad3, whereas Ad5 of species C, Ad17 of species D, and Ad4 of species E all have higher GC content than Ad3 (Table 2). Nucleotide sequence comparison between Ad3 and other human Ads revealed the highest degree of identity to species B1 Ad7, amounting to 96.43% (Table 3 ) followed by 82.56% identity to Ad11p and 82.45% to Ad35p, both of species B2. Sequence identity to the species C Ad5, species D Ad17, species E Ad4, and species F Ad40 amounted to 62.75, 65.74, 73.18, and 59.44%, respectively. Thus, Ad3 differs distinctly from the other human Ad species, with the exception of B1 Ad7. The difference among the B1 serotypes is comparable to the 98.14% identity found among the B2 Ad11/35 serotypes or the 94.7% found among the Ad2/5 serotypes (Chroboczek et al., 1992). The next closely related sequence to Ad3 is the species E Ad4 (73.18% identity), in agreement with the earlier notion that Ad4 exhibits close evolutionary relationship to the species B viruses (Bailey and Mautner, 1994). On the other side, Ad4 revealed a closer phylogenetic relationship with chimpanzee Ads (simian Ads) rather than with other human Ads, which let to the speculation that human Ad4, the single member of species E, emerged through a recent zoonosis (Purkayastha et al., 2005a).
Table 2.
Base composition and GC contents of selected human adenovirus genomes
| Ad species | Ad serotype | GenBank Accession No. | Mol% |
% GC content |
|||
|---|---|---|---|---|---|---|---|
| A | C | G | T | ||||
| A | Ad12 | NC001460 | 27.34 | 23.48 | 23.04 | 26.14 | 46.52 |
| B1 | Ad3 | DQ086466 | 25.29 | 25.72 | 25.33 | 23.66 | 51.05 |
| B1 | Ad7 | AY594255 | 25.37 | 25.78 | 25.25 | 23.61 | 51.03 |
| B2 | Ad11p | AF532578, AY163756, AY598970a | 26.04 | 24.42 | 24.46 | 25.08 | 48.88 |
| B2 | Ad35p | AY271307 | 26.04 | 24.40 | 24.49 | 25.08 | 48.88 |
| C | Ad5 | AC000008 | 23.29 | 28.03 | 27.16 | 21.51 | 55.20 |
| D | Ad17 | AF108105 | 22.76 | 28.14 | 28.44 | 20.66 | 56.58 |
| E | Ad4 | AY458656 | 21.95 | 28.93 | 28.72 | 20.39 | 57.65 |
| F | Ad40 | L19443 | 24.87 | 26.21 | 25.01 | 23.91 | 51.22 |
Table 3.
Percent genome homology among selected human adenovirus genomes
| Type | Species |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A |
B1 |
B1 |
B2 |
B2 |
C |
D |
E |
F |
|
| Ad12 (NC001460) | Ad3 | Ad7 (AY594255) | Ad11p (AF532578) | Ad35p (AY271307) | Ad5 (AC000008) | Ad17 (AF108105) | Ad4 (AY458656) | Ad40 (L19443) | |
| Ad12 | 100 | 60.14 | 60.35 | 60.17 | 59.99 | 59.78 | 58.94 | 58.77 | 61.52 |
| Ad3 | 100 | 96.43 | 82.56 | 82.45 | 62.75 | 65.74 | 73.18 | 59.44 | |
| Ad7 | 100 | 83.65 | 82.84 | 62.94 | 65.55 | 72.99 | 59.36 | ||
| Ad11p | 100 | 98.14 | 61.54 | 63.62 | 70.17 | 59.16 | |||
| Ad35p | 100 | 61.59 | 63.69 | 70.22 | 59.15 | ||||
| Ad5 | 100 | 62.71 | 64.23 | 58.94 | |||||
| Ad17 | 100 | 68.86 | 59.69 | ||||||
| Ad4 | 100 | 60.17 | |||||||
| Ad40 | 100 | ||||||||
Subsequent analysis of the translational features of Ad3 led to the identification of potential open reading frames (ORFs) with sequences in general longer than 300 bp, comparison of the deduced amino acid sequences to other members of the human Mastadenovirus, in particular Ad5, 7, 11, and 35 (Chroboczek et al., 1992, Gao et al., 2003, Mei et al., 2003, Purkayastha et al., 2005b, Stone et al., 2003). Fig. 1 summarizes the Ad3 genome organization containing the 39 identified putative coding regions, the seven hypothetical coding regions, and the two virus-associated (VA) RNAs. Similar to previously reported sequences from other human Ad serotypes, Ad3 shows highly organized genome structure with five early transcription units, E1A, E1B, E2, E3, and E4, two delayed early units IX and IVa2, and one late unit, the major late unit MLTU. Several of these potential Ad3 ORFs are novel, including seven hypothetical ORFs that are conserved among other serotypes, but for which no biological function has been reported. Their putative function needs to be confirmed independently.
Fig. 1.
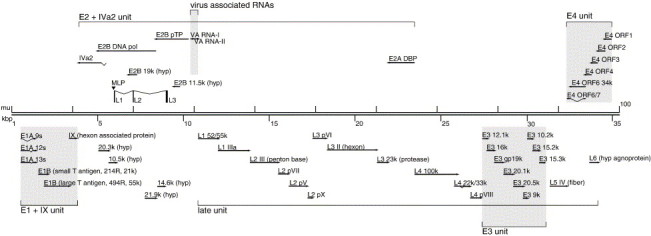
Genome organization of human Ad3. The map of the Ad3 genome (35,343 bp) is divided into 100 map units (mu). The genome consists of five early transcription units, E1A, E1B, E2, E3, and E4, two delayed early units IX and IVa2, and one late unit, the major late unit. 39 potential protein-coding regions and the direction of transcription are shown as solid arrows, relative to their position and orientation. Most late gene transcripts contain the tripartite leader consisting of three spliced leader sequences (L1–L3) at their 5′ ends. Splice events contributing to the transcript generation are indicated by diagonal thin lines. In addition, the Ad3 genome encodes VA RNA I and II transcription units and seven hypothetical (hyp) ORFs with unknown biological functions.
Noncoding motifs
Table 4 lists the predicted noncoding motifs of the Ad3 genome. The ITRs were confirmed to be 136 bp in length, ending with the sequence CTATCTATAT as for Ad7. This sequence differs from the consensus motif CATCATCAAT found in most other human Ads sequenced so far, including species B2 serotypes (Mei et al., 2003, Seshidhar Reddy et al., 2003, Stone et al., 2005). The 136 bp ITR sequence reported here is identical to the sequence reported earlier for human Ad3 (Tolun et al., 1979), except for a G–C mismatch at position 91. Two further sequence determinations (Kosturko et al., 1982, Cogan et al., 1992) confirmed our sequencing at this location (note that the sequence reported by Cogan is not available in electronic format). The entire Ad3 ITR sequence is very similar to the 136 bp ITR of the closely related Ad7 (Greiner strain) (Shinagawa and Padmanabhan, 1980), except for seven mismatches. Interestingly, recent sequencing of the Ad7 (Gomen strain) suggested an ITR of only 108 bp (Purkayastha et al., 2005b) and ten mismatched nucleotides compared to Ad3. The terminal 22 nucleotides of Ad3 and Ad7 are identical to human Ad4, in agreement with the earlier notion that Ad4 exhibits very high sequence similarity in its E1A (0 to 5.5 map units) and E2 regions (62 to 66 map units) to group B Ads (Kitchingman, 1985, Tokunaga et al., 1986). The Ad3 ITR contains the conserved ATAATATACC motif (base pairs 9 to 18), the minimum sequence necessary to interact with a complex of pTP and DNA polymerase during viral DNA replication (Chen et al., 1990). The consensus binding motifs for the host factors NFI/CTFI (bp 26 to 39) and NFIII/Oct-1 (bp 40 to 50) were found to be located at positions similar to Ad5 (Hay et al., 1995).
Table 4.
Predicted noncoding motifs of the adenovirus 3 genome
| Motif | Description | Position |
|---|---|---|
| CTATCT..TGACGT | ITR | 1–136 |
| ATAATATACC | DNApol-pTP binding site | 9–18 (ITR) |
| TGGAATGGTGCCAA | NFI binding motif | 26–39 (ITR) |
| CATGTAAATGA | NFIII binding motif | 40–50 (ITR) |
| TATTTA | TATA box for E1A | 480–485 |
| AATAAA | PolyA signal for E1Aa | 1494–1499 |
| TATATA | TATA box for E1B | 1549–1554 |
| TAAAGT | TATA box for pIX | 3384–3389 |
| AATAAA | PolyA signal pIXa | 3909–3914 |
| AATAAA | PolyA signal for E2Ba (overlapping with end of cds) | 3947–3952c |
| TGATTGGCTT | Inverted CAAT box for MLP | 5821–5830 |
| GGCCACGTGACC | Upstream element for MLP | 5840–5851 |
| GCCGGGGGGG | MAZ binding motif for MLP | 5862–5871 |
| TATAAAAG | TATA box for MLP | 5872–5879 |
| GGGGGCGGGCC | MAZ/SP1 binding motif for MLP | 5879–5889 |
| TCACTGT | Initiator element for MLP | 5901–5907 |
| TTGTCAGTTTC | DE1 for MLP | 5988–5998 |
| AACGAGGAGGATTTGA | DE2a and DE2b for MLP | 6003–6018 |
| AATAAA | PolyA signal for L1 | 13,830–13,835 |
| AATAAA | PolyA signal for L2a | 17,496–17,501 |
| AATAAA | PolyA signal for L3a | 21,938–21,943 |
| AATAAA | PolyA signal for E2Aa | 21,950–21,955c |
| TTAA | E2 TATA-box-like element | 26,639–26,642c |
| TATAA | TATA box for E3 | 27,085–27,089 |
| AATAAA | PolyA signal for L4 | 27,712–27,717 |
| AATAAA | PolyA signal for E3A | 29,001–29,006 |
| AATAAA | PolyA signal for E3Ba | 31,181–31,186 |
| AATAAA | PolyA signal for L5a | 32,335–32,340 |
| AATAAA | PolyA signal for L6a | 34,866–34,871 |
| AATAAA | PolyA signal for E4a | 32,352–32,357c |
| TATATATT | TATA box for E4 | 35,034–35,040c |
| ATAATATACC | DNApol-pTP binding site | 35,305–35,318c |
| CTATCT..TGACGT | ITR | 35,208–35,343c |
The nucleotide positions of putative motifs are noted in the 5′ to 3′ orientation.
cds, coding sequence; ITR, inverted terminal repeat; MLP, major late promoter; c, complementary.
Splice site predicted using ERPIN RNA structure prediction (see Materials and methods) cds, coding sequence; ITR, inverted terminal repeat; MLP, major late promoter; c, complementary.
Four canonical TATA box sequences conforming to the consensus TATAWADR (W designating A or T, D designating G, A, or T, and R designating G or A, respectively) were identified, including those for E1B, E3, E4, and MLP (Table 4). Furthermore, the E1A and E2A promoters contain the TATA-like box elements TATTA and TTAA, respectively, as described previously (Heysen et al., 1991, Kosturko et al., 1982). The Ad3 E2A promoter contains one TATA-like box less than Ad5. It was suggested that the lack of the second TATA-like box together with the absence of a minor cap site indicated that E2A and E2B transcripts both start at the same major cap site (Heysen et al., 1991). The upstream sequence of the Ad2 IVa2 promoter lacks a TATA element but contains an unusual TATAGAAA element downstream of the transcription start site at position +21 to +14 of the transcribed strand (Carcamo et al., 1990). More distal IVa2 promoter elements overlapping with the control region of the MLP may provide additional transcriptional activations. Similarly, we identified a TATA-like element TAAAGT about 40 nucleotides upstream of the Ad3 protein IX promoter region apparently lacking a TATA box as Ad5 (Engler, 1981). Our upstream element is identical to the one recently described for Ad7 (Purkayastha et al., 2005b). Furthermore, we identified thirteen polyadenylation signal sequences, but we found no site for the E1B region. As for Ad5, we found two polyadenylation signals in the E3 region, potentially giving rise to E3A and E3B transcripts. This feature is absent in Ad4, Ad7, and Ad11, where only one polyadenylation signal in E3 is known. A polyadenylation signal was also found for the L6 region encoding the putative agnoprotein (Mei et al., 2003).
Comparing ORFs of early regions
Table 5 lists the predicted sizes and positions of translation initiation and termination for the 46 identified putative Ad3 ORFs. The early E1A gene gives rise to the first viral transcript after DNA arrival in the nucleus (Shenk, 2001). The E1A proteins are the major transactivators controlling viral replication. The putative splice variants of Ad3 13S, 12S, and 9S of E1A were mapped according to previously published sequences for the Ad3 E1 region (Avvakumov et al., 2002, Cogan et al., 1992), the latter perfectly matching the herein described sequence (Table 1). Similar E1A ORFs have been described for other B species serotypes including Ad7 (Dijkema et al., 1980, Purkayastha et al., 2005b), Ad11 (Mei et al., 2003, Stone et al., 2005), and Ad35 (Gao et al., 2003). In the case of Ad5, the 13S, 12S, and 9S E1A proteins of 32, 26, and 6 kDa are thought to arise by differential splice site selection modulated by virus-induced stress signaling (Suomalainen et al., 2001, van der Houven van Oordt et al., 2000).
Table 5.
Forty-six predicted translation products and VA RNA genes of the Ad3 genome
| Feature | MW (kDa)a | Ad5 (others) equivalent | ATG | STOP |
|---|---|---|---|---|
| E1 region | ||||
| E1A 13s protein | 28.4 | E1A 13s mRNA 32-kDa | 576 j1250 | 1155 1455 |
| E1A 12s protein | 24.6 | E1A 12s mRNA 26-kDa | 576 j1250 | 1062 1455 |
| E1A 9s protein | 6.7 | E1A 9s mRNA 6-kDa | 576 j1250 | 647 1351 |
| E1B 21-kDa | 20.5 | E1B 19-kDa | 1603 | 2139 |
| E1B 55-kDa | 54.7 | E1B 55-kDa | 1908 | 3386 |
| Intermediate transcription regions IX and IVa2 | ||||
| Protein IX | 14.1 | IX | 3480 | 3896 |
| Protein IVa2 | 50.6 | IVa2 | 5572c j5281c | 5560c 3948c |
| E2 region | ||||
| E2B DNA pol | 128.6 | DNA pol | 8419c | 5051c |
| E2B pTP | 73.8 | Terminal protein | 10,344c | 8422c |
| E2A DBP | 58.3 | DBP | 23,557c | 22,004c |
| E3 region | ||||
| E3 12.1-kDa | 12.1 | E3 12.5-kDa | 27,403 | 27,723 |
| E3 16-kDa | 16.0 | 27,677 | 28,117 | |
| E3 gp19-kDa | 19.0 | gp19-kDa | 28,102 | 28,620 |
| E3 20.1-kDa | 20.0 | 28,650 | 29,189 | |
| E3 20.5-kDa | 20.5 | 29,202 | 29,771 | |
| E3 9-kDa | 9.0 | 29,786 | 30,019 | |
| E3 10.2-kDa | 10.3 | 30,061 | 30,336 | |
| E3 15.2-kDa | 15.2 | 30,341 | 30,745 | |
| E3 15.3-kDa | 15.2 | 30,738 | 31,148 | |
| E4 region | ||||
| E4 ORF1 | 14.1 | E4 ORF1 | 34,953c | 34,576c |
| E4 ORF2 | 16.0 | E4 ORF B | 34,579c | 34,145c |
| E4 ORF3 | 13.6 | E4 ORF3 | 34,148c | 33,795c |
| E4 ORF4 | 14.3 | E4 ORF4 | 33,786c | 33,418c |
| E4 ORF6 34-kDa | 34.7 | E4 34-kDa | 33,515c | 32,616c |
| E4 ORF6/7 | 16.0 | E4 ORF6/7 | 33,515c j32,619c | 33,342c 32,368c |
| VA RNA region | ||||
| VA RNA I | – | VA I | 10,422 | 10,591 |
| VA RNA II | – | VA II | 10,666 | 10,837 |
| L region | ||||
| L1 52/55-kDa | 43.8 | 52/55-kDa protein | 10,869 | 12,026 |
| L1 pIIIa | 65.7 | Pro-IIIa | 12,051 | 13,817 |
| L2 III (penton base) | 61.8 | Penton protein | 13,905 | 15,539 |
| L2 pVII | 21.2 | Pro-VII | 15,551 | 16,129 |
| L2 pV | 40.1 | Pro-V | 16,172 | 17,221 |
| L2 pX | 8.3 | Pro-X | 17,250 | 17,477 |
| L3 pVI | 27.1 | Pro-VI | 17,553 | 18,305 |
| L3 II (hexon) | 106.2 | Hexon | 18,418 | 21,252 |
| L3 23-kDa (protease) | 23.8 | 23-kDa (protease) | 21,289 | 21,918 |
| L4 100-kDa | 92.3 | L4 100-kDa protein | 23,588 | 26,074 |
| L4 22-kDa | 22.5 | 22-kDa | 25,776 | 26,375 |
| L4 33-kDa | 30.9 | 33-kDa | 25,776 j26,294 | 26,244 26,649 |
| L4 pVIII | 24.9 | Pro-VIII | 26,720 | 27,403 |
| L5 IV (fiber) | 34.8 | L5 IV fiber | 31,368 | 32,327 |
| L6 (agnoprotein, hyp) | 18.8 | – | 33,641 | 34,150 |
| Miscellaneous proteins | ||||
| 20.3-kDa (hyp) | 20.3 | (Ad7/Ad4) 20.6-kDa/19.4-kDa (hyp) | 5123 | 5692 |
| 11.5-kDa (hyp) | 11.5 | (Ad7/Ad11/Ad35/Ad4) 11.5-kDa (hyp) | 6144 | 6464 |
| 21.9-kDa (hyp) | 21.9 | (Ad7/Ad4/Ad11) (hyp) | 7829 | 8425 |
| 14.6-kDa (hyp) | 14.6 | (Ad7/Ad4) 14.5-kDa (hyp) | 8548 | 8949 |
| 19-kDa (hyp) | 19k | (Ad7) E2B 19-kDa (hyp) | 7389c | 6868c |
| 11.5-kDa (hyp) | 11.5 | (Ad7) E2B 11.3-kDa (hyp) | 9857c | 9543c |
C, complementary strand; j, join; hyp, hypothetical.
Sizes determined from our sequences may deviate from previously determined values.
The E1B region of Ad3 gives rise to two predicted proteins of 21 and 55 kDa corresponding to the small and large T antigen, respectively, representing the first report of an Ad3 E1B sequence. Similar E1B ORFs have been described for other B species serotypes including Ad7 (Dijkema et al., 1982), Ad11 (Mei et al., 2003, Stone et al., 2005), and Ad35 (Gao et al., 2003). The 21- and 55-kDa proteins of Ad3 show identities of 50 and 53% to Ad5 E1B proteins but show 99 and 98% identity to Ad7 and 88 and 85% identity to Ad11, respectively (Table 6 ). In the species C serotypes, both proteins are mainly involved in counteracting apoptosis.
Table 6.
Comparison of primary amino-acid sequences of predicted Ad3 proteins with corresponding proteins of other human adenoviruses
| Species |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B |
C |
D |
E | F |
|||||||||||
| B1 | B2 | |||||||||||||||
| Serotypes | Ad12 | Ad7 | Ad16 | Ad21 | Ad11 | Ad14 | Ad35 | Ad2 | Ad5 | Ad8 | Ad9 | Ad17 | Ad36 | Ad4 | Ad40 | Ad41 |
| E1 region | ||||||||||||||||
| E1A 12s protein | 39 | 98 | 76 | 76 | 36 | 41 | 53 | 56 | 33 | 35 | ||||||
| E1A 13s protein | 42 | 98 | 98 | 96 | 78 | 88 | 77 | 36 | 36 | 58 | 43 | 44 | 58 | 57 | 40 | 42 |
| E1A 9s protein | 98 | 72 | 72 | 49 | ||||||||||||
| E1B 21-kDa | 41 | 98 | 88 | 87 | 50 | 48 | 52 | 53 | 61 | 44 | ||||||
| E1B 55-kDa | 47 | 98 | 85 | 85 | 53 | 53 | 57 | 62 | 71 | 47 | ||||||
| Intermediate transcription regions | ||||||||||||||||
| Protein IX | 58 | 100 | 88 | 89 | 51 | 51 | 61 | 78 | 57 | 57 | ||||||
| Protein IVa2 | 78 | 99 | 93 | 93 | 82 | 82 | 85 | 85 | 91 | 80 | ||||||
| E2 region | ||||||||||||||||
| E2B DNA pol | 73 | 98 | 91 | 91 | 76 | 76 | 81 | 80 | 88 | 72 | ||||||
| E2B pTP | 77 | 99 | 94 | 94 | 80 | 80 | 80 | 79 | 89 | 73 | ||||||
| E2A DBP | 50 | 96 | 83 | 83 | 54 | 54 | 60 | 59 | 74 | 51 | 51 | |||||
| E3 region | ||||||||||||||||
| E3 12.1-kDa | 67 | 99 | 100 | 99 | 87 | 88 | 88 | 55 | 55 | 66 | 66 | 66 | 79 | |||
| E3 16-kDa | 97 | 98 | 94 | 62 | 61 | 62 | 30 | 27 | 35 | 28 | 33 | 54 | ||||
| E3 gp19-kDa | 23 | 98 | 98 | 98 | 80 | 70 | 80 | 32 | 36 | 35 | 35 | 34 | 63 | |||
| E3 20.1-kDa | 24 | 97 | 97 | 97 | 75 | 73 | 75 | 32 | 32 | 32 | 31 | |||||
| E3 20.5-kDa | 96 | 97 | 98 | 62 | 61 | 62 | 33 | 27 | 33 | 33 | ||||||
| E3 9-kDa | 66 | 81 | 98 | 40 | 42 | 42 | 48 | |||||||||
| E3 10.2-kDa | 52 | 100 | 98 | 100 | 93 | 95 | 93 | 51 | 47 | 65 | 64 | 75 | 42 | 43 | ||
| E3 15.2-kDa | 29 | 98 | 95 | 96 | 64 | 64 | 63 | 37 | 31 | 43 | 44 | 49 | 48 | 32 | 32 | |
| E3 15.3-kDa | 56 | 87 | 87 | 88 | 86 | 86 | 86 | 49 | 53 | 74 | 77 | 76 | 79 | 51 | 51 | |
| E4 region | ||||||||||||||||
| E4 ORF1 | 50 | 97 | 96 | 95 | 46 | 46 | 45 | 68 | ||||||||
| E4 ORF2 | 30 | 97 | 93 | 92 | 32 | 32 | 51 | 39 | 58 | 28 | ||||||
| E4 ORF3 | 61 | 100 | 98 | 98 | 49 | 49 | 72 | 72 | 86 | 57 | ||||||
| E4 ORF4 | 37 | 95 | 92 | 92 | 45 | 45 | 43 | 53 | 33 | |||||||
| E4 ORF6 34-kDa | 52 | 97 | 98 | 97 | 59 | 59 | 69 | 67 | 71 | 71 | ||||||
| E4 ORF6/7 | 42 | 97 | 97 | 97 | 47 | 47 | 66 | 34 | ||||||||
| L region | ||||||||||||||||
| L1 52/55-kDa | 73 | 100 | 94 | 95 | 70 | 69 | 79 | 79 | 84 | 77 | ||||||
| L1 IIIa | 73 | 99 | 93 | 93 | 75 | 75 | 79 | 79 | 88 | 72 | ||||||
| L2 III (penton base) | 73 | 99 | 85 | 85 | 70 | 70 | 76 | 76 | 84 | 72 | 73 | |||||
| L2 pVII | 74 | 98 | 91 | 91 | 69 | 69 | 81 | 87 | 69 | |||||||
| L2 pV | 59 | 99 | 84 | 84 | 61 | 60 | 65 | 64 | 79 | 57 | ||||||
| L2 pX | 62 | 100/99 | 92 | 92 | 69 | 69 | 77 | 88 | 67 | |||||||
| L3 pVI | 64 | 99 | 86 | 86 | 63 | 63 | 70 | 70 | 79 | 61 | ||||||
| L3 II (hexon) | 78 | 95 | 86 | 85 | 86 | 85 | 77 | 77 | 84 | 83 | 84 | 79 | 78 | |||
| L3 23-kDa (protease) | 81 | 98 | 89 | 89 | 80 | 80 | 83 | 79 | 89 | 81 | 83 | |||||
| L4 100-kDa | 64 | 97 | 85 | 85 | 64 | 69 | 62 | 76 | 65 | 63 | ||||||
| L4 22-kDa | 43 | 98 | 78 | 77 | 50 | 51 | 53 | 63 | 73 | 45 | ||||||
| L4 33-kDa | 98 | 69 | 69 | 46 | 47 | 62 | 40 | 38 | ||||||||
| L4 pVIII | 76 | 98 | 99 | 99 | 94 | 93 | 94 | 79 | 79 | 82 | 83 | 82 | 90 | 79 | 81 | |
| L5 IV (fiber) | 33 | 57 | 62 | 58 | 58 | 57 | 58 | 29 | 29 | 28 | 28 | 31 | 31 | 31 | 27 | |
| L6 (agnoprotein) | 97 | 94 | 95 | |||||||||||||
The DNA-binding protein (DBP) of E2A together with the two genes of the early E2B region encoding the DNA polymerase and the pre-terminal protein (pTP) provide the machinery for viral replication. A detailed characterization of the Ad3 E2A promoter (Heysen et al., 1991) and partial sequences of the pTP and DNA polymerase are available from the databank, but the sequence of the Ad3 DBP has not been reported previously. The Ad3 58.3-kDa DBP, the 73.8-kDa pTP, and the 128.6-kDa DNA polymerase revealed 54%, 80%, and 76% identity to the corresponding Ad5 proteins. The two zinc-binding domains in DBP were found to be fully conserved among Ad3, Ad7, Ad11, and Ad5. The first zinc-binding domain characterized by the consensus sequence HXCX8CXH is located at amino acid positions 259 to 273. Four distributed cystein residues at amino acid positions 383, 385, 437, and 454 characterize the second domain. The putative bipartite nuclear localization signal 44PPKRN48 and 84PPKKKP89 in the N-terminal domain of Ad11 DBP (Mei et al., 2003) is conserved in Ad3 and in Ad7 (Purkayastha et al., 2005b).
The E3 region together with the fiber gene is the most divergent part of the Ad genome (Bailey and Mautner, 1994). The E3 polymorphism has been suggested to be in part responsible for clinical manifestations, different for Ads of different subspecies (Wold and Gooding, 1991). Depending on the virus species, the E3 region encodes five to nine proteins that are all involved in antagonizing the host immune response to the viral infection (Horwitz, 2004). Although all seven Ad2/5 E3 proteins and the unique Ad3 20.1- and 20.5-kDa E3 proteins are expressed during infection in cultured cells (Burgert and Blusch, 2000), the E3 genomic region is entirely dispensable for virus growth in cell culture. Our sequence analysis of E3 agrees with previous data by Signas et al. (Table 1, Table 5, Fig. 1), which also identified nine putative Ad3 ORFs (Signas et al., 1986). These include the RID α/β 10.3- and 15.2-kDa proteins and the 15.3-kDa protein, all three of which are functionally conserved among all Ad species and are homologues of the Ad5 10.4-, 14.5-, and 14.7-kDa proteins. The Ad3 homologues of the Ad5 12.5- and gp19-kDa proteins are 12.1- and gp19-kDa, respectively. They are conserved in B, C, D, and E but not in F species. Two potential Ad3 ORFs encode the 20.1- and 20.5-kDa proteins which are of interest because their genes are absent in species C (Signas et al., 1986). They are absent or strongly diverged in species A Ad12 and species F Ad40. The Ad3 9-kDa transmembrane protein shares high homology only to the carboxy-terminal 20 amino acid end of the Ad2/5 11.6-kDa adenovirus death protein (ADP). The function of this protein is unknown, but it has high polymorphism among Ad3/7 field isolates (Kajon et al., 2005), and notably, species B2 and F viruses lack the potential 9-kDa/ADP ORF completely. The Ad7 homologue appears to have diverged from the Ad3 E3 9-kDa sequence in an unusual way (Hong et al., 1988) as it lacks the otherwise conserved carboxy-terminal 20 amino acids but has an insertion of 17 amino acids near the beginning of the potential ORF, completely unrelated to Ad3 or Ad2/5. This leaves only a central sequence with notable homology between Ad3 and Ad7 with a protein sequence identity of only 66%. Likewise, the potential Ad3 ORF encoding the 16-kDa protein differs substantially from Ad5 6.7 kDa equivalent. The proteins share some conserved amino acids in the carboxy terminus and the hydrophobic domain, but the Ad5 6.7-kDa protein lacks an 83-residue extension at the amino-terminal end present in Ad3 (Wilson-Rawls et al., 1990).
The E4 unit is well conserved among the human Ads and encodes at least six proteins involved in transcriptional regulation, mRNA transport, DNA replication, and enhancement or inhibition of apoptosis (Tauber and Dobner, 2001). A blast search revealed no published sequences of the Ad3 E4 region. Six potential Ad3 ORFs were identified based on sequence homology to Ad5. These include a 14.1-kDa protein related to the Ad5 ORF 1, a 16-kDa ORF2 protein, a 13.6-kDa ORF3 protein, a 14.3-kDa ORF4 protein, a 34.7-kDa ORF6 protein, and 16-kDa ORF6/7 protein. Their sequence identities to the Ad5 sequences amount to 46, 32, 49, 45, 59, and 47%, respectively, well within the range of similarity reported for E4 sequences of human species A–D (Tauber and Dobner, 2001). The best-characterized E4 protein is ORF6, a multifunctional protein participating in all of the abovementioned processes. It contains several conserved motifs such as a cystein-rich domain in the center with an HCHC motif critical for the zinc-binding in vitro (Boyer and Ketner, 2000). We found a conservation of the HCHC motif in Ad3 and also a conservation of an arginine-rich amphipathic α-helix domain overlapping with a nuclear retention signal of amino acids at position 236. Interestingly, the Ad9 E4-ORF1 was recently reported to be an oncogenic determinant and found to stimulate PI3-kinase via interaction with a PDZ protein leading to mammary gland tumor in female rats (Frese et al., 2003, Javier et al., 1992). The critical PDZ-binding domain is conserved in the Ad3 sequence, but it is unknown if this domain contributes to oncogenesis.
Seven hypothetical proteins were identified in the Ad3 genome as homologues of previously determined Ad2/5, Ad4, Ad7, or Ad11 sequences. Six of these sequences are on a 5-kbp stretch corresponding to the E2B region (Fig. 1). Four potential ORFs are embedded on the sense strand and two on the complementary strand. Due to a lack of biological data, no annotation of these sequences to any of the transcription units has been made yet.
Comparing ORFs of intermediate genes and VA RNA regions
The viral genes encoding the proteins IVa2 and IX are transcribed from their own promoters at an intermediate time point of infection. Protein IVa2 has two important functions. It is involved in packaging newly replicated viral DNA and has transactivating properties on the MLP promoter, where it either binds as a homodimer or a heterodimer (Young, 2003). Functional interaction between IVa2 and the rest of the packaging machinery appears to be serotype-specific. For example, the species B1 Ad7 IVa2 protein cannot package an Ad5 packaging sequence-containing chimeric virus (Zhang et al., 2001). The sequence identity of 82% between Ad3 and Ad5 IVa2 is surprisingly high, suggesting additional functions of the protein perhaps involved in housekeeping functions of adenoviral infections. As IVa2, protein IX has multiple functions (reviewed in (Parks, 2005), including stabilization of the capsid, transcriptional activation, and nuclear reorganization during viral DNA replication and packaging of full-length viral DNA genomes. A carboxy-terminal leucine repeat or coiled-coil domain, conserved in the Ad3 sequence, is important for the transcriptional activity of protein IX (Rosa-Calatrava et al., 2001). Furthermore, Ads encode one or two small virus-associated (VA) RNAs transcribed by the RNA polymerase III, depending on the serotype. VA RNAs counteract the activity of RNA-dependent protein kinase, up-regulated by the virus-induced interferon response (Kitajewski et al., 1986). All serotypes of species B1, including Ad3, Ad7, Ad16, and Ad21, contain two segments encoding VA RNA I and II, in contrast to species B2 or species A serotypes which only contain one VA RNA (Kidd et al., 1995). In Ad3, the VA RNA I and II were found to be located in tandem at nucleotide position 10,422 to 10,837 of the genome, comprising 416 nucleotides.
ORFs of late region
The Ad3 genome has six potential polyadenylation signals similar to Ad11, suggesting that the late genes of Ad3 are organized into six regions (L1–L6) expressed from the common major late promoter MLP (Mei et al., 2003). As in other human Ads, the MLP is located at 16.6 map units from the conventional left hand end of the genome (Fig. 1, for review on MLP, see Young, 2003). Several elements have been described to control the MLP function of Ad2/5 and were found here also for Ad3 (Table 4). These include the TATA box and the initiator element INR and, in addition, upstream elements such as the inverted CAAT box, upstream promoter element UPR, and two downstream adjacent elements (DE1 and DE2b/a). The DE elements are binding sites for the IVa2 protein complex. Furthermore, two conserved GC-rich sequences flanking the TATA box presumably functioning as MAZ-, SP1- or TFIIB-binding sites were found in Ad3. A surprisingly high 80% identity of the MLP and the first leader segment of late RNAs had been noticed before between Ad3/7 and Ad2 (Engler et al., 1981), presumably reflecting the fact that the DNA polymerase encoding sequence is located on the opposite strand.
Most viral late mRNAs contain a spliced tripartite leader (TPL) sequence of about 200 nucleotides, largely lacking secondary structure. This feature allows the translation of the downstream mRNA without the help of helicase functions, which are impaired late in infection due to dephosphorylation of eIF-4F (Huang and Schneider, 1991). eIF-4F helicase is necessary for scanning of the 40S ribosomal subunit on most cellular mRNAs. The TPL sequences thus give the viral transcripts a selective translation advantage. The TPLs of Ad3 are situated at nucleotide position 5903 to 5943 (41 nucleotides), nucleotide position 6962 to 7033 (72 nucleotides), and nucleotide position 9477 to 9563 (87 nucleotides) (Fig. 1). The overall length amounts to 200 nucleotides, identical to other human Ads such as Ad5 and Ad11. The different MLP transcription units encode ten of the eleven structural proteins and four helper proteins. The Ad3 L1 encodes two potential ORFs with homology to the 52/55-kDa protein and the precursor of protein IIIa (pIIIa). This precursor is cleaved by the viral 23-kDa protease, and the resulting pIIIa functions as a cementing protein linking adjacent hexon proteins at the icosahedral surface (Fabry et al., 2005). The conserved cleavage site 568LGGR↓G572 matches the substrate consensus motif (M,L,I)XGX↓G or (M,L,I)XGG↓X of the Ad2/5 protease (Tong, 2002). The identity between Ad3 and Ad5 pIIIa is 75% and 99% between Ad3 and Ad7. Similar relationships of Ad3 to Ad5 and Ad7 are found for the 52/55-kDa protein, 69% and 100%, respectively.
Four Ad3 ORFs were identified corresponding to homologues of the Ad5 L2 region. These include protein III (penton base), the precursor proteins pVII, pV, and pX (also called μ). Penton base is one of three major capsid proteins, and it is involved in virus uptake following binding of its RGD motif to several αv integrins (Wickham et al., 1993). The RGD motif was found to be conserved in Ad3 penton base like in most human Ad serotypes, as judged from partial sequence information (Mathias et al., 1994). Our full-length Ad3 penton base sequence revealed a 100% match to an earlier published sequence (Cuzange et al., 1994). The Ad3 penton base protein consists of 544 residues with 70% identity to Ad5 and 99% identity to Ad7. The other proteins VII, V, and X are rich in arginine and lysine, reflecting their function as DNA-binding core proteins. Protein VII is the major core protein and serves as a histone-like protein condensing the viral DNA. The arginine/lysine content of Ad5 and Ad3 pVII is very similar, 24% and 23%, respectively, reflecting their similar functions. The identities to the Ad5 homologues of pVII, pV, and pX are 69, 60, and 69%, respectively, whereas 98, 99, and 100 or 98.7% identities (for two different pX sequences) to the Ad7 homologues were found. Potential protease cleavage sites recognized by the viral 23-kDa protease are conserved in pVII and pX. The Ad3 pVII contains one potential cleavage site (21MYGG↓A25) and the pX two (24MLGR↓G28 and 42LGGG↓F46). No protease consensus motif was found in Ad3 protein V, in agreement with Ad5.
The L3 region of Ad3 encodes three proteins with homology to Ad5 proteins, the hexon (protein II), the 23-kDa cysteine protease, the sequence of which was published before (Houde and Weber, 1988), and the precursor of protein VI. The major capsid protein hexon is the most abundant structural protein (San Martin and Burnett, 2003). It carries the serotype- and species-specific determinants that are distributed over seven hypervariable regions. The Ad3 hexon sequence has been reported earlier (Table 1 and Pring-Akerblom et al., 1995), and it is 95% identical to Ad7 and 77% to Ad5. P23 is highly conserved among the different Ads. It catalyzes the maturational processing of six proteins, the precursors of the terminal protein (TP), protein IIIa, protein VI, protein VII, protein VIII, and protein X (Mangel et al., 2003). Virions with a defective protease assemble but fail to disassemble upon internalization and are not infectious (Greber et al., 1996). P23 is activated by an 11-residue carboxy-terminal peptide cleaved off from the precursor of VI (Mangel et al., 2003). This is thought to happen during virion assembly in the vicinity of the replicated viral DNA and might help recruiting protein VI to the inner surface of the capsid, thus connecting hexon and the viral DNA core. Similar to Ad2/5, two conserved viral protease recognition sites near the N- and the C-terminus 30LNGG↓A34 and 235IVGL↓G240 are present in the Ad3 pVI precursor. In the case of Ad2/5, the cleaved C-terminal peptide harbors nuclear import and export signals, allowing the protein to shuttle between the cytoplasm and link hexon to the nuclear import machinery (Wodrich et al., 2003). This C-terminal peptide also serves to activate the p23 protease switching protein VI function from regulation to assembly. The function of the N-terminal peptide is not known, although it has been suggested that endosomal membrane disruption occurs by virtue of an N-terminal amphipatic helix exposed upon proteolytic maturation (Wiethoff et al., 2005). A putative amphipatic helix sequence with hydrophobic residues is also conserved in Ad3. Precisely how membrane lysis of Ad occurs and how this is controlled by critical host factors (Meier et al., 2002, Meier et al., 2005) is not known.
In the L4 region, four potential Ad3 ORFs with homology to Ad5 genes were identified, the 100-kDa, 23-kDa, and 33-kDa protein and the pVIII precursor protein. The 100-kDa protein is not found in mature virions but acts as a scaffold to facilitate assembly of hexon trimers and is involved in the regulation of late viral protein synthesis (Shenk, 2001). For Ad2, the latter has been suggested to depend on RNA-binding motifs and a coiled-coiled motif at the amino acid position 280 to 345 allowing interactions with the eukaryotic initiation factor eIF4G (Cuesta et al., 2004). A critical 329RRK331-motif was found to be conserved in the Ad3 sequence. Ad3 100-kDa protein is 97% identical to the Ad7 and 64% to the Ad5 protein. The 33-kDa phosphoprotein is involved in viral assembly and shutoff of host cell translation (Fessler and Young, 1999). The potential ORF for 33-kDa protein arises by splicing of an intron sequence. In Ad3, an alternative potential ORF encoding a 22-kDa protein with identical 156 amino-terminal residues and a different carboxy-terminus of 43 residues is encoded by the first exon. The only biological evidence for the existence of the smaller protein comes from the finding that a virus mutant containing an early stop codon in the first exon lost expression of two (faint) protein bands of the expected sizes (Fessler and Young, 1999). The identity to the Ad7 homologue is 98% and 46% to the Ad5 homologue. The pVIII precursor protein reveals a 98% identity to the Ad7 protein and 79% identity to the Ad5 protein. Within the 227 amino acid sequence, three viral protease cleavage sites are conserved, starting at amino acid position 108, 128, and 154, respectively. pVIII is thought to stabilize the capsid, but it is not clear if it has additional functions and if all of the precursor polypeptides are incorporated into virions (Fabry et al., 2005).
L5 and L6 each contain only one potential ORF, encoding the fiber protein and the hypothetical unidentified agnoprotein. The carboxy-terminal knob of the trimeric fiber protein determines receptor binding, whereas the amino terminal domain attaches noncovalently to the penton base protein. The length of the shaft domains of species C and B viruses differs considerably due to variation of the number of shaft repeats. Like all other B species serotypes, Ad3 has short fiber shafts. The Ad3 shaft lacks the KKTK sequence, a motif suggested to bind heparan sulfate proteoglycans (Smith et al., 2003). The Ad3 fiber sequence has a 29% identity to Ad5 fiber and a 57% identity to Ad7 fiber, consistent with earlier sequencing data (Signas et al., 1985). This confirms the notion that the fiber proteins are the least conserved of all proteins among the human Ad serotypes in terms of nucleic acid sequence but not in terms of structure (Cusack, 2005). As the putative ORF encoding the 169 amino acid residues agnoprotein is highly conserved in the species B genomes and is followed by a polyadenylation signal not present in the Ad5 genome, it is likely to be expressed during infection (Mei et al., 2003, Purkayastha et al., 2005b). It was speculated that this hypothetical protein is a putative DNA-binding protein (Purkayastha et al., 2005b).
Construction of an E1-deleted recombinant Ad3 vector expressing eGFP
To establish a recombinant E1-deleted Ad3 vector expressing the eGFP reporter protein, we initially employed traditional cloning in E. coli as described earlier (Chartier et al., 1996, Sirena et al., 2004). This attempt was based on recABCD using moderate copy number entry vectors, and it was not successful (data not shown). We thus employed the single copy BAC vector pKSB2 as an acceptor, together with the recently described ET recombination system (Zhang et al., 2000). The pKSB2 plasmid supports stable maintenance of large DNA fragments in E. coli. For the Ad3CMV-eGFP construction, left and right terminal Ad3 sequences, separated by a zeocin selection cassette, were first cloned into pBluescript (Fig. 2A, for details see Materials and methods) followed by subcloning of the Ad3LRzeo-cassette into pKSB2 (Fig. 2B). The resulting pKSB2Ad3LRzeo was linearized and co-transformed together with wt Ad3 genomic DNA into E. coli DH10B pre-transformed with pKD46, which encodes the arabinose-inducible phage lambda recombinases (Fig. 2C). Homologous recombination in these cells resulted in the plasmid pKSB2Ad3wt containing the complete Ad3 genome. In contrast to our initial attempts, the efficiency of this novel recombination strategy was unexpectedly high as 9 out of 10 recovered clones carried full-length Ad3 genomes. Species C and B derived recombinant Ad genomes have been reported to be maintained as high or moderately high copy plasmids in E. coli (Chartier et al., 1996, Hitt et al., 1997, Seshidhar Reddy et al., 2003). Probably, this is not applicable for all Ad genomes, including Ad3. It is known that some viral genomes are unstable in E. coli as traditional plasmids. For example, genomes from large DNA viruses, such as herpesviruses, exceed the cloning capacity of normal cloning vectors and are thus often manipulated in BACs (Messerle et al., 1997). Besides the genome size, genome instability is another critical factor as reported for coronaviruses which are only stable in E. coli as BACs (Almazan et al., 2000). Whether the genome of Ad3 is particularly unstable or whether the efficiency of the ET recombination in our hands was much higher than the efficiency of the recABCD system is not known yet.
Fig. 2.
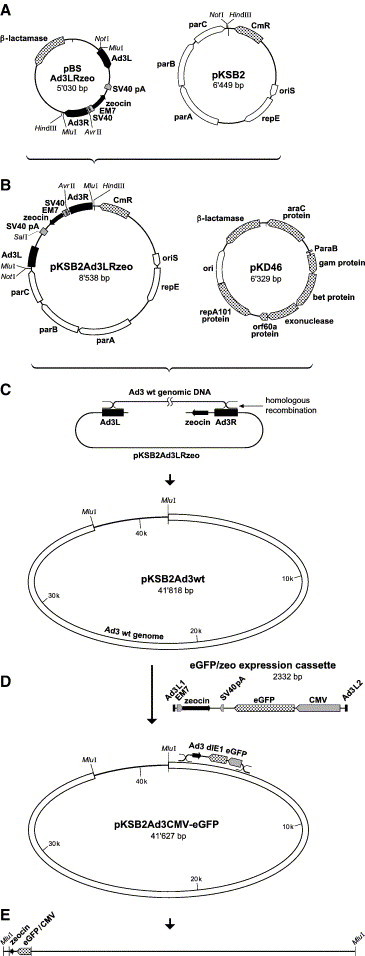
Generation of a BAC of E1-deleted Ad3 encoding the eGFP reporter gene. (A) The plasmid pBSAd3LRzeo contains a zeocin selection cassette flanked by a 468-bp fragment of the Ad3 left end sequence and a 528-bp fragment of the Ad3 right end sequence. (B) Transfer of the NotIB–HindIII fragment containing the Ad3LRzeo cassette to the single copy pKSB2 plasmid. (C) Transformation of DH10B bacteria expressing the phage lambda recombinases allows homologous recombination between the SalI-linearized pKSB2Ad3LRzeo and Ad3 genomic DNA, resulting in pKSBAd3wt. (D) Subsequent homologous recombination between pKSBAd3wt and a CMV-eGFP/zeo cassette flanked by short homologous sequences of 40 bp results in pKSB2 Ad3CMV-eGFP, which contains the eGFP expression cassette in reverse orientation. (E) Release of the viral genome by the flanking unique MluI endonuclease sites followed by transfection of helper 911-Ad3E1B cells yielding Ad3CMV-eGFP. (For details of the procedure, see Materials and methods.)
In order to introduce the CMV-driven eGFP expression cassette via a second homologous recombination, the PCR-generated eGFP/zeo cassette flanked by short homologous sequences was transformed into recombinase-expressing DH10B cells harboring the pKSB2Ad3wt plasmid (Fig. 2D). Insertion of the inverse-oriented expression cassette into the Ad3 sequence resulted in a 2469-bp deletion of the E1 region encompassing the sequence from nucleotide 561 to 3029. This deletion comprised the ORFs for E1A and most of E1B but left intact a residual 409-bp stretch of the pIX promoter. Similar E1 deletions had been engineered in an E1-deleted Ad35 vector (Gao et al., 2003). The resulting BACmid pKSB2Ad3CMV-eGFP was digested with MluI to release the genomic DNA (Fig. 2E), which was subsequently transfected into helper 911-Ad3E1B cells. GFP-positive plaques were detected under the fluorescence microscope about 10 days post-transfection. Plaque-purified clones were amplified and grown to high titers. The predicted genome structure of Ad3CMV-eGFP was confirmed by restriction analysis of isolated viral DNA (data not shown).
In order to grow E1-deleted Ad3-based vectors, a modified complementation cell line was produced that expressed the Ad3 E1B 55-kDa protein. Expression of additional species B E1B 55-kDa protein was previously demonstrated to be sufficient to complement the E1 deletion of species B Ads, when expressed in helper cell lines such as PER.C6 (Holterman et al., 2004, Vogels et al., 2003) or HER293 (Gao et al., 2003). We stably transfected 911 helper cells using the pPGK-Ad3E1B-neo plasmid expressing the Ad3 E1B 55-kDa gene under the housekeeping phosphoglycerate kinase (PGK) promoter. Several clones were isolated and characterized for their expression of Ad3 E1B 55-kDa protein and ability to support the growth of Ad3CMV-eGFP virus. The results indicated that the Ad3 E1B 55-kDa protein was expressed at low levels sufficient to grow plaqueing Ad3CMV-eGFP (data not shown). For batch purification of Ad3CMV-eGFP, the 911-Ad3E1B cells were additionally transiently transfected with the pPGK-Ad3E1B-neo plasmid increasing the expression of the limiting Ad3 E1B55K protein.
Transduction of CD46-positive target cells
We next measured Ad3CMV-eGFP-mediated gene delivery to CD46-positive cells, including the stably transfected baby hamster kidney cells BHK-CD46 (Sirena et al., 2004), the human hematopoietic cell lines K562 and THP-1, and the human lung carcinoma cell line A549, which expressed both CD46 and the Coxsackie virus B Ad receptor CAR binding to Ad2/5 vectors (Nagel et al., 2003). We compared the transduction efficiency of Ad3CMV-eGFP, a chimeric Ad3 fiber-Ad5-based vector in which the fiber protein of Ad5 was swapped with Ad3 (AdCMV-eGFP-5/F3) (Sirena et al., 2004), and Ad5CMV-eGFP (Nagel et al., 2003). In CAR negative, CD46-positive BHK-CD46 cells, both Ad3CMV-eGFP and AdCMV-eGFP-5/F3 yielded dose-dependent eGFP expression, about 75- and 39-fold higher than eGFP expression from the Ad5 vector and 97- and 25-fold higher than CD46 negative parental cells (MOI 30, Fig. 3 ). These results confirmed our earlier notion that CD46 efficiently mediates uptake of both Ad3 and Ad5/F3-chimeric viruses (Sirena et al., 2004). A similar boost of transgene expression was found in CD46-positive K562 cells and, to a lesser extent, in CD46-expressing THP-1 cells. For CAR- and CD46-positive A549 cells, all three recombinant vectors induced a robust transgene expression, reaching highest levels with Ad3CMV-eGFP followed by about 1.7-fold lower levels with AdCMV-eGFP-5/F3 and by 2.5-fold less eGFP with Ad5CMV-eGFP at MOI 3.3. Notably, the transduction levels of CD46-positive cells with Ad3CMV-eGFP were consistently higher with the chimeric AdCMV-eGFP-5/F3. This effect was particularly pronounced with K562 cells, although the fractions of eGFP-positive cells were comparable between the two viruses (data not shown). Similar results were reported in a study of an Ad35 and Ad5/F35-chimeric vector, where the Ad35 vector yielded more than 3-fold higher transduction of human hematopoietic stem cells than the chimeric vector, measuring the mean fluorescence intensity of GFP or luciferase transgenes (Sakurai et al., 2003). Although the luciferase cassette was inserted in the reverse orientation, the authors speculated that this difference could result from the different make up of the native Ad35 and the Ad5 capsid bearing the Ad35 fibers, affecting virus entry. Another study with an Ad11-eGFP and an Ad11 fiber swapped Ad5, in contrast, reported that the percentage of transgene expressing cells from both vectors were similar except in K562 cells where the chimeric vector was apparently superior (Stone et al., 2005). These results may not be comparable since the eGFP cassette of the Ad5/F11-chimeric vector was in the E3 region and in the E1 region of the Ad11-based vector. In our case, it is possible that slight differences in expression levels are due to the reverse orientation of the reporter cassette in Ad3CMV-eGFP, as opposed to the Ad5-based vectors carrying a forward orientated eGFP expression cassette.
Fig. 3.
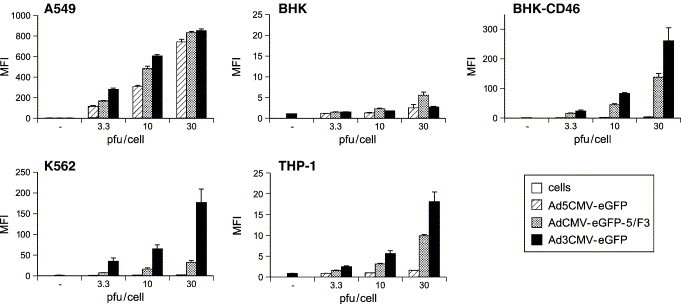
Ad3-mediated e-GFP expression in permissive human cells and CD46-transfected rodent cells. The indicated cells were incubated with eGFP expressing Ad3, Ad5, or Ad5/F3 at different MOIs. eGFP expression was analyzed 2 days post-infection by flow cytometry. Results are expressed as means of fluorescence intensity (MFI).
Conclusions
We have determined the complete Ad3 sequence of 35,343 base pairs. Ad3 together with other B1 serotypes and species E Ad4 belong to the causative agents of epidemic outbreaks of acute respiratory disease. The complementation of the Ad3 sequence reported here may help to develop improved high-throughput assays for genome typing, allowing to monitor infectious diseases caused by this agent more accurately. The transduction results obtained using the E1-deleted Ad3-based eGFP vector revealed efficient entry of Ad3 into CD46-positive cells. Future use of this vector may allow answering specific questions concerning the entry pathway of Ad3 both in vivo and in cultured cells.
Materials and methods
Cells and viruses
Most cells used here were described previously (Sirena et al., 2004) and were grown in Dulbecco's modified Eagle's medium (DMEM) plus 8% FCS. The molecular identity of the Ad3 stock (prototype strain GB, kindly provided by the late T. Adrian, Medizinische Hochschule Hannover, Germany) was verified by DNA restriction analysis (Adrian et al., 1986). Ad3 was grown in A549 cells, and DNA was isolated following the protocol of Hardy et al. (1997).
Nucleotide sequence, gene organization, and sequence comparison
Using isolated viral DNA, the nucleotide sequence of both strands of the Ad3 genome was determined (GenBank accession no. DQ086466) starting from the published sequences for hexon (accession X76549 (Pring-Akerblom et al., 1995)), E3 region (accession M15952 (Signas et al., 1986)), penton base (accession Z29487 (Cuzange et al., 1994)), fiber (accession X01998 (Signas et al., 1985)), E1A (accession AF492352 (Avvakumov et al., 2002)), ITR and left end (accession J01963 (Kosturko et al., 1982)), pIX (accession J01962 (Engler, 1981)), and pTP (accession U52534; unpublished). To confirm the exact ends of the ITR sequence, an NheI 794 bp left end fragment was subcloned into XbaI–EcoRV restricted pBluescript (ks+) (Stratagene). Sequencing reactions were performed using primers of 18 to 22 nucleotides and the chain termination kit ABI Big Dye Terminator v.3.1 (Applied Biosystems, Foster City, CA, USA) and subsequently analyzed on a 310 Genetic Analyzer (Perkin-Elmer). The raw data were assembled and verified using the sequence analysis software Sequencer (version 4.1, Gene Codes Corporation, Ann Arbor, MI, USA). Identification of putative open reading frames and the corresponding amino acid sequences, as well as general sequence comparisons, were performed using the GENtle GNU software (Mr. Magnus Manske, Germany, http://gentle.magnusmanske.de). Identification and sequence comparisons of the determined putative proteins were performed with blastp from NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). Potential splice sites of E1A were identified according to earlier published information (accession AF492352 (Avvakumov et al., 2002)). Splice sites of L4 22/23k IVa2 and E4 ORF6/7 genes were identified using Wise2 (http://www.ebi.ac.uk/Wise2/) or NNSPLICE version 0.9 (http://www.fruitfly.org/seq_tools/splice.html) (Reese et al., 1997). PolyA signals were either predicted using ERPIN (http://tagc.univ-mrs.fr/erpin/) (Gautheret and Lambert, 2001) (marked with asterisks in Table 4) or alternatively identified based on polyA signals described for human Ad7 by Purkayastha et al. (2005a). The TATA box consensus sequence was taken from: http://www.epd.isb-sib.ch/promoter_elements.
Construction of an E1-deleted recombinant Ad3CMV-eGFP
The recombinant Ad3 bacterial artificial chromosome (BAC) was constructed by ET recombination (Zhang et al., 2000) of Ad3 genomic DNA with a BAC-vector carrying subcloned left and right terminal Ad3 fragments (Fig. 2). We constructed the pBSAd3LRzeo plasmid containing the left and right end of Ad3 genome as follows. Two PCR-generated fragments were cloned into pBluescript (ks+). The first fragment encompassed 468 bp of the Ad3 left end sequence and was PCR-amplified using the forward primer 5′-GATCTCTAGACGCGTCTATCTATATAATATACCTTATAG-3′ (inserting the restriction sites XbaI and MluI) and the reverse primer 5′-GATCGAATTCGGTACCACTAGTATTTAAATTAGCGATCAGCTGACACC-3′ (inserting the restriction sites SwaI and EcoRI). This first fragment was cloned into pBluescript by XbaI and EcoRI restriction sites. An additional cloning step was used to introduce an AvrII site downstream of the EcoRI site of this intermediate product. The second fragment encompassed 528 bp of the Ad3 right end sequence and was PCR-amplified using the forward primer 5′-AGAGCCTAGGAGCAAAGCCACCCCTCGCGG-3′ (adding the restriction site AvrII) and the reverse primer 5′-AAAAGGATCCACGCGTCTATCTATATAATATACCTTATAG-3′ (adding the sites MluI and BamHI). This second fragment was cloned by AvrII and BamHI restriction sites. An AvrII–PvuII fragment containing the zeocine resistance marker from pcDNA3.1 zeo (Invitrogen) was cloned into the AvrII and SwaI site, connecting the two Ad3 arms and resulting in pBSAd3LRzeo. In order to transfer the Ad3LRzeo cassette to the BACmid pKSB2, the NotI–HindIII fragment containing this sequence was ligated with the NotI–HindIII-restricted pKSB2 vector. The pKSB2 is a single copy plasmid that supports stable maintenance of large DNA fragments in E. coli and a derivative of the BAC vector pKSO (Messerle et al., 1997) with a modified polylinker (PmeI–NsiI–PacI–NotI–SfiI–ClaI–BamHI–SacII–HindIII). Colonies containing pKSB2Ad3LRzeo were selected using chloramphenicol and zeocin™ at concentrations of 10 μg/ml and 25 μg/ml, respectively.
In order to generate pKSB2Ad3wt, which contained the complete Ad3 genome, homologous recombination in DH10B (Invitrogen) was performed. For this, chemically competent DH10B cells were first transformed with the pKD46 plasmid, which encodes the arabinose-inducible phage lambda recombinases redα (exo), redβ (bet), and redγ (gam) (Datsenko and Wanner, 2000). For recombinase induction, DH10B bacteria were grown at a reduced temperature of 30 °C (which is crucial for maintenance of pKD46) in LB ampicillin (50 μg/ml) and 0.1% (w/v) arabinose. Arabinose-induced electrocompetent cells were co-transformed with SalI-linearized pKSB2Ad3LRzeo and genomic wt Ad3 DNA and were selected on chloramphenicol plates. Nine of ten colonies revealed correct integration in pKSB2Ad3wt, which was verified by BamHI restriction analysis.
In order to insert the eGFP/zeo expression cassette into pKSB2Ad3wt, DH10B bacteria containing the BACmid pKSB2 Ad3wt were re-transformed with pKD46 and re-induced followed by transformation with a PCR-amplified fragment containing an eGFP/zeo cassette. The eGFP/zeo cassette contained the CMV-driven eGFP gene and an EM7-driven zeocin resistance gene derived from pcDNA3.1 zeo (Invitrogen). The expression cassette was flanked by short homologous Ad3 sequences of 40 bp, which were introduced using the PCR forward primer 5′-TGCCAGCGAGAAGAGTTTTCTCCTCCGCGCCGCAAGTCAGTCGGATCTGATCAGCACGTG-3′ and the reverse primer 5′-TTGGTAATCACATTATGTTCAAATACAGGCCATTTTTTGCGAATTCATGTTCTTTCCTGC-3′. The sequence of the forward primer was identical to Ad3 sequence 521 to 560, whereas the second primer was identical to sequence 3030 to 3070, resulting in a 2469-bp deletion of the Ad3 E1 region. This procedure deleted the ORFs encoding E1A and most of E1B but left intact a residual 409-bp stretch of the pIX promoter, similar as described for the Ad35-based vector by Gao et al. (2003). The integrity of the resulting pKSBAd3CMV-eGFP was confirmed by digesting isolated DNAs with restriction enzymes BamHI and HindIII. Release of the viral genome by the flanking unique MluI endonuclease sites was followed by transfection of helper 911-Ad3E1B cells. Viral genomic DNAs from rescued virus particles were analyzed using the above restriction enzymes. For large batch preparation of Ad3CMV-eGFP, helper 911-Ad3E1B cells were transiently transfected with the E1B-expression plasmid pcDNA3PGK-Ad3E1B 55 kDa. For this, 2 μg of the pcDNA3PGK-Ad3E1B 55 kDa was calcium-phosphate-precipitated and added to 106 911-Ad3E1B cells (or multiples thereof with adjusted DNA amounts). The medium was replaced after 8 to 12 h with DMEM 2% HS, and the cells were infected with the recombinant Ad3CMV-eGFP. Virus was amplified using methods described previously (Hemmi et al., 1998). CsCl purification of Ad3CMV-eGFP yielded 1.6 × 108 fluorescence focus forming units. This rather low yield was likely due to the transient nature of the complementing cells and may be overcome in the future by using a stable cell line expressing higher levels of Ad3 E1B 55-kDa protein.
Establishment of an Ad3 E1B-expressing packaging cell line
To allow the generation of E1-deleted Ad3-based vectors, a modified 911 helper cell line (Fallaux et al., 1996) was produced and characterized, similar as described for other serotype B-based vectors (Gao et al., 2003, Holterman et al., 2004, Vogels et al., 2003). For this, 106 911 cells were transfected with 2 μg of the pcDNA3PGK-Ad3E1B 55-kDa plasmid using calcium phosphate precipitation. Following transfection, G418-resistant 911-Ad3E1B cell clones were established and analyzed by Western blotting for protein expression. The pcDNA3PGK-Ad3E1B 55-kDa plasmid was constructed in two steps. First, the CMV promoter in pcDNA3-turbo-neo (Fleischli et al., 2005) was removed with MfeI and KpnI and replaced with an EcoRI–KpnI fragment of the mouse phosphoglycerate kinase (PGK) promoter, excised from the pPKGneo plasmid. In a second step, the Ad3 E1B 55-kDa sequence was PCR-amplified using the forward primer 5′-AAAGAATTCGGTGGCATGAGGTTCAGAGCG-3′ in combination with the reverse primer 5′-AAATCTAGATTACTTATCGTCGTCATCCTTGTAATCGTCAGTTTCTTCACCACTAGAACC-3′, which introduced a carboxy-terminal Flag-tag. Following digestion with EcoRI and XbaI, the fragment was cloned in the accordingly restricted pcDNA3PGK vector.
Flow cytometry
For eGFP expression analysis, triplicates of 1 × 105 cells were infected at multiplicities of infection (MOIs) of 3.3, 10, and 30, washed 2 h post-infection (p.i.) with RPMI 2% HS and analyzed 2 days p.i. by flow cytometric analysis (Cytomics FC500; Beckman Coulter, Fullerton, CA, USA). Mean fluorescence intensities and the overall proportion of infected viable cells were scored.
Acknowledgments
We thank C. Leta Fuchs for technical assistance and F. Ochsenbein for help with graphic designs. This work has been supported by the Cancer Society of the Kanton Zürich (SH), Oncoswiss (UFG), and the Kanton Zürich (SH, UFG).
References
- Abrahamsen K., Kong H.L., Mastrangeli A., Brough D., Lizonova A., Crystal R.G., Falck-Pedersen E. Construction of an adenovirus type 7a E1A-vector. J. Virol. 1997;71(11):8946–8951. doi: 10.1128/jvi.71.11.8946-8951.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian T., Wadell G., Hierholzer J.C., Wigand R. DNA restriction analysis of adenovirus prototypes 1 to 41. Arch. Virol. 1986;91(3–4):277–290. doi: 10.1007/BF01314287. [DOI] [PubMed] [Google Scholar]
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U.S.A. 2000;97(10):5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N., Sahbegovic M., Zhang Z., Shuen M., Mymryk J.S. Analysis of DNA binding by the adenovirus type 5 E1A oncoprotein. J. Gen. Virol. 2002;83(Pt 3):517–524. doi: 10.1099/0022-1317-83-3-517. [DOI] [PubMed] [Google Scholar]
- Bailey A., Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205(2):438–452. doi: 10.1006/viro.1994.1664. [DOI] [PubMed] [Google Scholar]
- Bergelson J.M., Cunningham J.A., Droguett G., Kurt-Jones E.A., Krithivas A., Hong J.S., Horwitz M.S., Crowell R.L., Finberg R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275(5304):1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- Boyer J.L., Ketner G. Genetic analysis of a potential zinc-binding domain of the adenovirus E4 34k protein. J. Biol. Chem. 2000;275(20):14969–14978. doi: 10.1074/jbc.M000566200. [DOI] [PubMed] [Google Scholar]
- Burgert H.G., Blusch J.H. Immunomodulatory functions encoded by the E3 transcription unit of adenoviruses. Virus Genes. 2000;21(1–2):13–25. [PubMed] [Google Scholar]
- Carcamo J., Maldonado E., Cortes P., Ahn M.H., Ha I., Kasai Y., Flint J., Reinberg D. A TATA-like sequence located downstream of the transcription initiation site is required for expression of an RNA polymerase II transcribed gene. Genes Dev. 1990;4(9):1611–1622. doi: 10.1101/gad.4.9.1611. [DOI] [PubMed] [Google Scholar]
- Chartier C., Degryse E., Gantzer M., Dieterle A., Pavirani A., Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996;70(7):4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Mermod N., Horwitz M.S. Protein–protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J. Biol. Chem. 1990;265(30):18634–18642. [PubMed] [Google Scholar]
- Chroboczek J., Bieber F., Jacrot B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology. 1992;186(1):280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- Cogan J.D., Jones S.N., Hall R.K., Tibbetts C. Functional diversity of E1A gene autoregulation among human adenoviruses. J. Virol. 1992;66(6):3833–3845. doi: 10.1128/jvi.66.6.3833-3845.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimeen-Irwin B., Ellis S., Christiansen D., Ludford-Menting M.J., Milland J., Lanteri M., Loveland B.E., Gerlier D., Russell S.M. Ligand binding determines whether CD46 is internalized by clathrin-coated pits or macropinocytosis. J. Biol. Chem. 2003;278(47):46927–46937. doi: 10.1074/jbc.M308261200. [DOI] [PubMed] [Google Scholar]
- Cuesta R., Xi Q., Schneider R.J. Structural basis for competitive inhibition of eIF4G–Mnk1 interaction by the adenovirus 100-kilodalton protein. J. Virol. 2004;78(14):7707–7716. doi: 10.1128/JVI.78.14.7707-7716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S. Adenovirus complex structures. Curr. Opin. Struct. Biol. 2005;15(2):237–243. doi: 10.1016/j.sbi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Cuzange A., Chroboczek J., Jacrot B. The penton base of human adenovirus type 3 has the RGD motif. Gene. 1994;146(2):257–259. doi: 10.1016/0378-1119(94)90302-6. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defer C., Belin M.T., Caillet-Boudin M.L., Boulanger P. Human adenovirus–host cell interactions: comparative study with members of subgroups B and C. J. Virol. 1990;64(8):3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Guilmi A.M., Barge A., Kitts P., Gout E., Chroboczek J. Human adenovirus serotype 3 (Ad3) and the Ad3 fiber protein bind to a 130-kDa membrane protein on HeLa cells. Virus Res. 1995;38(1):71–81. doi: 10.1016/0168-1702(95)00043-p. [DOI] [PubMed] [Google Scholar]
- Dijkema R., Dekker B.M., van Ormondt H., de Waard A., Maat J., Boyer H.W. Gene organization of the transforming region of weakly oncogenic adenovirus type 7: the E1a region. Gene. 1980;12(3–4):287–299. doi: 10.1016/0378-1119(80)90112-2. [DOI] [PubMed] [Google Scholar]
- Dijkema R., Dekker B.M., van Ormondt H. Gene organization of the transforming region of adenovirus type 7 DNA. Gene. 1982;18(2):143–156. doi: 10.1016/0378-1119(82)90112-3. [DOI] [PubMed] [Google Scholar]
- Dudding B.A., Wagner S.C., Zeller J.A., Gmelich J.T., French G.R., Top F.H., Jr. Fatal pneumonia associated with adenovirus type 7 in three military trainees. N. Engl. J. Med. 1972;286(24):1289–1292. doi: 10.1056/NEJM197206152862403. [DOI] [PubMed] [Google Scholar]
- Engler J.A. The nucleotide sequence of the polypeptide IX gene of human adenovirus type 3. Gene. 1981;13(4):387–394. doi: 10.1016/0378-1119(81)90018-4. [DOI] [PubMed] [Google Scholar]
- Engler J.A., Chow L.T., Broker T.R. Sequences of human adenovirus Ad3 and Ad7 DNAs encoding the promoter and first leader segment of late RNAs. Gene. 1981;13(2):133–143. doi: 10.1016/0378-1119(81)90002-0. [DOI] [PubMed] [Google Scholar]
- Fabry C.M., Rosa-Calatrava M., Conway J.F., Zubieta C., Cusack S., Ruigrok R.W., Schoehn G. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005;24(9):1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallaux F.J., Kranenburg O., Cramer S.J., Houweling A., Van Ormondt H., Hoeben R.C., Van Der Eb A.J. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 1996;7(2):215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- Fessler S.P., Young C.S. The role of the L4 33K gene in adenovirus infection. Virology. 1999;263(2):507–516. doi: 10.1006/viro.1999.9951. [DOI] [PubMed] [Google Scholar]
- Fleischli C., Verhaag S., Havenga M., Sirena D., Schaffner W., Cattaneo R., Greber U.F., Hemmi S. The distal short consensus repeats 1 and 2 of the membrane cofactor protein CD46 and their distance from the cell membrane determine productive entry of species B adenovirus serotype 35. J. Virol. 2005;79(15):10013–10022. doi: 10.1128/JVI.79.15.10013-10022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese K.K., Lee S.S., Thomas D.L., Latorre I.J., Weiss R.S., Glaunsinger B.A., Javier R.T. Selective PDZ protein-dependent stimulation of phosphatidylinositol 3-kinase by the adenovirus E4-ORF1 oncoprotein. Oncogene. 2003;22(5):710–721. doi: 10.1038/sj.onc.1206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggar A., Shayakhmetov D.M., Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 2003;9(11):1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- Gall J., Kass-Eisler A., Leinwand L., Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 1996;70(4):2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Robbins P.D., Gambotto A. Human adenovirus type 35: nucleotide sequence and vector development. Gene Ther. 2003;10(23):1941–1949. doi: 10.1038/sj.gt.3302097. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001;313(5):1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- Greber U.F., Webster P., Weber J., Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15(8):1766–1777. [PMC free article] [PubMed] [Google Scholar]
- Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M.L. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 1997;71(3):1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenga M.J., Lemckert A.A., Grimbergen J.M., Vogels R., Huisman L.G., Valerio D., Bout A., Quax P.H. Improved adenovirus vectors for infection of cardiovascular tissues. J. Virol. 2001;75(7):3335–3342. doi: 10.1128/JVI.75.7.3335-3342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenga M.J., Lemckert A.A., Ophorst O.J., van Meijer M., Germeraad W.T., Grimbergen J., van Den Doel M.A., Vogels R., van Deutekom J., Janson A.A., de Bruijn J.D., Uytdehaag F., Quax P.H., Logtenberg T., Mehtali M., Bout A. Exploiting the natural diversity in adenovirus tropism for therapy and prevention of disease. J. Virol. 2002;76(9):4612–4620. doi: 10.1128/JVI.76.9.4612-4620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.T., Freeman A., Leith I., Monaghan A., Webster A. Molecular interactions during adenovirus DNA replication. Curr. Top. Microbiol. Immunol. 1995;199(Pt 2):31–48. doi: 10.1007/978-3-642-79499-5_2. [DOI] [PubMed] [Google Scholar]
- Hemmi S., Geertsen R., Mezzacasa A., Peter I., Dummer R. The presence of human coxsackievirus and adenovirus receptor is associated with efficient adenovirus-mediated transgene expression in human melanoma cell cultures. Hum. Gene Ther. 1998;9(16):2363–2373. doi: 10.1089/hum.1998.9.16-2363. [DOI] [PubMed] [Google Scholar]
- Heysen A., Verwaerde P., D'Halluin J.C. Nucleotide sequence and regulation of the adenovirus type 3 E2A early promoter. Virology. 1991;181(1):241–250. doi: 10.1016/0042-6822(91)90489-x. [DOI] [PubMed] [Google Scholar]
- Hitt M.M., Addison C.L., Graham F.L. Human adenovirus vectors for gene transfer into mammalian cells. Adv. Pharmacol. 1997;40:137–206. doi: 10.1016/s1054-3589(08)60140-4. [DOI] [PubMed] [Google Scholar]
- Holterman L., Vogels R., van der Vlugt R., Sieuwerts M., Grimbergen J., Kaspers J., Geelen E., van der Helm E., Lemckert A., Gillissen G., Verhaagh S., Custers J., Zuijdgeest D., Berkhout B., Bakker M., Quax P., Goudsmit J., Havenga M. Novel replication-incompetent vector derived from adenovirus type 11 (Ad11) for vaccination and gene therapy: low seroprevalence and non-cross-reactivity with Ad5. J. Virol. 2004;78(23):13207–13215. doi: 10.1128/JVI.78.23.13207-13215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.S., Mullis K.G., Engler J.A. Characterization of the early region 3 and fiber genes of Ad7. Virology. 1988;167(2):545–553. [PubMed] [Google Scholar]
- Horwitz M.S. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 2004;6(Suppl 1):S172–S183. doi: 10.1002/jgm.495. [DOI] [PubMed] [Google Scholar]
- Houde A., Weber J.M. Sequence of the human adenovirus type 3 protease. Nucleic Acids Res. 1988;16(23):11374. doi: 10.1093/nar/16.23.11374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.T., Schneider R.J. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell. 1991;65(2):271–280. doi: 10.1016/0092-8674(91)90161-q. [DOI] [PubMed] [Google Scholar]
- Javier R., Raska K., Jr., Shenk T. Requirement for the adenovirus type 9 E4 region in production of mammary tumors. Science. 1992;257(5074):1267–1271. doi: 10.1126/science.1519063. [DOI] [PubMed] [Google Scholar]
- Kajon A.E., Xu W., Erdman D.D. Sequence polymorphism in the E3 7.7K ORF of subspecies B1 human adenoviruses. Virus Res. 2005;107(1):11–19. doi: 10.1016/j.virusres.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kanerva A., Wang M., Bauerschmitz G.J., Lam J.T., Desmond R.A., Bhoola S.M., Barnes M.N., Alvarez R.D., Siegal G.P., Curiel D.T., Hemminki A. Gene transfer to ovarian cancer versus normal tissues with fiber-modified adenoviruses. Mol. Ther. 2002;5(6):695–704. doi: 10.1006/mthe.2002.0599. [DOI] [PubMed] [Google Scholar]
- Kidd A.H., Garwicz D., Oberg M. Human and simian adenoviruses: phylogenetic inferences from analysis of VA RNA genes. Virology. 1995;207(1):32–45. doi: 10.1006/viro.1995.1049. [DOI] [PubMed] [Google Scholar]
- Kitajewski J., Schneider R.J., Safer B., Munemitsu S.M., Samuel C.E., Thimmappaya B., Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986;45(2):195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- Kitchingman G.R. Sequence of the DNA-binding protein of a human subgroup E adenovirus (type 4): comparisons with subgroup A (type 12), subgroup B (type 7), and subgroup C (type 5) Virology. 1985;146(1):90–101. doi: 10.1016/0042-6822(85)90055-8. [DOI] [PubMed] [Google Scholar]
- Knaan-Shanzer S., Van Der Velde I., Havenga M.J., Lemckert A.A., De Vries A.A., Valerio D. Highly efficient targeted transduction of undifferentiated human hematopoietic cells by adenoviral vectors displaying fiber knobs of subgroup B. Hum. Gene Ther. 2001;12(16):1989–2005. doi: 10.1089/104303401753204562. [DOI] [PubMed] [Google Scholar]
- Kosturko L.D., Sharnick S.V., Tibbetts C. Polar encapsidation of adenovirus DNA: cloning and DNA sequence of the left end of adenovirus type 3. J. Virol. 1982;43(3):1132–1137. doi: 10.1128/jvi.43.3.1132-1137.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.G., Wadell G. Comparison of 17 genome types of adenovirus type 3 identified among strains recovered from six continents. J. Clin. Microbiol. 1988;26(5):1009–1015. doi: 10.1128/jcm.26.5.1009-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck M.D., Davis A.R., Chengalvala M., Natuk R.J., Morin J.E., Molnar-Kimber K., Mason B.B., Bhat B.M., Mizutani S., Hung P.P. Immunogenicity and efficacy testing in chimpanzees of an oral hepatitis B vaccine based on live recombinant adenovirus. Proc. Natl. Acad. Sci. U.S.A. 1989;86(17):6763–6767. doi: 10.1073/pnas.86.17.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel W.F., Baniecki M.L., McGrath W.J. Specific interactions of the adenovirus proteinase with the viral DNA, an 11-amino-acid viral peptide, and the cellular protein actin. Cell. Mol. Life Sci. 2003;60(11):2347–2355. doi: 10.1007/s00018-003-2318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias P., Wickham T., Moore M., Nemerow G. Multiple adenovirus serotypes use alpha v integrins for infection. J. Virol. 1994;68(10):6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y.F., Skog J., Lindman K., Wadell G. Comparative analysis of the genome organization of human adenovirus 11, a member of the human adenovirus species B, and the commonly used human adenovirus 5 vector, a member of species C. J. Gen. Virol. 2003;84(Pt 8):2061–2071. doi: 10.1099/vir.0.19178-0. [DOI] [PubMed] [Google Scholar]
- Meier O., Boucke K., Hammer S.V., Keller S., Stidwill R.P., Hemmi S., Greber U.F. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 2002;158(6):1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier O., Gastaldelli M., Boucke K., Hemmi S., Greber U.F. Early steps of clathrin-mediated endocytosis involved in phagosomal escape of Fcgamma receptor-targeted adenovirus. J. Virol. 2005;79(4):2604–2613. doi: 10.1128/JVI.79.4.2604-2613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier O., Greber U.F. Adenovirus endocytosis. J. Gene Med. 2003;5(6):451–462. doi: 10.1002/jgm.409. [DOI] [PubMed] [Google Scholar]
- Messerle M., Crnkovic I., Hammerschmidt W., Ziegler H., Koszinowski U.H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U.S.A. 1997;94(26):14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel H., Maag S., Tassis A., Nestle F.O., Greber U.F., Hemmi S. The alphavbeta5 integrin of hematopoietic and nonhematopoietic cells is a transduction receptor of RGD-4C fiber-modified adenoviruses. Gene Ther. 2003;10(19):1643–1653. doi: 10.1038/sj.gt.3302058. [DOI] [PubMed] [Google Scholar]
- Nan X., Peng B., Hahn T.W., Richardson E., Lizonova A., Kovesdi I., Robert-Guroff M. Development of an Ad7 cosmid system and generation of an Ad7deltaE1deltaE3HIV(MN) env/rev recombinant virus. Gene Ther. 2003;10(4):326–336. doi: 10.1038/sj.gt.3301903. [DOI] [PubMed] [Google Scholar]
- Parks R.J. Adenovirus protein IX: a new look at an old protein. Mol. Ther. 2005;11(1):19–25. doi: 10.1016/j.ymthe.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Pring-Akerblom P., Trijssenaar F.E., Adrian T. Sequence characterization and comparison of human adenovirus subgenus B and E hexons. Virology. 1995;212(1):232–236. doi: 10.1006/viro.1995.1474. [DOI] [PubMed] [Google Scholar]
- Purkayastha A., Ditty S.E., Su J., McGraw J., Hadfield T.L., Tibbetts C., Seto D. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J. Virol. 2005;79(4):2559–2572. doi: 10.1128/JVI.79.4.2559-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A., Su J., Carlisle S., Tibbetts C., Seto D. Genomic and bioinformatics analysis of HAdV-7, a human adenovirus of species B1 that causes acute respiratory disease: implications for vector development in human gene therapy. Virology. 2005;332(1):114–129. doi: 10.1016/j.virol.2004.10.041. [DOI] [PubMed] [Google Scholar]
- Rea D., Havenga M.J., van Den Assem M., Sutmuller R.P., Lemckert A., Hoeben R.C., Bout A., Melief C.J., Offringa R. Highly efficient transduction of human monocyte-derived dendritic cells with subgroup B fiber-modified adenovirus vectors enhances transgene-encoded antigen presentation to cytotoxic T cells. J. Immunol. 2001;166(8):5236–5244. doi: 10.4049/jimmunol.166.8.5236. [DOI] [PubMed] [Google Scholar]
- Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4(3):311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Roelvink P.W., Lizonova A., Lee J.G., Li Y., Bergelson J.M., Finberg R.W., Brough D.E., Kovesdi I., Wickham T.J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 1998;72(10):7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Calatrava M., Grave L., Puvion-Dutilleul F., Chatton B., Kedinger C. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 2001;75(15):7131–7141. doi: 10.1128/JVI.75.15.7131-7141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W.C. Update on adenovirus and its vectors. J. Gen. Virol. 2000;81(Pt 11):2573–2604. doi: 10.1099/0022-1317-81-11-2573. [DOI] [PubMed] [Google Scholar]
- Ryan M.A., Gray G.C., Smith B., McKeehan J.A., Hawksworth A.W., Malasig M.D. Large epidemic of respiratory illness due to adenovirus types 7 and 3 in healthy young adults. Clin. Infect. Dis. 2002;34(5):577–582. doi: 10.1086/338471. [DOI] [PubMed] [Google Scholar]
- Sakurai F., Mizuguchi H., Hayakawa T. Efficient gene transfer into human CD34+ cells by an adenovirus type 35 vector. Gene Ther. 2003;10(12):1041–1048. doi: 10.1038/sj.gt.3301959. [DOI] [PubMed] [Google Scholar]
- San Martin C., Burnett R.M. Structural studies on adenoviruses. Curr. Top. Microbiol. Immunol. 2003;272:57–94. doi: 10.1007/978-3-662-05597-7_3. [DOI] [PubMed] [Google Scholar]
- Schmitz H., Wigand R., Heinrich W. Worldwide epidemiology of human adenovirus infections. Am. J. Epidemiol. 1983;117(4):455–466. doi: 10.1093/oxfordjournals.aje.a113563. [DOI] [PubMed] [Google Scholar]
- Segerman A., Atkinson J.P., Marttila M., Dennerquist V., Wadell G., Arnberg N. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 2003;77(17):9183–9191. doi: 10.1128/JVI.77.17.9183-9191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshidhar Reddy P., Ganesh S., Limbach M.P., Brann T., Pinkstaff A., Kaloss M., Kaleko M., Connelly S. Development of adenovirus serotype 35 as a gene transfer vector. Virology. 2003;311(2):384–393. doi: 10.1016/s0042-6822(03)00161-2. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov D.M., Papayannopoulou T., Stamatoyannopoulos G., Lieber A. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J. Virol. 2000;74(6):2567–2583. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. Adenoviridae: the viruses and their replication. In: Fields B.N., Howley P.M., Griffin D.E., editors. Virology. 4th ed. Lippincott-Raven Publisher; Philadelphia: 2001. pp. 2265–2300. [Google Scholar]
- Shinagawa M., Padmanabhan R. Comparative sequence analysis of the inverted terminal repetitions from different adenoviruses. Proc. Natl. Acad. Sci. U.S.A. 1980;77(7):3831–3835. doi: 10.1073/pnas.77.7.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signas C., Akusjarvi G., Pettersson U. Adenovirus 3 fiber polypeptide gene: implications for the structure of the fiber protein. J. Virol. 1985;53(2):672–678. doi: 10.1128/jvi.53.2.672-678.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signas C., Akusjarvi G., Pettersson U. Region E3 of human adenoviruses; differences between the oncogenic adenovirus-3 and the non-oncogenic adenovirus-2. Gene. 1986;50(1–3):173–184. doi: 10.1016/0378-1119(86)90322-7. [DOI] [PubMed] [Google Scholar]
- Sirena D., Lilienfeld B., Eisenhut M., Kalin S., Boucke K., Beerli R.R., Vogt L., Ruedl C., Bachmann M.F., Greber U.F., Hemmi S. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J. Virol. 2004;78(9):4454–4462. doi: 10.1128/JVI.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.A., Idamakanti N., Rollence M.L., Marshall-Neff J., Kim J., Mulgrew K., Nemerow G.R., Kaleko M., Stevenson S.C. Adenovirus serotype 5 fiber shaft influences in vivo gene transfer in mice. Hum. Gene Ther. 2003;14(8):777–787. doi: 10.1089/104303403765255165. [DOI] [PubMed] [Google Scholar]
- Stecher H., Shayakhmetov D.M., Stamatoyannopoulos G., Lieber A. A capsid-modified adenovirus vector devoid of all viral genes: assessment of transduction and toxicity in human hematopoietic cells. Mol. Ther. 2001;4(1):36–44. doi: 10.1006/mthe.2000.0410. [DOI] [PubMed] [Google Scholar]
- Stevenson S.C., Rollence M., White B., Weaver L., McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J. Virol. 1995;69(5):2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson S.C., Rollence M., Marshall-Neff J., McClelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J. Virol. 1997;71(6):4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D., Furthmann A., Sandig V., Lieber A. The complete nucleotide sequence, genome organization, and origin of human adenovirus type 11. Virology. 2003;309(1):152–165. doi: 10.1016/s0042-6822(02)00085-5. [DOI] [PubMed] [Google Scholar]
- Stone D., Ni S., Li Z.Y., Gaggar A., DiPaolo N., Feng Q., Sandig V., Lieber A. Development and assessment of human adenovirus type 11 as a gene transfer vector. J. Virol. 2005;79(8):5090–5104. doi: 10.1128/JVI.79.8.5090-5104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M., Nakano M.Y., Keller S., Boucke K., Stidwill R.P., Greber U.F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999;144(4):657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M., Nakano M.Y., Boucke K., Keller S., Greber U.F. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 2001;20(6):1310–1319. doi: 10.1093/emboj/20.6.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber B., Dobner T. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene. 2001;278(1–2):1–23. doi: 10.1016/s0378-1119(01)00722-3. [DOI] [PubMed] [Google Scholar]
- Tokunaga O., Yaegashi T., Lowe J., Dobbs L., Padmanabhan R. Sequence analysis in the E1 region of adenovirus type 4 DNA. Virology. 1986;155(2):418–433. doi: 10.1016/0042-6822(86)90204-7. [DOI] [PubMed] [Google Scholar]
- Tolun A., Alestrom P., Pettersson U. Sequence of inverted terminal repetitions from different adenoviruses: demonstration of conserved sequences and homology between SA7 termini and SV40 DNA. Cell. 1979;17(3):705–713. doi: 10.1016/0092-8674(79)90277-0. [DOI] [PubMed] [Google Scholar]
- Tong L. Viral proteases. Chem. Rev. 2002;102(12):4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- van der Houven van Oordt W., Diaz-Meco M.T., Lozano J., Krainer A.R., Moscat J., Caceres J.F. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 2000;149(2):307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels R., Zuijdgeest D., van Rijnsoever R., Hartkoorn E., Damen I., de Bethune M.P., Kostense S., Penders G., Helmus N., Koudstaal W., Cecchini M., Wetterwald A., Sprangers M., Lemckert A., Ophorst O., Koel B., van Meerendonk M., Quax P., Panitti L., Grimbergen J., Bout A., Goudsmit J., Havenga M. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 2003;77(1):8263–8271. doi: 10.1128/JVI.77.15.8263-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Seggern D.J., Huang S., Fleck S.K., Stevenson S.C., Nemerow G.R. Adenovirus vector pseudotyping in fiber-expressing cell lines: improved transduction of Epstein–Barr virus-transformed B cells. J. Virol. 2000;74(1):354–362. doi: 10.1128/jvi.74.1.354-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell G. Adenoviruses. In: Zuckerman A.J., Banatvala J.E., Pattison J.R., editors. Principles and Practice of Clinical Virology. 4th ed. John Wiley and Sons; 2000. pp. 307–327. [Google Scholar]
- Wickham T.J., Mathias P., Cheresh D.A., Nemerow G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73(2):309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- Wiethoff C.M., Wodrich H., Gerace L., Nemerow G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 2005;79(4):1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Rawls J., Saha S.K., Krajcsi P., Tollefson A.E., Gooding L.R., Wold W.S. A 6700 MW membrane protein is encoded by region E3 of adenovirus type 2. Virology. 1990;178(1):204–212. doi: 10.1016/0042-6822(90)90395-8. [DOI] [PubMed] [Google Scholar]
- Wodrich H., Guan T., Cingolani G., Von Seggern D., Nemerow G., Gerace L. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003;22(23):6245–6255. doi: 10.1093/emboj/cdg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W.S., Gooding L.R. Region E3 of adenovirus: a cassette of genes involved in host immunosurveillance and virus–cell interactions. Virology. 1991;184(1):1–8. doi: 10.1016/0042-6822(91)90815-s. [DOI] [PubMed] [Google Scholar]
- Yotnda P., Onishi H., Heslop H.E., Shayakhmetov D., Lieber A., Brenner M., Davis A. Efficient infection of primitive hematopoietic stem cells by modified adenovirus. Gene Ther. 2001;8(12):930–937. doi: 10.1038/sj.gt.3301488. [DOI] [PubMed] [Google Scholar]
- Young C.S. The structure and function of the adenovirus major late promoter. Curr. Top. Microbiol. Immunol. 2003;272:213–249. doi: 10.1007/978-3-662-05597-7_8. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Muyrers J.P., Testa G., Stewart A.F. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 2000;18(12):1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- Zhang W., Low J.A., Christensen J.B., Imperiale M.J. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 2001;75(21):10446–10454. doi: 10.1128/JVI.75.21.10446-10454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


