Summary
Background
Lower respiratory tract infections (LRTIs) are a leading cause of childhood morbidity and mortality. Potentially pathogenic organisms are present in the respiratory tract in both symptomatic and asymptomatic children, but their presence does not necessarily indicate disease. We aimed to assess the concordance between upper and lower respiratory tract microbiota during LRTIs and the use of nasopharyngeal microbiota to discriminate LRTIs from health.
Methods
First, we did a prospective study of children aged between 4 weeks and 5 years who were admitted to the paediatric intensive care unit (PICU) at Wilhelmina Children's Hospital (Utrecht, Netherlands) for a WHO-defined LRTI requiring mechanical ventilation. We obtained paired nasopharyngeal swabs and deep endotracheal aspirates from these participants (the so-called PICU cohort) between Sept 10, 2013, and Sept 4, 2016. We also did a matched case-control study (1:2) with the same inclusion criteria in children with LRTIs at three Dutch teaching hospitals and in age-matched, sex-matched, and time-matched healthy children recruited from the community. Nasopharyngeal samples were obtained at admission for cases and during home visits for controls. Data for child characteristics were obtained by questionnaires and from pharmacy printouts and medical charts. We used quantitative PCR and 16S rRNA-based sequencing to establish viral and bacterial microbiota profiles, respectively. We did sparse random forest classifier analyses on the bacterial data, viral data, metadata, and the combination of all three datasets to distinguish cases from controls.
Findings
29 patients were enrolled in the PICU cohort. Intra-individual concordance in terms of viral microbiota profiles (96% agreement [95% CI 93–99]) and bacterial microbiota profiles (58 taxa with a median Pearson's r 0·93 [IQR 0·62–0·99]; p<0·05 for all 58 taxa) was high between nasopharyngeal and endotracheal aspirate samples, supporting the use of nasopharyngeal samples as proxy for lung microbiota during LRTIs. 154 cases and 307 matched controls were prospectively recruited to our case-control cohort. Individually, bacterial microbiota (area under the curve 0·77), viral microbiota (0·70), and child characteristics (0·80) poorly distinguished health from disease. However, a classification model based on combined bacterial and viral microbiota plus child characteristics distinguished children with LRTIs from their matched controls with a high degree of accuracy (area under the curve 0·92).
Interpretation
Our data suggest that the nasopharyngeal microbiota can serve as a valid proxy for lower respiratory tract microbiota in childhood LRTIs, that clinical LRTIs in children result from the interplay between microbiota and host characteristics, rather than a single microorganism, and that microbiota-based diagnostics could improve future diagnostic and treatment protocols.
Funding
Spaarne Gasthuis, University Medical Center Utrecht, and the Netherlands Organization for Scientific Research.
Introduction
Lower respiratory tract infections (LRTIs) are a major cause of morbidity and mortality in children worldwide.1, 2 Although multiple host, environmental, and lifestyle factors are known to increase susceptibility to LRTIs,3 why some children remain asymptomatic after exposure to pathogens and others develop severe disease remains unclear. Classically, LRTIs are caused by acquisition in the upper respiratory tract of pathogenic viruses and bacteria, which replicate and spread towards the lower respiratory tract, where they invade the mucosa, leading to inflammation and clinical disease.4 Many of these microorganisms are, however, pathobionts, and are frequently encountered in the upper respiratory tract of healthy children too.5 We therefore hypothesised that a balanced microbial community protects pathobionts from causing LRTIs.
Previous studies6, 7 in children have shown a relation between the bacterial composition of the nasopharynx microbiota and susceptibility to upper or lower respiratory infectious episodes over time. We have reported that oral microbes such as Prevotella and Leptotrichia spp in the nasopharyngeal niche were strongly associated with subsequent development of upper respiratory tract infections in children and were more abundant during these infections.8 By contrast, Corynebacterium and Dolosigranulum spp were associated with resistance to symptomatic respiratory disease during the first year of life and were less abundant during upper respiratory tract infections.8 Additionally, in infants with LRTIs caused by respiratory syncytial virus, increased presence of Haemophilus influenzae and Streptococcus pneumoniae was strongly associated with increased severity of host inflammation, suggesting an important role for the complete microbiota of the upper respiratory tract and the symptomology of clinical disease.9
Research in context.
Evidence before this study
We hypothesised that the entire nasopharyngeal microbiota might have a role in susceptibility to, and severity of, lower respiratory tract infection (LRTIs). We searched PubMed with the terms “(child, preschool[mh] OR infant[mh] NOT infant, newborn[mh]) AND (respiratory tract infections[mh] OR pneumonia[tiab] OR bronchiolitis[tiab] OR wheezing[tiab]) AND (microbiota[mh] OR microbiome[tiab]) AND (case-control studies[mh] OR prospective[tiab])” for articles published in any language up to May 1, 2018. We identified 14 publications, three of which pertained to the role of the microbiota in acute LRTIs in children. One study, which focused only on infants at high risk of atopy, showed that specific microbiota profiles were associated with the development of respiratory infections. A second small (n=100) matched case-control study showed that Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis were associated with cases, whereas no specific taxon was associated with controls. However, the study was underpowered to provide conclusive results. The third study included infants younger than 1 year only, and did not include a control group of healthy children. It showed that Moraxella was associated with less severe bronchiolitis and Streptococcus was associated with more severe bronchiolitis.
Added value of this study
In our study, we show that the nasopharyngeal viral and bacterial microbiota can be used as proxy for lung microbiota in childhood LRTIs. We also showed the relationship between microbial community composition and susceptibility to, and severity of, LRTIs in children. Because we used a strictly matched case-control design in a cohort of 461 children, our study is the first to confidently show the association between microbiota and LRTIs in children. To our knowledge, the accuracy of the model we developed to discriminate LRTIs from health is unprecedented. Furthermore, the phenotype-independent nature of the associations between respiratory microbiota and childhood LRTIs has not previously been reported.
Implications of all the available evidence
The complete nasopharyngeal ecosystem has an important role in the development and severity of LRTIs in young children. The excellent accuracy of our classifier model provides a basis for future microbiota-based diagnostic tools, which could have major implications for future treatment protocols. Our data provide insights that could be crucial for establishing optimal therapeutic strategies, including targeted antibiotic treatment. The phenotype-independent associations during acute disease that we identified challenge conventional views about the role of viruses and bacteria in LRTI pathogenesis, especially the dichotomy between bronchiolitis (viral origin) and pneumonia (bacterial origin).
No case-control study has yet addressed the relation between the nasopharyngeal microbiota and the presence, clinical symptoms, and severity of childhood LRTIs. Furthermore, the profiling of nasopharyngeal microbiota has never been studied in the context of classification of states of health and disease. We aimed to investigate the association between upper and lower respiratory tract microbiota during childhood LRTI, the use of microbiota to predict the presence and severity of LRTIs, and the associations between microbiota and disease across different clinical presentations of LRTI.
Methods
Study design and participants
We did a prospective strictly matched case-control study in young children hospitalised for LRTIs. First, we did a prospective study from Sept 10, 2013, to Sept 4, 2016 to assess whether, during acute paediatric LRTIs, the nasopharynx microbiota serves as a valid proxy for lower respiratory tract microbiota. Eligible children were aged between 4 weeks and 5 years and were admitted to the paediatric intensive care unit (PICU) at Wilhelmina Children's Hospital (Utrecht, Netherlands) for a WHO-defined LRTI requiring mechanical ventilation (we refer to this group as the PICU cohort; appendix).10 Exclusion criteria are detailed in the appendix.
Over the same period, we also did a prospective, matched case-control study with the same inclusion and exclusion criteria used for the PICU cohort. Cases were recruited from three Dutch teaching hospitals (Spaarne Hospital, Hoofddorp; Kennemer Gasthuis, Haarlem; and St Antonius Hospital, Nieuwegein). For each case, two age-matched, sex-matched, and time-matched healthy controls were recruited from a pool of 1052 healthy children aged 4 weeks–5 years who were recruited through well-baby clinics and the local municipalities (appendix). When a healthy child seemed to be a proper match for a new case, they were contacted and visited at home within 2 weeks of admission of the case.
Both studies were approved by the Dutch National Ethics Committee. The case-control study conformed to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines for reporting of case-control studies (appendix).11 Written informed consent was obtained from the parents of all participants. Further details about both studies are in the appendix.
Procedures
In the PICU cohort, nasopharyngeal swabs and endotracheal aspirates were obtained within 4 h of intubation by trained nurses. In the case-control study, nasopharyngeal swabs were taken in cases generally within 1 h of admission to the paediatric ward, and during home visits to controls. Extensive medical histories were taken and demographic, lifestyle, and environmental data were collected for all children in both studies. Two expert paediatricians (MAvH and MEM) independently classified all cases in the case-control study as one of three major disease phenotypes: pneumonia, bronchiolitis, or wheezing illness. Cases with an unclear phenotype were classified as mixed. Disagreement between the paediatricians was solved by consensus. These classifications were based on the entire medical record, including all clinical notes at and during admission, and the results of any laboratory assessments or imaging that were done.
Bacterial DNA was isolated from samples as previously described.12 Amplification of the V4 hypervariable region of the 16S rRNA gene was done with barcoded universal primer pair 533F/806R. Amplicon pools were sequenced with the Illumina MiSeq platform (San Diego, CA, USA) and processed in our bioinformatics pipeline as previously described.8 To avoid operational taxonomic units (OTUs) with identical annotations, we refer to OTUs by their taxonomical annotations combined with a rank number based on the abundance of each one. In case when an OTU could not be confidently annotated as either of two species, both species are indicated and separated by a solidus. Viruses were genetically profiled with qualitative multiplex real-time PCR (RespiFinder SMARTfast 22 [Maastricht, Netherlands]). S pneumoniae, Staphylococcus aureus, H influenzae, and Moraxella catarrhalis were identified by quantitative PCR.
Statistical analysis
All analyses of matched samples accounted for the matched nature of the samples. A p value of less than 0·05 or a Benjamini-Hochberg adjusted q value of less than 0·05 was considered significant. Significance of differences in baseline characteristics and viral detection were calculated with conditional logistic regression. Non-metric multidimensional scaling plots were based on Bray-Curtis dissimilarity matrices; significance was calculated with adonis (vegan). To assess concordance between the bacterial microbiota composition of the nasopharynx and endotracheal aspirates, we compared the intra-individual and inter-individual Bray-Curtis similarity, which was calculated as 1–the Bray-Curtis dissimilarity. Statistical significance was established with the Wilcoxon rank-sum test. The correlation of individual bacterial taxa in paired samples was determined by Pearson's correlation.
We assessed host characteristics associated with microbiota composition with a stepwise selected distance-based redundancy analysis,13 and projected them in non-metric multidimensional scaling plots by using envfit (vegan). We did hierarchical clustering as described previously.9 Random forest analyses were used to establish biomarker species that most discriminate between clusters (VSURF).14 We used metagenomeSeq and cross-validated VSURF analyses to identify specific microbial taxa associated with cases or controls.15 Sparse random forest classifier analyses were done on the bacterial data, viral data, metadata, and the combination of all three datasets. We assessed the performance of these classifiers by calculating the area under the receiver operating characteristic curve with the out-of-bag predictions for classification as previously described.16 Because the potential real-world application of these classification models requires robust determination of biomarker bacteria, we also built the classification models with OTUs merged at the genus level. These analyses were done for the entire case-control cohort and were repeated in part for each of the phenotypes independently.
We built a cross-validated sparse random forest prediction model to investigate the extent to which duration of hospitalisation as a measure of disease severity could be predicted with all available data (caret). As a second measure of disease severity, we stratified cases according to physicians' decision to treat with antibiotics during admission (Dutch paediatricians generally reserve antibiotic treatment for children with clinically severe LRTIs) and did separate post-hoc analyses accordingly. As a third measure of severity, we did a post-hoc analysis of the nasopharyngeal data of our PICU cohort in relation to an age-matched and season-matched subset from our case-control cohort. Data analyses were done in R (version 3.2).
Role of the funding source
The study funders had no role in study design; data collection, analysis, or interpretation; or writing of the report. The corresponding author had full access to all study data and final responsibility for the decision to submit for publication.
Results
29 patients were enrolled in the PICU cohort. Viral presence in paired nasopharyngeal and endotracheal aspirates was almost in full agreement (96% [95% CI 93–99]). Bacterial microbiota of paired samples showed good concordance in composition (median within Bray-Curtis similarity 0·61) and low inter-participant concordance (0·10; p<0·0001; appendix). Furthermore, we noted a significantly correlated Shannon diversity (Pearson's r 0·66; p<0·0001). 58 taxa (combined relative abundance of 80·1%) were strongly correlated in the paired samples (median Pearson's r 0·93 [IQR 0·62–0·99]; p<0·05 for all 58 taxa individually). Only three common constituents of the nasopharyngeal microbiota—Staphylococcus, Corynebacterium, and Dolosigranulum spp—were almost exclusively present in nasopharyngeal samples and absent from endotracheal aspirates (Pearson's r for all three <0·20; p>0·50 for all three individually; appendix). We identified no taxa in endotracheal samples that were not present in nasopharynx samples (appendix). When assessing whether there were differences in the relative abundance for individual taxa between nasopharyngeal samples and endotracheal aspirates, we found a significant result only for Corynebacterium propinquum (Kruskal-Wallis test; Benjamini-Hochberg adjusted q=0·004), Corynebacterium macginleyi/accolens (q=0·019), Dolosigranulum pigrum (q=0·003), and three very low abundant taxa (median relative abundance <0·1%). 20 (69%) people in the PICU cohort had suspected bacterial infection, and 16 (55%) had a culture-confirmed bacterial infection. The other nine (31%) had suspected viral infection. Five (27%) received antibiotic treatment before sampling. There was no difference in overall microbiota composition between those in the PICU cohort with or without bacterial infection, or between those who did and did not receive antibiotics before sampling (appendix). Furthermore, stratified analysis did not show any difference between subcohorts in the results of the concordance analyses (appendix).
We recruited 154 cases and 307 controls (for one case only one matched healthy control could be recruited from the database; table ) for our case-control study. 40% of cases and controls were female (table). Median age of cases was 13·6 months (IQR 4·9–27·4). Cases were significantly more likely to have a history of respiratory tract infections, wheezing symptoms, used antibiotics in the past 6 months, and been exposed to tobacco smoke than controls (table). Controls were breastfed for at least 3 months more often than were cases, and the education level of parents of controls was higher than that of cases (table).
Table.
Baseline characteristics of cases and matched controls
| Cases (n=154) | Controls (n=307) | p value* | ||
|---|---|---|---|---|
| Sex | ||||
| Female | 61 (40%) | 122 (40%) | .. | |
| Male | 93 (60%) | 185 (60%) | .. | |
| Age, months | 13·6 (4·9–27·4) | 14·1 (5·3–28·4) | .. | |
| Born at term | 142 (92%) | 294 (96%) | 0·111 | |
| Mode of delivery | .. | .. | 0·457 | |
| Vaginal | 124 (81%) | 260 (85%) | .. | |
| Elective caesarean section | 15 (10%) | 26 (8%) | .. | |
| Emergency caesarean section | 15 (10%) | 21 (7%) | .. | |
| Season of sampling | ||||
| Spring | 50 (32%) | 91 (30%) | .. | |
| Summer | 22 (14%) | 44 (14%) | .. | |
| Autumn | 8 (5%) | 19 (6%) | .. | |
| Winter | 74 (48%) | 153 (50%) | .. | |
| Medical history | ||||
| Lower respiratory tract infection† | 38 (25%) | 22 (7%) | <0·0001 | |
| Wheezing | 41 (27%) | 22 (7%) | <0·0001 | |
| Otitis | 38 (25%) | 46 (15%) | 0·008 | |
| Hospitalisation for respiratory tract infection | 33 (21%) | 10 (3%) | <0·0001 | |
| Antibiotics during previous 6 months | 41 (27%) | 19 (6%) | <0·0001 | |
| Breastfeeding >3 months‡ | 58 (38%) | 169 (55%) | 0·0003 | |
| Parents' education level§ | .. | .. | <0·0001 | |
| High | 99 (64%) | 262 (85%) | .. | |
| Intermediate | 49 (32%) | 42 (14) | .. | |
| Low | 6 (4%) | 3 (1%) | .. | |
| Number of siblings | 1·0 (1·0–2·0) | 1·0 (0·0–1·0) | 0·002 | |
| Tobacco smoke exposure | 36 (23%) | 44 (14%) | 0·015 | |
Data are n (%) or median (IQR). Data for medication use were acquired from pharmacy printouts, whereas the rest of the data were acquired from parent questionnaires. Matching factors were not tested.
Calculated by univariate conditional logistic regression.
Included any parent-reported lower respiratory tract infections.
Breastfeeding did not have to be exclusive.
Education level was classed as low (primary school education or pre-vocational education as highest qualification), intermediate (selective secondary education or vocational education), or high (a degree from a university of applied sciences or an academic university).
In our control cohort, the composition of the respiratory microbiota was significantly associated with month of sampling (R2 6·2%), age (R2 4·2%), day-care attendance, breastfeeding, parent-reported history of respiratory tract infections in the participant, and previous antibiotic treatment within the past 6 months, among others (all p<0·05; appendix). Sex was not correlated with microbiota composition in the control cohort (R2 0·5%; p=0·35).
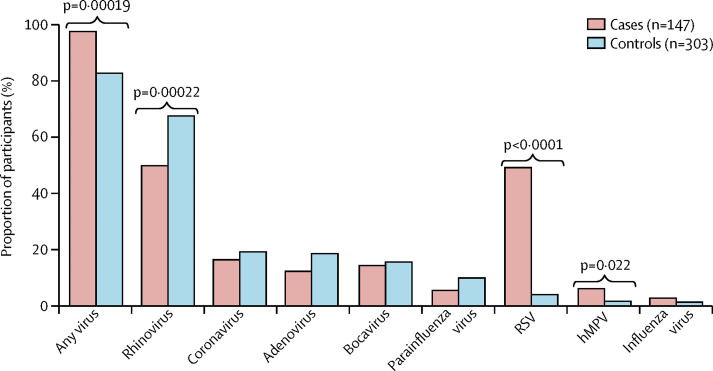
Viral data were available for 147 cases and 303 controls (appendix). We detected one or more viruses in 143 (97%) cases and 250 (83%) controls (p<0·00019; figure 1 ). There was a mean of 1·6 viruses per case sample and of 1·4 per control sample (p=0·04). The most commonly detected viruses overall were rhinovirus, coronaviruses, respiratory syncytial virus, and adenoviruses (figure 1). Influenza was relatively rare in both groups (figure 1). Respiratory syncytial virus (72 [49%] vs 12 [4%]; p<0·0001), and human metapneumovirus (9 [6%] vs 5 [2%]; p=0·022) were present significantly more often in LRTI cases than in controls (figure 1). Rhinovirus was detected more often in controls than in cases (204 [67%] vs 73 [50%]; p=0·00022; figure 1).
Figure 1.
Virus detected by quantitative PCR in cases and matched controls
RSV=respiratory syncytial virus. hMPV=human metapneumovirus.
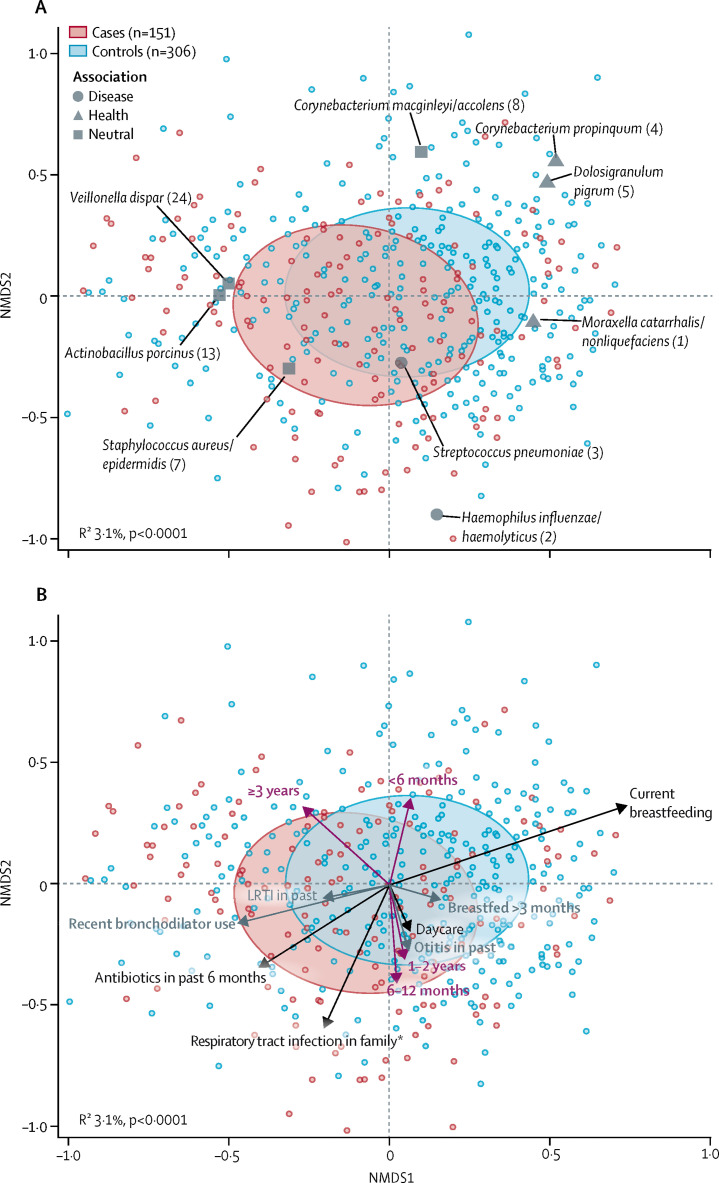
With respect to bacterial microbiota, although cases did not have a higher median bacterial biomass than controls (54·1 pg/μL [IQR 9·8–147·4] vs 54·5 pg/μL [16·7–165·2]; p=0·28), microbiota composition differed significantly between groups (R2 3·1%; p<0·0001; figure 2A ). Projection of the vectors for host characteristics associated with microbiota composition showed that previous antibiotic use in the past 6 months, bronchodilator use in the past 3 months, and a parent-reported history of respiratory tract infection in the family pointed in the direction of disease (figure 2B).
Figure 2.
NMDS biplots of individual nasopharyngeal microbiota composition at admission in cases with LRTIs and in matched controls
(A) shows the nine bacterial species biomarkers determined by random forest analysis on hierarchical clustering results, whereas (B) shows a posteriori projection of covariates that significantly explained the compositional variation between cases and controls (grey represents significance in univariable analysis, and black significance in multivariable analysis) and the association with age (purple). Ellipses represent the SD for all points within each cohort. Stress=0·269. In (A), operational taxonomic units of bacterial species are referred to by their taxonomical annotations and a rank number (shown in parentheses), which is based on the abundance of each given operational taxonomic unit. For readability, only a selection of the covariates explaining the largest variations between cases and controls are displayed in (B). In (B), the age effect (vertical orientation for younger vs older participants) was roughly perpendicular to the disease–health axis (horizontal orientation), showing that age-related differences in microbiota composition per se are not associated with disease. NMDS=non-metric multidimensional scaling. LRTIs=lower respiratory tract infections. *At time of sampling of the participant, at least one family member was experiencing a respiratory tract infection.
Bacterial datasets were available for 151 cases and 306 controls (appendix). We noted seven distinct microbiota profiles—ie, Staphylcoccus aureus/epidermidis, C macginleyi/accolens, Haemophilus influenzae/haemolyticus, Moraxella catarrhalis/nonliquefaciens; Veillonella dispar and Actinobacillus porcinus, S pneumoniae; and C propinquum and D pigrum—within the cases and controls (hierarchical clustering; appendix). The profiles dominated by H influenzae/haemolyticus (137 [30%] of 457 samples) or by S pneumoniae (28 [6%]) were significantly associated with LRTI cases, whereas those dominated by M catarrhalis/nonliquefaciens (216 [47%]), or by C propinquum and D pigrum (44 [10%]) were significantly associated with health (χ2 test p<0·05 for all; appendix). A posteriori plotting of the biomarker species of these clusters in the non-metric multidimensional scaling ordination further supported the associations between profiles and health or disease (figure 2A). The profile dominated by H influenzae/haemolyticus (median bacterial load 126 pg/μL; appendix) had a significantly higher bacterial load than all other profiles (Wilcoxon rank-sum test p<0·05 for all comparisons) except for that dominated by S aureus/epidermidis (p=0·25). The median bacterial load of the S-pneumoniae-dominated profile (67 pg/μL) was significantly higher than that of the profile dominated by C propinquum and D pigrum (15 pg/μL; p=0·002), but did not differ from that of the profile dominated by M catarrhalis/nonliquefaciens (35 pg/μL; p=0·40).
On an individual bacterial taxon level, 49 taxa differentiated cases from controls (combined relative abundance 83·5%). This between-group difference was confirmed for 17 of these bacteria by cross-validated random forest analysis (appendix). Among the differentially abundant taxa, we noted a significantly higher abundance of H influenzae/H haemolyticus, S pneumoniae, Actinomyces spp, and Prevotella spp in LRTI cases than in controls, and a significantly higher abundance of different Moraxella spp, C propinquum, D pigrum, and Helcococcus spp in controls than in cases (appendix).
MAvH and MEM classified 37 cases as pneumonia, 57 as bronchiolitis, 34 as wheezing illness, and 26 cases as mixed (appendix). Respiratory syncytial virus was more common in cases with bronchiolitis (34 [62%] of 55 vs four [4%] of 112; p<0·0001), cases with pneumonia (18 [56%] of 32 vs three [4%] of 73; p=0·00057), and cases with mixed phenotypes (15 [58%] of 26 vs 1 [2%] of 52; p=0·001) than in controls (appendix). Rhinovirus was significantly less common in cases of pneumonia and bronchiolitis than in controls (appendix). Human metapneumovirus was detected only in pneumonia and bronchiolitis cases (appendix).
When we stratified by clinical phenotype, the overall bacterial microbiota composition again differed significantly between cases and controls for each presentation (p≤0·004 for all; appendix). The differential abundance of individual microbes between cases and controls was highly similar for each phenotype (appendix). In all phenotypes, Haemophilus, Neisseria, and oral taxa (eg, Actinomyces) were overrepresented and multiple Moraxella, Dolosigranulum, and Helcococcus spp were underrepresented in cases compared with controls (appendix). The differences in microbiota composition between cases and controls in the mixed-phenotype group largely overlapped with those for the three other phenotypes (appendix).
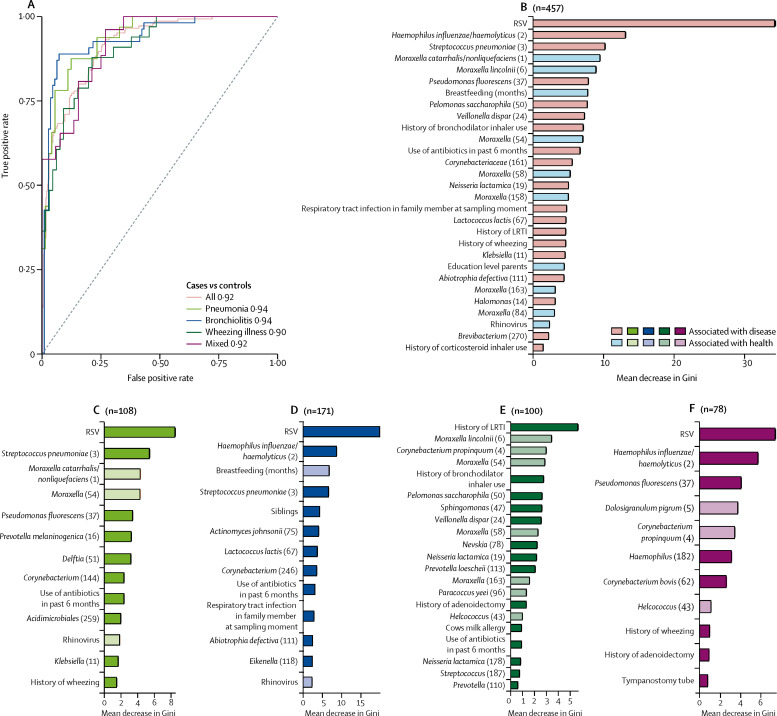
When we combined viral and bacterial biomarkers with host factors in a sparse random forest analysis, the accuracy of classification of LRTI versus health was very high (area under the curve [AUC] 0·92; figure 3A ).
Figure 3.
ROC curves for distinguishing disease from health for unstratified and stratified sparse random forest classifying models on the basis of 16S rRNA data, viral presence, and patient characteristics (A), and the disease-discriminatory variables that these models encompass (B–F)
The random forest models include all cases (B), pneumonia cases (C), bronchiolitis cases (D), wheezing illness cases (E), and mixed cases (F) versus healthy controls. In (B)–(F), the x-axis shows the importance of the variable to the accuracy of the model, which was estimated by calculating the mean decrease in Gini after randomly permuting the values of each given variable (mean and SD, 100 replicates); the direction of the associations was estimated post hoc with point biserial correlations. Because multiple OTUs of individual bacterial species were identified, we refer to OTUs by their taxonomical annotations and a rank number (shown in parentheses), which is based on the abundance of each given OTU. ROC=receiver operating characteristic. RSV=respiratory syncytial virus. LRTIs=lower respiratory tract infection. OTU=operational taxonomic unit.
Important predictors of disease were, among others, the presence of respiratory syncytial virus, a high abundance of H influenzae/haemolyticus, S pneumoniae, and Pseudomonas fluorescens, and low abundance of several Moraxella spp, antibiotic treatment in the past 6 months, and lack of breastfeeding (figure 3B). This combined classification system outperformed the models based on bacterial microbiota alone (AUC 0·77), viral microbiota alone (0·70), child characteristics alone (0·80), or the model in which only the two classically most important pathobionts—ie, respiratory syncytial virus and S pneumoniae—were included (AUC 0·75). External validation of our classifier model in the PICU cohort showed correct classification in 92% of nasopharyngeal samples and 100% of endotracheal aspirates. Separate models for each of the phenotypes showed equally high accuracy in classification of LRTIs (AUC 0·90–0·94; figure 3A, C–F). To test more broad and universally applicable classification models with bacterial microbiota data clustered on a genus level instead of an OTU level, classification of the presence of LRTI versus health was very high (entire cohort AUC 0·92, phenotype-specific AUC 0·86–0·94; appendix).
Physicians prescribed antibiotic treatment after sampling of the nasopharynx in 43 (28%) of 154 cases (29 [78%] of 37 pneumonia cases, four [7%] of 57 bronchiolitis cases, four [11%] of 37 cases of wheezing illness, and six [23%] of 26 cases of mixed infection). In post-hoc analyses, viral presence in the microbiota at admission did not differ between infants prescribed antibiotics and those not prescribed antibiotics (appendix). With respect to bacterial ecology, we noted similar but slightly more pronounced differences in microbiota compositions between children prescribed antibiotics and matched controls (R2 5·8%; p<0·0001) than between those not prescribed antibiotics and matched controls (2·6%; p<0·0001; appendix). Children prescribed antibiotics did not have increased abundances of pathobionts such as H influenzae/haemolyticus, or S pneumoniae, but had increased abundances of oral taxa, such as Veillonella, Prevotella, and Actinomyces spp (appendix).
Duration of hospitalisation could be predicted fairly accurately using data available at admission by a random forest model including 14 viral, bacterial, and host characteristics (Pearson's r 0·50, p<0·0001; appendix). Predictors from highest to lowest importance were younger age and abundance of C propinquum, Neisseria, and S aureus/epidermidis (appendix). When only children not prescribed antibiotics were included, prediction of hospitalisation duration at admission became stronger (Pearson's r 0·55, p<0·0001), whereas predictive ability was lost when only those prescribed antibiotics were included (p=0·73; appendix).
The overall microbiota compositions of the PICU cases was more substantially different from those of healthy controls (R2 5·6%; p<0·0001) than from those of cases from the case-control cohort (R2 4·2; p<0·0001; appendix). Furthermore, the PICU cases had more pronounced overrepresentation of several Haemophilus, Streptococcus (including S pneumoniae), Veillonella, and Actinomyces spp, and more pronounced underrepresentation of multiple Moraxella spp, and especially of Dolosigranulum and Corynebacterium spp, than the cases compared with healthy controls (appendix).
Discussion
In this matched case-control study, we showed a strong association between upper respiratory tract microbiota and the presence and severity of childhood LRTIs. We also showed that LRTIs can be uniquely differentiated from health by combined viral, bacterial, and lifestyle or environmental predictors, underlining the multifactorial pathophysiology of childhood LRTI. Furthermore, we showed that this predictive ability is largely independent from the clinical phenotype.
The upper respiratory tract microbiome is generally thought to be the source of LRTIs in childhood,5 although evidence supporting this link is scarce (particularly in young children). Here, we showed that, in line with previous work, there is high intra-individual concordance of viral17, 18 and bacterial19, 20 microbiota profiles between nasopharyngeal and endotracheal aspirate samples in patients with LRTI admitted to a PICU. The Bray-Curtis similarity of 0·61 approximates that of biological replicates (ie, two sequentially obtained lavages from the same lung lobe of the same child) of microbiota profiles of the lungs.20 This finding suggests not only that the upper respiratory microbiota is the source for the lower respiratory tract, but also that, except for a few commensal species, microbial colonisation and proliferation in the nasopharynx parallels that in the lower airways during childhood LRTIs. Therefore, our findings support the idea that upper respiratory tract samples can be used as a proxy for lung microbiota in childhood LRTIs.
Next, in our unselected, strictly matched case-control cohort, we showed a strong association between nasopharyngeal microbiota composition and the presence of childhood LRTIs. Viral presence was ubiquitous in both cases and controls, with respiratory syncytial virus and, to a lesser extent, human metapneumovirus, highly overrepresented in cases, in line with the results of studies of the viral causes of childhood LRTIs.21 The presence and abundance of Haemophilus spp, S pneumoniae, and oral species were strongly associated with disease, in line with previous reports linking these taxa to susceptibility to, and severity of, respiratory tract infections in children.6, 7, 22, 23 By contrast, the abundance of potentially beneficial bacteria like Moraxella, Corynebacterium, Dolosigranulum, and Helcococcus spp were underrepresented in cases, in line with previous reports connecting these genera with prevention of infections.6, 7, 12, 24 By combining viral, bacterial, and host-related predictors, we were able to differentiate children with LRTIs from strictly matched healthy controls. The accuracy of prediction of infection was greatly diminished when individual predictors were used, underlining the multifactorial pathophysiology of childhood LRTIs. The contribution of the nasopharyngeal microbiota, both bacterial and viral, seems to be largely independent of clinical presentation, and even holds for bronchiolitis and wheezing illness, which are generally assumed to have a viral cause.25
Results from our case-control study were confirmed independently in a second cohort, which showed that Corynebacterium and Dolosigranulum were nearly absent in children with LRTIs admitted to the PICU, suggesting that these children especially had reduced resistance to overgrowth and dissemination of pathobionts to the lungs.5 Furthermore, in post-hoc analyses, oral species were associated with both the decision to treat with antibiotics and with duration of hospitalisation, which suggests that the abundance of these species is associated with the severity of LRTIs.26 A possible mechanism is that Gram-negative oral bacteria promote a pro-inflammatory mucosal response,27 leading to an increase in catecholamines that in turn accelerates the growth of these same Gram-negative oral species and of potential pathogens such as Haemophilus spp and S pneumoniae.28, 29 Therefore, it would be interesting to study whether prescribing antibiotics on the basis of the abundance of oral bacteria in respiratory specimens would improve outcomes.
Our findings have three implications. First, the accuracy of our model in discriminating LRTIs from health suggests that microbiota-based diagnostics might have potential clinical application. Diagnostics for the detection of potentially pathogenic viruses and bacteria cover only some pathobionts and discriminate poorly between asymptomatic colonisation and causes of symptomatic disease. If a microbiota-based diagnostic or classification tool could improve accuracy and discriminatory power, it would have major implications for treatment protocols. A proof-of-principle study of rapid microbiota-based diagnosis (<12 h) of severe pneumonia in adults showed that such diagnostic tools improve diagnostic accuracy and could be clinically applied.30 If the cost of such technology decreases further and becomes available for paediatric use, use of broad-spectrum antibiotics could be avoided more often, and the most abundant or overgrowing species could be targeted with narrow-spectrum drugs.31 Although our microbiota-based approach has to be validated in independent cohorts, the similar performance of the genus-level model suggests potential for future development of universal or country-based or region-based models for the prediction of severity and duration of disease by combined microbiota and host characteristics. Such a model would potentially allow physicians to increase or decrease the threshold for antimicrobial treatment depending on the predicted outcome.
Second, the finding that specific groups of microorganisms are associated with health, in line with data from studies from around the world,6, 8, 32, 33, 34 suggests that studies should be done to obtain mechanistic insight into the potential role of these species in disease prevention. Corynebacterium spp have been reported to reduce virulence of S aureus and inhibit growth of S pneumoniae in vitro.35, 36 Furthermore, nasal application of Corynebacterium spp induced resistance against respiratory syncytial virus and secondary pneumococcal pneumonia in infant mice.37 These findings emphasise the need for future research efforts to assess the combined effects of these commensal bacteria in modulation of the respiratory ecosystem, especially the containment of potential pathogens such as respiratory syncytial virus, Haemophilus spp, and Streptococcus spp, and of host immune responses underlying respiratory symptoms.
Third, the phenotype-independent association of viral and bacterial microbiota with LRTIs parallels the highly overlapping clinical presentation of disease in children, which means that a robust gold standard for accurate classification and treatment of LRTIs is not available.38 Our findings contribute to an emerging body of evidence suggesting that viruses contribute to presumed bacterial pneumonia39 and that bacteria seem to have an important role in the pathogenesis and severity of presumed viral bronchiolitis9 and wheezing illness.40 These findings show the inappropriateness of conventional single-bacteria and single-virus causation per Koch's postulates. Our findings also allude to the hypothesis that there is a universal pathway for the development of clinical LRTIs, linked to microbial dysbiosis, whereby clinical phenotypes are driven more by host (eg age, anatomy, baseline mucosal inflammation, status of innate and adaptive immunity, genetic background) and environmental characteristics rather than by single pathogen characteristics. Thus, treatment decisions should not be based on clinical phenotype but rather on disease severity. This scientific debate is only beginning, and, in addition to confirmatory studies of our results, many discussions among and between clinicians, microbiologists, and biologists need to take place. However, diagnostic and treatment protocols could be adapted within the next 5 years if such changes are judged to be appropriate.
The major strength of our study is the strictly matched case-control design, which precludes bias as a result of the confounding effects of age, time, and sex. Furthermore, the unselected recruitment of cases should have given us a cohort that is highly representative of the patients treated by paediatric clinicians, and the consistent patterns in our unsupervised (ie, hierarchical clustering) and supervised (ie, metagenomeSeq) analyses contribute to the robustness of our results. Our study also has limitations. First, case-control designs could theoretically introduce selection bias that could affect the validity and reproducibility of our results. Second, quantitative PCR-based assays detect only known respiratory viruses rather than the entire respiratory virome. However, virome studies report a high concordance between the results of metagenomic sequencing and quantitative PCR-based assays.41 Third, as with any observational study, our findings do not necessarily prove causality. Longitudinal analyses are underway to address the causality of respiratory microbiota in respiratory disease. Fourth, endotracheal aspirate might not perfectly reflect the lower respiratory tract microbiota extending into the bronchi and alveoli. That said, clinical evidence based on conventional microbiology data has suggested that tracheal aspirates are a good proxy for the lower respiratory tract, and therefore an appropriate proxy for the clinical diagnosis of cause of disease in children with severe LRTIs.42 Furthermore, data published in 2017 showed a strong concordance with negligible differences between bacterial microbiota from endotracheal samples and those from bronchial lavages.43 Finally, 16S rRNA sequencing permits annotation up to the in between genus-level and species-level identification of bacteria, but does not provide the resolution of metagenomic techniques (eg, shotgun sequencing), especially for closely related species, such as streptococcal species. We tried to provide some species-level data by using quantitative PCR to confirm four common and potentially pathogenic OTUs, and these findings supported our conclusions. However, future studies might be needed on multiple levels to further confirm our data and refine the conclusions.
Overall, our findings suggest that microbiota-based diagnostics should be further explored. Additionally, our prediction model for severity of disease should be validated in different settings and countries to explore its usefulness for treatment optimisation and antimicrobial stewardship.
Data sharing
Sequence data that support the findings of this study have been deposited in the US National Center for Biotechnology Information Sequence Read Archive database (BioProject ID PRJNA428382).
Acknowledgments
Acknowledgments
We thank the participating children and their families, all the members of the research team at Spaarne Gasthuis Academy, the laboratory staff, the Streeklaboratorium Haarlem, the GGD Kennemerland, the JGZ Kennemerland, and Olga Krabbe for her primary exploration of the data. This work is dedicated to the memory of Dr Reinier H Veenhoven, who contributed to the planning and design of the study.
Contributors
MAvH, EAMS, and DB conceived and designed the study. WHM, MAvH, MEM, AMV, and NJGJ enrolled participants. MLJNC was responsible for execution and quality control of laboratory work. WHM and DB analysed the data. WHM, MAvH, EAMS, and DB wrote the paper. All authors interpreting results, critically revised the Article for important intellectual content, and approved the final version.
Declaration of interests
EAMS has received unrestricted research support from Pfizer, and grant support for vaccine studies from Pfizer and GlaxoSmithKline. DB has received unrestricted fees (paid to her institution) for advisory work for Friesland Campina, and research support from Nutricia and MedImmune. All other authors report no competing interests.
Supplementary Material
References
- 1.Liu L, Oza S, Hogan D. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet. 2016;388:3027–3035. doi: 10.1016/S0140-6736(16)31593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Study 2013 Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudan I. Epidemiology and etiology of childhood pneumonia. Bull World Health Organ. 2008;86:408–416. doi: 10.2471/BLT.07.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsolia MN, Psarras S, Bossios A. Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin Infect Dis. 2004;39:681–686. doi: 10.1086/422996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Man WH, de Steenhuijsen Piters WAA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teo SM, Mok D, Pham K. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biesbroek G, Tsivtsivadze E, Sanders EAM. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 8.Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med. 2017;196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 9.De Steenhuijsen Piters WAA, Heinonen S, Hasrat R. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO IMCI chart booklet. http://www.who.int/maternal_child_adolescent/documents/IMCI_chartbooklet/en/
- 11.von Elm E, Altman DG, Egger M. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. PLoS Med. 2007;4:e296. doi: 10.1371/journal.pmed.0040296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prevaes SMPJ, de Winter-de Groot KM, Janssens HM. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med. 2016;193:504–515. doi: 10.1164/rccm.201509-1759OC. [DOI] [PubMed] [Google Scholar]
- 13.Blanchet FG, Legendre P, Borcard D. Forward selection of explanatory variables. Ecology. 2008;89:2623–2632. doi: 10.1890/07-0986.1. [DOI] [PubMed] [Google Scholar]
- 14.Genuer R, Poggi J-M, Tuleau-Malot C. Variable selection using random forests. Pattern Recognit Lett. 2010;31:2225–2236. [Google Scholar]
- 15.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vatanen T, Kostic AD, D'Hennezel E. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van de Pol AC, Wolfs TFW, van Loon AM. Molecular quantification of respiratory syncytial virus in respiratory samples: reliable detection during the initial phase of infection. J Clin Microbiol. 2010;48:3569–3574. doi: 10.1128/JCM.00097-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins SM, Webb DL, Torrance SA. Comparison of a real-time reverse transcriptase PCR assay and a culture technique for quantitative assessment of viral load in children naturally infected with respiratory syncytial virus. J Clin Microbiol. 2005;43:2356–2362. doi: 10.1128/JCM.43.5.2356-2362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassis CM, Erb-Downward JR, Dickson RP. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037–e00045. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marsh RL, Kaestli M, Chang AB. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. 2016;4:37. doi: 10.1186/s40168-016-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain S, Self WH, Wunderink RG. Community-acquired pneumonia requiring hospitalization among US adults. N Engl J Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. MBio. 2011;2:e00245–e00e10. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brook I. Prevotella and Porphyromonas infections in children. J Med Microbiol. 1995;42:340–347. doi: 10.1099/00222615-42-5-340. [DOI] [PubMed] [Google Scholar]
- 24.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012;78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO . 2nd edn. World Health Organization; Geneva: 2013. Pocket book of hospital care for children: guidelines for the management of common illnesses with limited resources. [PubMed] [Google Scholar]
- 26.Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. 2014;2:238–246. doi: 10.1016/S2213-2600(14)70028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson LR, Sanders JG, McDonald D. A communal catalogue reveals Earth's multiscale microbial diversity. Nature. 2017;551:457. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marks LR, Davidson BA, Knight PR, Hakansson AP. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. MBio. 2013;4:e00438–e00513. doi: 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Donnell PM, Aviles H, Lyte M, Sonnenfeld G. Enhancement of in vitro growth of pathogenic bacteria by norepinephrine: importance of inoculum density and role of transferrin. Appl Environ Microbiol. 2006;72:5097–5099. doi: 10.1128/AEM.00075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pendleton KM, Erb-Downward JR, Bao Y. Rapid pathogen identification in bacterial pneumonia using real-time metagenomics. Am J Respir Crit Care Med. 2017;196:1610–1612. doi: 10.1164/rccm.201703-0537LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogaert D, van Belkum A. Antibiotic treatment and stewardship in the era of microbiota-oriented diagnostics. Eur J Clin Microbiol Infect Dis. 2018;37:795–798. doi: 10.1007/s10096-018-3198-6. [DOI] [PubMed] [Google Scholar]
- 32.Biesbroek G, Bosch AATM, Wang X. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190 doi: 10.1164/rccm.201401-0073OC. 140612135546007. [DOI] [PubMed] [Google Scholar]
- 33.Luna PN, Hasegawa K, Ajami NJ. The association between anterior nares and nasopharyngeal microbiota in infants hospitalized for bronchiolitis. Microbiome. 2018;6:2. doi: 10.1186/s40168-017-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salter SJ, Turner C, Watthanaworawit W. A longitudinal study of the infant nasopharyngeal microbiota: the effects of age, illness and antibiotic use in a cohort of South East Asian children. PLoS Negl Trop Dis. 2017;11:e0005975. doi: 10.1371/journal.pntd.0005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol. 2016;7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio. 2016;7:e01725–e01815. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanmani P, Clua P, Vizoso-Pinto MG. Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front Microbiol. 2017;8:1613. doi: 10.3389/fmicb.2017.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott JAG, Wonodi C, Moïsi JC. The definition of pneumonia, the assessment of severity, and clinical standardization in the Pneumonia Etiology Research for Child Health study. Clin Infect Dis. 2012;54(suppl 2):S109–S116. doi: 10.1093/cid/cir1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosch AATM, Biesbroek G, Trzcinski K, Sanders EAM, Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9:e1003057. doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beigelman A, Bacharier LB. Early-life respiratory infections and asthma development. Curr Opin Allergy Clin Immunol. 2016;16:172–178. doi: 10.1097/ACI.0000000000000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lysholm F, Wetterbom A, Lindau C. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS One. 2012;7:e30875. doi: 10.1371/journal.pone.0030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCauley LM, Webb BJ, Sorensen J, Dean NC. Use of tracheal aspirate culture in newly intubated patients with community-onset pneumonia. Ann Am Thorac Soc. 2016;13:376–381. doi: 10.1513/AnnalsATS.201506-368BC. [DOI] [PubMed] [Google Scholar]
- 43.Dickson RP, Erb-Downward JR, Freeman CM. Bacterial topography of the healthy human lower respiratory tract. MBio. 2017;8:e02287–e02316. doi: 10.1128/mBio.02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the US National Center for Biotechnology Information Sequence Read Archive database (BioProject ID PRJNA428382).