To the Editor,
When facing the global health emergency of COVID-19 (the illness caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)), as declared by the WHO at the end of January 2020 [1], clinical microbiology laboratories worldwide are ideally asked to refine their diagnostic paraphernalia to maximize direct detection of SARS-CoV-2 in patient samples. Reducing the rate of undocumented SARS-CoV-2 infections and their contagiousness is crucial to sustain COVID-19 containment measures [2]. Accordingly, ever-increasing numbers of SARS-CoV-2-infected people and asymptomatic carriers—in whom virus shedding may depend on their viral loads [3]—pose the need to adapt laboratory response capacity to this unprecedented situation [4].
Unlike for national reference laboratories, it is difficult for many routine laboratories to tailor and/or redirect daily activity to COVID-19 as rapidly as the person-to-person transmission of SARS-CoV-2 is occurring. Nevertheless, major efforts are required for those laboratories—such as ours—serving hospitals that are redesigning their configuration to convert ordinary wards into ‘COVID’ wards as much as possible. Consequently, laboratories are not only asked for early detection of patients infected with SARS-CoV-2 but are also asked to confirm COVID-19 cases so as to identify and quarantine patients as early as possible [4].
We describe a real-life experience at the Fondazione Policlinico Universitario A. Gemelli (FPG) IRCCS, a large teaching hospital in Rome (Italy), which underwent a progressive adaptation of our clinical microbiology laboratory in response to the rapidly evolving COVID-19 emergency in the Lazio region, during March 2020. This experience started on March 1—when our regional government authorized laboratories, other than the national reference laboratory in Rome, to perform real-time PCR assays to detect SARS-CoV-2 RNA in nasopharyngeal swab samples—and ended on March 24 (i.e. at the time of writing).
We identified three decisive interventions that contributed to optimizing our COVID-19 diagnostic laboratory workflow, and we assessed the turnaround time (TAT) values for positive or negative results as key indicators of efficiency. We performed viral RNA detection in all samples using the Korea Ministry of Food and Drug Safety approved Allplex 2019-nCoV assay (Arrow Diagnostics S.r.l., Genova, Italy)—a single-tube assay able to detect the three target genes (E gene, RdRP gene and N gene) as in the WHO recommended protocols. Hence, TAT (the time from sample receipt in the laboratory to release of PCR result) was dependent either on the time taken for pre-PCR steps or on laboratory working hours (12 h/day versus 24 h/day). Overall, we processed 5074 samples of patients suspected for SARS-CoV-2 infection—who were admitted to FPG and other Lazio hospitals—with a minimum of five to a maximum of 466 samples per day and with a median TAT value of 8 h (interquartile range (IQR) 7–10.75 h). The number of samples tested for SARS-CoV-2 greatly exceeded the number of samples routinely tested for most common respiratory viruses. For example, in February 2020, we processed 547 samples for viral agents other than SARS-CoV-2.
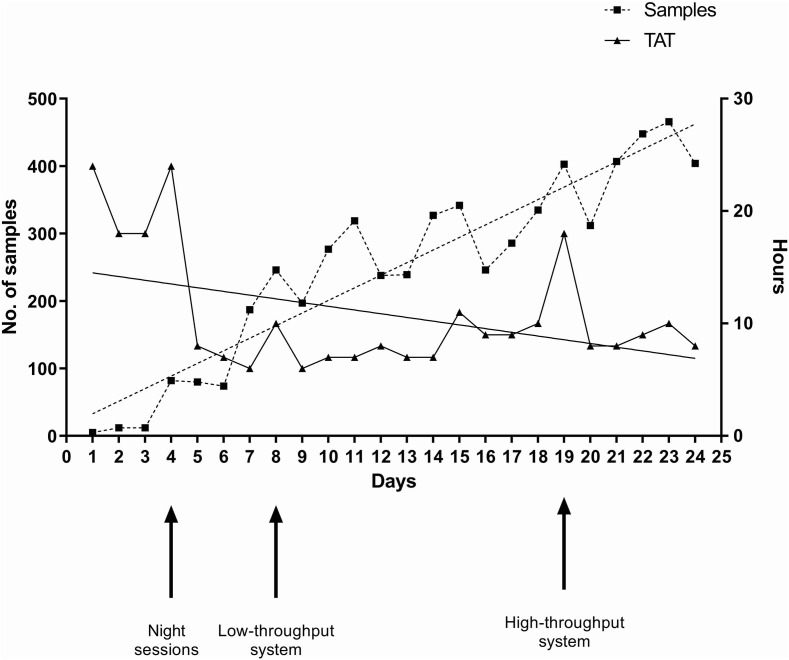
In particular, we started using the Qiagen EZ1 Advanced XL system (14 samples per run; Qiagen, Hilden, Germany) to automatically extract sample RNA before performing the Allplex 2019-nCoV assay (two runs/12 h). As shown in Fig. 1 , TAT increased (median values, 18 h (IQR 14.50–20.50 h) to 24 h (IQR 21–27 h)) up to 6 March, when we decided to extend working hours to night shifts (7 p.m. to 7 a.m.). Following this intervention (the first intervention), TAT decreased (median values, 6 h (IQR 4–8 h) to 8 h (IQR 6–9 h)) until 10 March, when we decided to use the Seegene Nimbus automated system—that uses the STARMag Universal Cartridge kit—(40 samples per run) for RNA extraction and PCR assay set up. This intervention (the second) allowed TAT to return to median values of 6 h (IQR 4–9 h) to 8 h (IQR 5–10 h). On 21 March, TAT increased (median value, 18 h (IQR 12.75–21 h)), so a further intervention (the third) became necessary. This involved using the Qiagen QIAsymphony SP/AS automated system (96 samples per run) for RNA extraction and PCR assay set up. Once again, TAT returned to the previous values (8 h) and remained stable for the following days. Of note, changing from the Seegene Nimbus to the Qiagen QIAsymphony SP/AS led to a decrease of samples needing repeat testing (3.2% (84/2618) versus 2.2% (48/2191)), which was a consequence of invalid/undetermined results obtained over time.
Fig. 1.
Timeline of median turnaround time (TAT) values for the samples tested for direct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection during March 2020. Arrows indicate the laboratory's adaptive interventions to the progressively increasing number of samples that took place in the study period (1 March to 24 March). One intervention (first arrow) was to extend the working to night hours. The subsequent interventions involved a change from low-throughput (second arrow) to high-throughput (third arrow) systems for viral RNA extraction and PCR assay setup.
We documented the successful implementation of the Allplex 2019-nCoV assay into our routine laboratory workflow, which was directed to diagnose other (albeit less relevant) microbial pathogens. Hence, as experienced in other European laboratories [5], increasing the number of samples to be processed daily necessitated substantial changes in laboratory work organization. These changes did not involve the purchase or loan of equipment—the Qiagen EZ1, Seegene Nimbus and Qiagen QIAsymphony instruments were already available in the laboratory—but involved redesigning the instruments' use. Consequently, while continuing to perform the routine diagnostic testing (albeit with delayed TAT; data not shown), the existing laboratory staff, in cooperation with newly (ad hoc) employed personnel, directed their skills toward a new, laborious and expensive diagnostic challenge. In particular, a qualified (no specific training was necessary), tireless (extra working hours including holidays were needed) and robust (no one developed COVID-19) staff was composed of eight senior and three junior workers who were almost exclusively committed to the performance of SARS-CoV-2 testing. Of note, the new personnel worked only during the night shifts.
In conclusion, our findings support the concept that the clinical microbiology laboratory can be extraordinarily responsive to emergencies like the one we are experiencing. While providing a concrete example on how to expand the laboratory response capacity, we are aware that we still have to optimize our response to the COVID-19 outbreak.
Transparency declaration
The authors report no conflicts of interest relevant to this letter. No external funding was received for this study.
Acknowledgements
The authors would like to thank all members of the FPG COVID Laboratory Group: Margherita Cacaci, Elena De Carolis, Tiziana D'Inzeo, Barbara Fiori, Simona Galuppi, Liliana Giordano, Rosalia Graffeo, Marilisa La Rosa, Marilena La Sorda, Flora Marzia Liotti, Cecilia Martini, Cosima Marturano, Luca Masucci, Giulia Menchinelli, Rosario Nicotra, Ivana Palucci, Creola Rocchetti, Michela Sali, Mara Salvioni, Riccardo Torelli and Antonietta Vella. The authors are grateful to Franziska Lohmeyer for her English language assistance.
Editor: L. Leibovici
References
- 1.World Health Organization . 2020. Coronavirus disease (COVID-2019) situation reports.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ Available from: [Google Scholar]
- 2.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020 doi: 10.1126/science.abb3221. eabb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuen K.S., Ye Z.W., Fung S.Y., Chan C.P., Jin D.Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konrad R., Eberle U., Dangel A., Treis B., Berger A., Bengs K. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Euro Surveill. 2020;25:2000173. doi: 10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed] [Google Scholar]