Graphical abstract
Highlights
► Two new trypanosome strains were discovered in British bats. ► The new strains are closely related to New World strains. ► Old World and New World strains diverged within the last 11 million years. ► The true date of divergence is likely to be considerably more recent. ► Parasites indirectly indicate recent intercontinental movement of bats.
Abbreviations: MYA, million years ago; ML, maximum likelihood; GTR, general time reversible; FFLB, fluorescent fragment length barcoding; gGAPDH, glycosomal glyceraldehyde phosphate dehydrogenase; BEAST, Bayesian evolutionary analysis by sampling trees
Keywords: Phylogeny, Biogeography, Trypanosomatid, Fluorescent fragment length barcoding, Ribosomal DNA, GAPDH
Abstract
The global distribution of bat taxa indicates that the Atlantic and Pacific Oceans are effective barriers to movement between the Old and New Worlds. For instance, one of the major suborders, Yinpterochiroptera, has an exclusively Old World distribution, and within the other, Yangochiroptera, no species and only five genera are common to both. However, as bats are sometimes blown out to sea, and have colonised isolated islands, occasional natural movement between the New and Old Worlds does appear to be possible. Here we identify new genotypes of a blood parasite, Trypanosoma dionisii, in Old World bats that are closely related to South American strains. Using highly conservative calibration points, divergence of Old and New World strains is estimated to have occurred 3.2–5.0 million years ago (MYA), depending on the method used (upper 95% CL for maximum time 11.4 MYA). The true date of divergence is likely to be considerably more recent. These results demonstrate that taxon-specific parasites can indicate historical movements of their hosts, even where their hosts may have left no lasting phylogenetic footprint.
1. Introduction
Bats originated approximately 65 million years ago (MYA) (Eick et al., 2005, Jones et al., 2005), probably in Africa (Eick et al., 2005). Endemism is high at each taxonomic level (Teeling et al., 2005), and the patterns of distribution suggest that the geographic separation of the Old and New Worlds has been an effective barrier to the movement of bats. For example, one of the two proposed suborders, Yinpterochiroptera (which includes three microbat families: Rhinopomatidae, Rhinolophidae and Megadermatidae, as well as the megabat family Pteropodidae), has an exclusively Old World distribution. Within the other proposed suborder, Yangochiroptera, no species and only five genera are common to both (Eick et al., 2005, Teeling et al., 2005). Evidence for movement of bats between these regions is limited. New World species of the genus Myotis are estimated to have initially diverged 12.2 ± 2.0 MYA, indicating that Myotis arrived from the Old World before this (Stadelmann et al., 2007). It is not known when the other genera found in both the Old and New Worlds made the crossing.
Occasionally, New World bats are reported in Europe. Some, such as Myotis lucifugus, have been accidentally transported onboard ships (Constantine, 2003). Others are thought to have arrived naturally, apparently blown off-course by more than 1000 km during migrations. Examples include Hoary Bats (Lasiurus cinereus) that are occasionally found in Iceland, the Galapagos Islands, and Orkney Islands (see Constantine (2003) and references therein), and bats found in Bermuda, over 1000 km from the United States (Constantine, 2003). In addition, some have colonised isolated islands; the Hawaiian Hoary Bat (Lasiurus cinereus semotus) is thought to be most closely related to a subspecies of Hoary Bat from North America, implying natural movement over a distance of over 4000 km (Morales and Bickham, 1995). However, such movements are difficult to study as the distances travelled, the infrequency of movements, and the unlikelihood of capturing one of the relevant individuals render traditional approaches such as banding, radiotracking, and stable isotope analysis unlikely to yield useful results.
Some species of trypanosomes, single-celled parasites that are found in the blood of vertebrates, cause important diseases in humans and livestock (Hoare, 1972). For example, in Africa, some variants of Trypanosoma brucei are responsible for Sleeping Sickness in humans, while a range of other species cause similar wasting diseases of livestock (known as nagana). In South and Central America, Trypanosoma cruzi causes Chagas Disease in humans and is also known to infect a wide range of reservoir animals (Hoare, 1972, Miles et al., 2009). Trypanosomes have been described from bats from all continents in which they occur (Hoare, 1972, Molyneux, 1991). Many bat trypanosomes (including T. dionisii and T. vespertilionis) are morphologically indistinguishable from T. cruzi. Therefore these bat trypanosomes can only be assigned to the T. cruzi subgenus T. (Schizotrypanum) sp. based on morphology alone. As little is known of the host specificity of most bat trypanosomes, their diversity may have been considerably over- or underestimated (Hoare, 1972). The objective of this study was to examine occurrence and diversity of trypanosome species in British bats. Because many parasites, including species of bat trypanosome, can persist in related groups of hosts, understanding their phylogeography can provide valuable clues to the historical movements of host organisms.
2. Materials and methods
2.1. PCR detection and identification of trypanosomes
One-hundred and eighty-eight blood samples from 13 species of bat were collected in southern Britain in 2006 (collected under Natural England licence 20082696 and preceding licences, and Home Office licence PPL30/2243). Bats were identified by F. Mathews (Exeter) using standard field guides (Greenaway and Hutson, 1990, Dietz and von Helversen, 2004). Genomic DNA was extracted from blood samples using the DNeasy Kit (Qiagen Inc.), using negative controls to test for contamination during DNA extraction. As blood samples were collected at different times, DNA quality was tested using primers 5′-GGCCGCGGTAYYHYRACC-3′ and 5′-GATKGCGCTGTTATCCCTRG-3′, designed to be specific for a ∼400 bp region of the vertebrate mitochondrial 16S ribosomal RNA gene, using a touchdown PCR program (Hamilton and Tyler, 2008), which indicated the presence of amplifiable DNA in 154 DNA samples. To test for the presence of trypanosomal DNA, PCR reactions were carried out using trypanosome-specific rDNA primers 18S-1f and 18S-4r (Hamilton et al., 2008) and trypanosome-positive samples were further analysed by fluorescent fragment length barcoding (FFLB), a method that discriminates between species using length differences of four ribosomal regions (Hamilton et al., 2008). FFLB profiles were also obtained from isolates of Trypanosoma dionisii and T. vespertilionis, two trypanosome species known from European bats, for comparison. Trypanosomal gGAPDH and partial 18S rDNA genes were amplified and sequenced as described previously (Hamilton et al., 2008) from two individual bats giving each novel FFLB profile. Attempts to amplify longer 18S rDNA regions from one genotype (New1) failed, likely due to limited DNA quantity/quality. GenBank accession numbers are: New 1 gGAPDH FN599056 (853 bp); 18S rDNA FN599057 (526 bp); New 2 gGAPDH FN599055 (858 bp); 18S rDNA FN599058 (1445 bp).
2.2. Phylogenetic analyses
2.2.1. 18S rDNA alignment
The two partial 18S rDNA sequences obtained in this study, which cover the most variable (V7–V8) region of this gene, were manually aligned with sequences from other T. dionisii isolates. The alignment contained 22 taxa, and all 541 characters in the V7–V8 region were included in subsequent analyses.
2.2.2. GAPDH alignment 1
The new gGAPDH gene sequences obtained in this study were manually aligned with those from other trypanosomes in the T. cruzi clade. This alignment contained 23 sequences and analyses were based on 836 characters. A trypanosome from a kangaroo, the most divergent taxon within this clade (Stevens et al., 1999, Hamilton et al., 2007), was used as an outgroup.
2.2.3. GAPDH alignment 2
For molecular dating analysis, the new gGAPDH sequences, and others from recent studies, were added to the alignment of Hamilton et al. (2007). Forty-nine trypanosomal and 23 other trypanosomatid sequences were selected from this alignment to represent known trypanosomatid diversity, and four bodonid sequences were used as outgroups. This analysis included 839 characters. Alignments are available as Supplementary online documentation. All alignments were analysed by maximum likelihood (ML), maximum parsimony, and ML distance analysis as implemented in the program PAUP version 4.0b10 (Swofford, 2003), essentially as described previously (Hamilton et al., 2007).
2.3. Molecular dating
Divergence dates were estimated from gGAPDH gene alignment 2. Although concatenated 18S rDNA and gGAPDH gene alignments generally provide increased resolution (Hamilton et al., 2007, McInnes et al., 2009), analyses were based on gGAPDH, as only short 18S rDNA sequences could be obtained from genotype New 1. Dates were calculated from the optimal ML tree using PATHd8 (Fig. 1 ) (Britton et al., 2007) and BEAST v. 1.6.1 using a Bayesian Markov chain Monte Carlo (MCMC) approach (Drummond and Rambaut, 2007). Posterior probability distributions of node ages were obtained using GTR with rate variation (six gamma categories) and an uncorrelated log-normal model of rate variation. As, under certain circumstances, BEAST may become stuck on local optima, monophyletic clades (indicated in Fig. 1) were predefined in order to obtain a topology that corresponded to the phylogeny obtained from more robust phylogenetic methods based on more comprehensive concatenated datasets (Hamilton et al., 2007, McInnes et al., 2009). A Yule tree prior (Yule, 1925) was employed, which assumes a constant speciation rate for each branch on the tree, although the population structures of T. dionisii and most other trypanosome species are not known. Two independent MCMC chains were run for a minimum of 20,000,000 generations (burn-in 20%), with parameters sampled every 1000 generations. In order to assess that the MCMC chains were had stabilized, lnL plots were examined using Tracer v. 1.4 (Rambaut and Drummond, 2007). Four host-restricted trypanosomatid clades, which received robust bootstrap support in trees based on concatenated 18S rDNA and gGAPDH genes (Hamilton et al., 2007, McInnes et al., 2009), were used for calibration. The oldest estimated divergence dates for their host groups were used as the maximum times of divergence of the parasite clades, as host-restricted parasites are likely to have diversified after diversification of their hosts (Stevens and Rambaut, 2001). We used 390 ± ½ million years ago (MYA) for the origin of trypanosomatids, based on the origin of insects (Gaunt and Miles, 2002); evidence suggests that the ancestral trypanosomatid was an insect parasite (Hamilton et al., 2004). Maximum dates were also applied to: (1) 100 MYA for the origin of the ‘mammalian clade’ (Hamilton et al., 2007), based on the first major diversification of mammals (Bininda-Emonds et al., 2007); (2) 77 MYA for the origin of the ‘avian clade’ (Hamilton et al., 2007); based on diversification of passerine birds (Barker et al., 2004, Ericson et al., 2006); and (3) 184 MYA for the origin of the crocodilian clade (Viola et al., 2009), based on diversification of extant crocodiles (Janke et al., 2005). To evaluate the effect of priors on the analysis, we conducted an analysis after removing all sequence data. This resulted in markedly different inferred divergence times, suggesting that the data are driving the results of these analyses rather than the priors.
Fig. 1.
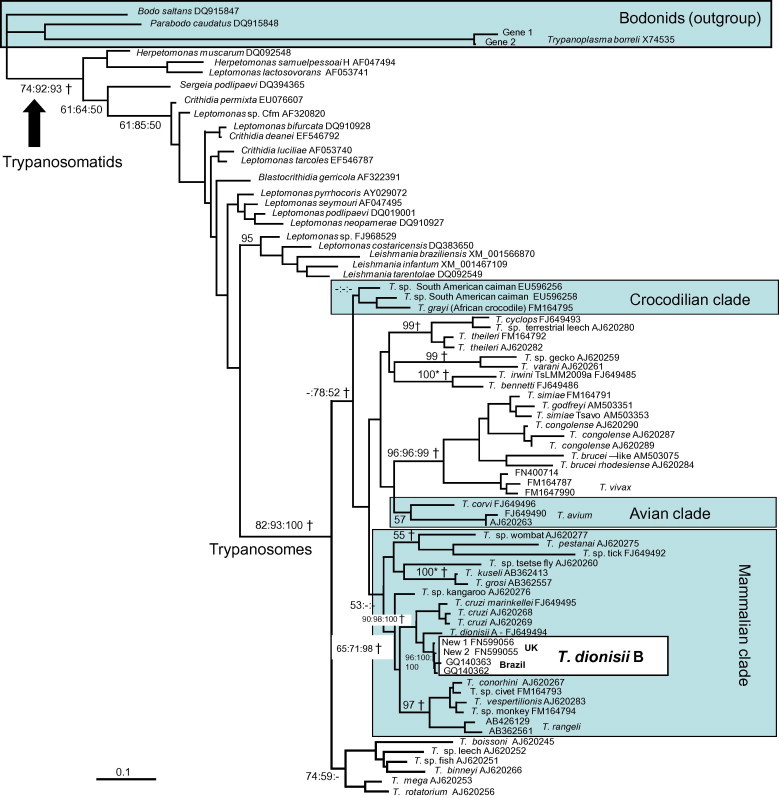
ML gene tree, based on gGAPDH alignment 2, used for molecular dating analyses. The model of nucleotide substitution was GTR + G. −Ln = 18000.59528. Highlighted clades are host-restricted clades used for calibration. Only bootstrap values for the main clades are given. †Indicates clades which were constrained for BEAST analysis. Single values at nodes are ML bootstrap values (%, based on 1000 replicates); multiple values are ML, MP and ML distance bootstrap values. 100* = Support values of 100% by all methods. – = Support value <50.
3. Results
We identified four distinct trypanosome genotypes in a survey of 154 British bats from which amplifiable DNA was obtained using fluorescent fragment length barcoding (Table 1 ). Two of the FFLB profiles from European isolates matched those of previously described bat trypanosomes, T. dionisii (hereafter T. dionisii A) and T. vespertilionis. The other two profiles were new: ‘New 1’ from Serotine Bats (Eptesicus serotinus) and Whiskered Bats (Myotis mystacinus), and ‘New 2’ from Noctule Bats (Nyctalus noctula) (Table 2 ). Phylogenetic analysis using 18S rRNA and gGAPDH gene sequences revealed that both are new strains of T. dionisii and are closely related to each other (Fig. 1, Fig. 2 ). Their gGAPDH and 18S rDNA (V7–V8 region) genes differed by 0.48% (4 out of 836 bp) and 1% (6 out of 541 bp), respectively, from each other compared to a minimum gGAPDH divergence of 4.4% from T. dionisii A. Phylogenetic analysis placed both new strains in T. dionisii group B, previously only known from South America (Maia da Silva et al., 2009, Cavazzana et al., 2010), rather than with European T. dionisii group A (Fig. 1, Fig. 2). In South America, T. dionisii group B has been found in 12 bat species from four families (Molossidae, Noctilionidae, Phyllostomidae, and Vespertilionidae) including insectivorous, frugivorous, and haematophagous species (Cavazzana et al., 2010). Within T. dionisii group B, sequences from European and South American isolates differed by a minimum of 0.2% (1 out of 541 bp) for V7–V8 18S rDNA and 0.72% (6 out of 836 bp) for the gGAPDH gene.
Table 1.
Trypanosome species found in British bats.
| Bat species | No. sampled a | Locations (No. sampled) b | No. +ve | Trypanosome species |
|---|---|---|---|---|
| Common Pipistrelle (Pipistrellus pipistrellus) | 1 | CA | 1 | T. dionisii A |
| Soprano Pipistrelle (Pipistrellus pygmaeus) | 25 | LA (12); WY (13) | 7 | T. dionisii A c |
| Barbastelle Bat (Barbastella barbastellus) | 1 | WY | 0 | |
| Bechstein’s Bat (Myotis bechsteinii) | 5 | BO (1); BF (1); GR (2); WY (1) | 0 | |
| Brown Long-eared Bat (Plecotus auritus) | 12 | WY (7); BO (2); BF (2); GR (1) | 0 | |
| Brandt’s Bat (Myotis brandtii) | 2 | BF (1); BO (1) | 0 | |
| BF (5); BO (4) | ||||
| Daubenton’s Bat (Myotis daubentonii) | 26 | GR (1); WY (18) | 0 | |
| Greater Horseshoe Bat (Rhinolophus ferrumequinum) | 15 | GR (14); BO (1) | 0 | |
| Lesser Horseshoe Bat (Rhinolophus hipposideros) | 5 | GR (4); BE (1) | 0 | |
| Natterer’s Bat (Myotis nattereri) | 32 | BF (8); BO (15); GR (5); WY (4) | 0 | |
| Noctule Bat (Nyctalus noctula) | 9 | AM (1); BIA (8) | 3 | T. dionisii B (New 2), T. vespertilionisd |
| Serotine Bat (Eptesicus serotinus) | 14 | AM (6); BA (2); BO (4); SL (1); WY (1) | 3 | T. dionisii B (New 1) |
| Whiskered Bat (Myotis mystacinus) | 7 | BO (6); BF (1) | 2 | T. dionisii B (New 1) |
Only includes bats which provided amplifiable DNA.
Codes for locations: AM: Amesbury – 51°10′11.95″N 1°47′14.62″W; BA: Bath – 51°23′11.28″N 2°24′08.78″W; BE: Belcombe – 51°36′46′79″N 2°39′63.93″W; BO: Box – 51°25′07.57″N 2°14′30.83″W; BIA: Bristol International Airport – 51°22′36.59″N 2°42′52.53″W; BF: Brown’s Folly – 51°23′30.35″N 2°17′45.66″W; LA: Lacock – 51.41′45.54″N 2.11′78.57″W; CA: Captive, origin unknown; GR: Gripwood – 51°20′20.17″N 2°15′24.82″W; SL: Slaughterford – 51°27′34.69″N 2°27′34.69″W; WY: Wytham – 51°46′31.20″N 1°20′25.84″W.
All infected bats were from WY.
All three bats were infected with New 2, of which one was co-infected with T. vespertilionis; all from BIA.
Table 2.
Fluorescent fragment length barcoding profiles of British bat trypanosomes.
| Fragment lengths (bp) |
||||
|---|---|---|---|---|
| 18S1 | 18S3 | 28S1 | 28S2 | |
| Trypanosoma vespertilionis | 256 | 222 | 286 | 185 |
| Trypanosoma dionisii A | 259 | 225 | 289 | 196 |
| Trypanosoma dionisii B (New 1) | 259 | 228 | 308 | 206 |
| Trypanosoma dionisii B (New 2) | 270 | 233 | 350 | 221 |
Fig. 2.
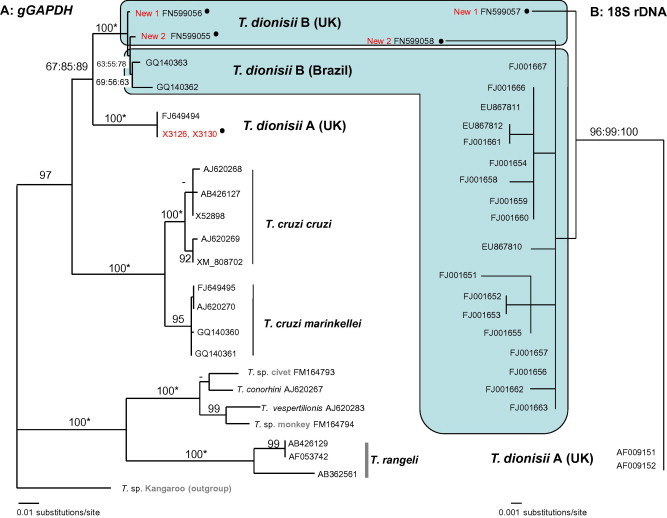
(A) ML gGAPDH gene tree of the T. cruzi clade. The model of nucleotide substitution was GTR + G. −Ln = 3026.91352. (B) ML tree, based on the D7–D8 region of 18S rDNA. −Ln = 3273.71272. The model of nucleotide substitution was K81 + G, with equal base frequencies. • = Sequences obtained in this study.
Maximum times of divergence between the sequences from South American and the new European T. dionisii B strains were estimated from the gGAPDH dataset, using maximum calibration points, under the assumption that divergences of host-restricted parasite clades post-date divergence of their hosts. Analysis using PATHd8 suggests that the most similar sequences from Old and New World strains diverged 3.2 MYA, whereas Bayesian analysis in BEAST suggested they diverged 5.0 MYA (95% CI for maximum time 11.4 MYA). Dates for divergence of the UK strains were 1.6 and 8.7 MYA (95% CI for maximum time 17.5 MYA) using these two methods, respectively. Given the conservative divergence dates used for the host taxa, and that host-restricted trypanosome clades may have evolved some time after diversification of their host groups, true divergence dates for parasites are likely to have been considerably more recent. Despite these limitations, the data clearly show that divergence must have occurred well after the separation of the Old and New Worlds, and possibly considerably more recently.
4. Discussion
This study has identified strains of bat parasite in the United Kingdom that are closely related to those from Brazil. We suggest that movement of bats between the Old and New Worlds within the last ∼5 million years (MY), certainly since geographical isolation of the Old and New Worlds, is the most likely explanation for the observed distribution of these trypanosomes. Of course, movement may also have been considerably more recent, but this is difficult to determine, as other calibration points are not available. We infer that migrant bats passed on their trypanosomes to native bat species, although the migrant bats themselves may not have established viable populations. The phylogenetic resolution provides insufficient evidence to determine whether movement occurred from Old World to New World or visa versa. Neither can the route of movement be inferred, as the wider distribution of these strains in either the Old or the New Worlds is currently unknown. Furthermore, although T. dionisii is a parasite specific to bats, a lack of specificity to particular species of bat host could be a critical factor in facilitating the spread of T. dionisii strains within continents. Trypanosoma (Schizotrypanum) sp. has been reported from ∼70 species and in all continents in which bats are found (Hoare, 1972), but molecular studies would be required to determine the true distribution of species and strains.
The most likely explanation for the presence of T. dionisii B in the New and Old Worlds is movement of an infected bat. Although T. dionisii can enter cells (Baker, 1985, Oliveira et al., 2009) and forms cysts in the muscular tissue of the heart (Gardner and Molyneux, 1988a), suggesting pathogenicity, British bats have been shown to live for over a year with T. (Schizotrypanum) sp. (Gardner and Molyneux, 1988a) with no obvious detrimental effects. Thus, bats are probably capable of normal flying and movement (including some long-distance flights) while infected. Movement of T. dionisii by birds is an unlikely explanation, since it falls in the T. cruzi clade that is only known in mammals (Stevens et al., 1999, Hamilton et al., 2007). Movement by the invertebrate vector in the absence of a vertebrate host is also unlikely. Cimicid bugs (the Cimicidae, or bed and bat bugs) are the only known vectors of T. dionisii and also transmit several other bat trypanosome species (Gardner and Molyneux, 1988b, Hamilton et al., 2007). Cimicid bugs are unable to fly; some species are associated with bat roosts where they associate with bats mostly during feeding, and then leave for substantial periods of time (Reinhardt and Siva-Jothy, 2007). Movement by an arthropod vector on bird hosts is possible, as some cimicid species can feed on both birds and bats (Reinhardt and Siva-Jothy, 2007), but this is an unlikely scenario as a bug would need to feed on an infected bat in one continent, transfer to a bird, migrate and then transfer to and infect a bat in the second continent. Additionally, the infection could not be maintained in generations of bugs feeding exclusively on birds, since birds are not known hosts of bat trypanosomes. The vectorial capacity of the other bug family that is parasitic on bats, Polyctenidae (polictenids, or “bat bugs”), is unknown, but could not move in the absence of a bat host. These wingless bugs live exclusively and permanently on bat hosts and are found on certain microchiropteran bats in tropical and subtropical regions of both the Old and New Worlds; of these, each of the two bug subfamilies is exclusive to one or other region, with no overlap between the Old and New Worlds. Moreover, while polictenid bugs can be transmitted experimentally between bats (Dick et al., 2009), bat–bird transmission has not been reported.
This raises the question of whether movement(s) of the infected bat(s) occurred naturally or was facilitated by humans. Constantine (2003) cited several reports of bats which had been inadvertently transported by humans, for example, by bats roosting in airplanes or ships. Presumably, most human-assisted transportations go unreported and these are likely to have occurred with increasing frequency since the 1800s, when ships increased significantly in size and speed, together with increased volumes of trade. In support of this idea, trypanosome infections were found in Eptesicus and Myotis in this study; there are reports of long-distance translocations of members of these genera that were mediated by humans, likely because these bats occasionally roost in ships (Constantine, 2003). However it seems that two separate introductions would have been required to produce the two distinct T. dionisii B genotypes we have identified within the UK. The gGAPDH genes of these strains differed by 4 bp, and there is even greater divergence between some of the South American T. dionisii strains. While it is difficult to date how recently this divergence occurred, it seems unlikely that such divergence could have occurred since any human-mediated introduction of their bat hosts. Trypanosoma vivax was introduced into Latin America in cattle imported from Africa, possibly in the late 19th century (Jones and Davila, 2001), yet gGAPDH sequences of South American and West African T. vivax are identical (Adams et al., 2010). Thus, natural movement of bats seems a more likely explanation, as this would provide sufficient time for either T. dionisii divergence within the Old or New Worlds and/or multiple introductions to occur. However, more evidence is required to definitively resolve this issue.
While the precise route of parasite movement cannot be inferred based on the current evidence, one possible route for movement of bats between the Old and New Worlds is between Alaska and Siberia, particularly across the Bering Land Bridge during a period of low sea level. Bats are known to occur throughout much of Alaska, some close to the Arctic circle (MacDonald and Cook, 2009). Phylogenetic evidence supports a Palaearctic origin of the New World Myotis (Stadelmann et al., 2007), suggesting this as a possible route, while within the New Word the relatively small sea barrier prior to closure of the Isthmus of Central America (2–3 MYA) is unlikely to have represented a major barrier to bat movement (Stadelmann et al., 2007).
More broadly, the T. cruzi clade includes several other species/lineages found in bats and also trypanosomes found in terrestrial mammals from Australia, the New World and Africa. Our results suggest that bats can disperse trypanosome genotypes across substantial geographical barriers, a mechanism previously proposed to explain the widespread distribution of the T. cruzi clade (Stevens et al., 1999, Hamilton et al., 2009).
In conclusion, the distribution of trypanosome strains in this study implies relatively recent bat movements that are not possible to infer from the direct examination of bat phylogenies. Given the importance of bats as hosts of zoonoses highly pathogenic to humans, understanding the historical patterns of parasite/host movements can provide useful insights into the contemporary distributions and potential spread of pathogens such as rabies and coronaviruses.
Acknowledgment
We thank Professor Wendy Gibson (University of Bristol) for providing DNA from bat trypanosomes. We thank two anonymous reviewers for their helpful comments on the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ympev.2012.01.007.
Appendix A. Supplementary material
References
- Adams E.R., Hamilton P.B., Rodrigues A.C., Malele I.I., Delespaux V., Teixeira M.M.G., Gibson W.C. New Trypanosoma (Duttonella) vivax genotypes from tsetse flies in East Africa. Parasitology. 2010;137:641–650. doi: 10.1017/S0031182009991508. [DOI] [PubMed] [Google Scholar]
- Baker J.R. Bat trypanosome models for Trypanosoma cruzi. Parasitol. Today. 1985;1:111. doi: 10.1016/0169-4758(85)90006-7. [DOI] [PubMed] [Google Scholar]
- Barker F.K., Cibois A., Schikler P., Feinstein J., Cracraft J. Phylogeny and diversification of the largest avian radiation. Proc. Natl. Acad. Sci. USA. 2004;101:11040–11045. doi: 10.1073/pnas.0401892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds O.R.P., Cardillo M., Jones K.E., MacPhee R.D.E., Beck R.M.D., Grenyer R., Price S.A., Vos R.A., Gittleman J.L., Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Britton T., Anderson C.L., Jacquet D., Lundqvist S., Bremer K. Estimating divergence times in large phylogenetic trees. Syst. Biol. 2007;56:741–752. doi: 10.1080/10635150701613783. [DOI] [PubMed] [Google Scholar]
- Cavazzana M., Jr., Marcili A., Lima L., da Silva F.M., Junqueira Â.C.V., Veludo H.H., Viola L.B., Campaner M., Nunes V.L.B., Paiva F., Coura J.R., Camargo E.P., Teixeira M.M.G. Phylogeographical, ecological and biological patterns shown by nuclear (ssrRNA and gGAPDH) and mitochondrial (Cyt b) genes of trypanosomes of the subgenus Schizotrypanum parasitic in Brazilian bats. Int. J. Parasitol. 2010;40:345–355. doi: 10.1016/j.ijpara.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Constantine D.G. Geographic translocation of bats: known and potential problems. Emerg. Infect. Dis. 2003;9:17–21. doi: 10.3201/eid1309.020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick C.W., Esberard C.E.L., Graciolli G., Bergallo H.G., Gettinger D. Assessing host specificity of obligate ectoparasites in the absence of dispersal barriers. Parasitol. Res. 2009;105:1345–1349. doi: 10.1007/s00436-009-1563-1. [DOI] [PubMed] [Google Scholar]
- Dietz C., von Helversen O. Tuebingen and Erlangen; Germany: 2004. Illustrated identification key to the bats of Europe. [Google Scholar]
- Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7 doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick G.N., Jacobs D.S., Matthee C.A. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera) Mol. Biol. Evol. 2005;22:1869–1886. doi: 10.1093/molbev/msi180. [DOI] [PubMed] [Google Scholar]
- Ericson P.G.P., Anderson C.L., Britton T., Elzanowski A., Johansson U.S., Kallersjo M., Ohlson J.I., Parsons T.J., Zuccon D., Mayr G. Diversification of Neoaves: integration of molecular sequence data and fossils. Biol. Lett. 2006;2:543–547. doi: 10.1098/rsbl.2006.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.A., Molyneux D.H. Schizotrypanum in British bats. Parasitology. 1988;97:43–50. doi: 10.1017/s0031182000066725. [DOI] [PubMed] [Google Scholar]
- Gardner R.A., Molyneux D.H. Trypanosoma (Megatrypanum) incertum from Pipistrellus pipistrellus: development and transmission by cimicid bugs. Parasitology. 1988;96(Pt 3):433–447. doi: 10.1017/s0031182000080082. [DOI] [PubMed] [Google Scholar]
- Gaunt M.W., Miles M.A. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Greenaway F., Hutson A.M. Bruce Coleman Books; Uxbridge: 1990. A Field Guide to British Bats. [Google Scholar]
- Hamilton P.B., Tyler C.R. Identification of microsatellite loci for parentage analysis in roach Rutilus rutilus and 8 other cyprinid fish by cross-species amplification, and a novel test for detecting hybrids between roach and other cyprinids. Mol. Ecol. Resour. 2008;8:462–465. doi: 10.1111/j.1471-8286.2007.01994.x. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Stevens J.R., Gaunt M.W., Gidley J., Gibson W.C. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int. J. Parasitol. 2004;34:1393–1404. doi: 10.1016/j.ijpara.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Gibson W.C., Stevens J.R. Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol. Phylogenet. Evol. 2007;43:15–25. doi: 10.1016/j.ympev.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Adams E.R., Malele I.I., Gibson W.C. A novel, high throughput technique for species identification reveals a new species of tsetse-transmitted trypanosome related to the Trypanosoma brucei subgenus, Trypanozoon. Infect. Genet. Evol. 2008;8:26–33. doi: 10.1016/j.meegid.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Adams E.R., Njiokou F., Gibson W.C., Cuny G., Herder S. Phylogenetic analysis reveals the presence of the Trypanosoma cruzi clade in African terrestrial mammals. Infect. Genet. Evol. 2009;9 doi: 10.1016/j.meegid.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Hoare C.A. Blackwell Scientific Publications; Oxford and Edinburgh: 1972. The Trypanosomes of Mammals. [Google Scholar]
- Janke A., Gullberg A., Hughes S., Aggarwal R.K., Arnason U. Mitogenomic analyses place the gharial (Gavialis gangeticus) on the crocodile tree and provide Pre-K/T divergence times for most crocodilians. J. Mol. Evol. 2005;61:620–626. doi: 10.1007/s00239-004-0336-9. [DOI] [PubMed] [Google Scholar]
- Jones T.W., Davila A.M. Trypanosoma vivax - out of Africa. Trends Parasitol. 2001;17:99–101. doi: 10.1016/s1471-4922(00)01777-3. [DOI] [PubMed] [Google Scholar]
- Jones K.E., Bininda-Emonds O.R.P., Gittleman J.L. Bats, clocks, and rocks: diversification patterns in chiroptera. Evolution. 2005;59:2243–2255. [PubMed] [Google Scholar]
- MacDonald, S.O., Cook, J.A., 2009 Recent Mammals of Alaska. University of Alaska Press.
- Maia da Silva F., Marcili A., Lima L., Cavazzana M., Jr., Ortiz P.A., Campaner M., Takeda G.F., Paiva F., Nunes V.L.B., Camargo E.P., Teixeira M.M.G. Trypanosoma rangeli isolates of bats from Central Brazil: genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Trop. 2009;109:199. doi: 10.1016/j.actatropica.2008.11.005. [DOI] [PubMed] [Google Scholar]
- McInnes L.M., Gillett A., Ryan U.M., Austen J., Campbell R.S.F., Hanger J., Reid S.A. Trypanosoma irwini n. sp (Sarcomastigophora: Trypanosomatidae) from the koala (Phascolarctos cinereus) Parasitology. 2009;136:875–885. doi: 10.1017/S0031182009006313. [DOI] [PubMed] [Google Scholar]
- Miles M.A., Llewellyn M.S., Lewis M.D., Yeo M., Baleela R., Fitzpatrick S., Gaunt M.W., Mauricio I.L. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology. 2009;136:1509–1528. doi: 10.1017/S0031182009990977. [DOI] [PubMed] [Google Scholar]
- Molyneux D.H. Trypanosomes of bats. In: Kreier J.P., Baker J.R., editors. Parasitic Protozoa. Academic Press; San Diego: 1991. pp. 195–224. [Google Scholar]
- Morales J.C., Bickham J.W. Molecular systematics of the genus Lasiurus (Chiroptera, Vespertilionidae) based on restriction-site maps of the mitochondrial ribosomal genes. J. Mammal. 1995;76:730–749. [Google Scholar]
- Oliveira M.P.D., Cortez M., Maeda F.Y., Fernandes M.C., Haapalainen E.F., Yoshida N., Mortara R.A. Unique behavior of Trypanosoma dionisii interacting with mammalian cells: invasion, intracellular growth, and nuclear localization. Acta Trop. 2009;110:65–74. doi: 10.1016/j.actatropica.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Rambaut, A., Drummond, A.J., 2007. Tracer v1.4. <http://beast.bio.ed.ac.uk/Tracer>.
- Reinhardt K., Siva-Jothy M.T. Biology of the bed bugs (Cimicidae) Annu. Rev. Entomol. 2007;52:351–374. doi: 10.1146/annurev.ento.52.040306.133913. [DOI] [PubMed] [Google Scholar]
- Stadelmann B., Lin L.K., Kunz T.H., Ruedi M. Molecular phylogeny of New World Myotis (Chiroptera, Vespertilionidae) inferred from mitochondrial and nuclear DNA genes. Mol. Phylogenet. Evol. 2007;43:32–48. doi: 10.1016/j.ympev.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Stevens J.R., Rambaut A. Evolutionary rate differences in trypanosomes. Infect. Genet. Evol. 2001;1:143–150. doi: 10.1016/s1567-1348(01)00018-1. [DOI] [PubMed] [Google Scholar]
- Stevens J.R., Noyes H.A., Dover G.A., Gibson W.C. The ancient and divergent origins of the human pathogenic trypanosomes, Trypanosoma brucei and T. cruzi. Parasitology. 1999;118:107–116. doi: 10.1017/s0031182098003473. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L., 2003. PAUP∗. Phylogenetic Analysis Using Parsimony (∗and Other Methods). Sinauer Associates, Sunderland, MA.
- Teeling E.C., Springer M.S., Madsen O., Bates P., O’Brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Viola L.B., Almeida R.S., Ferreira R.C., Campaner M., Takata C.S.A., Rodrigues A.C., Paiva F., Camargo E.P., Teixeira M.M.G. Evolutionary history of trypanosomes from South American caiman (Caiman yacare) and African crocodiles inferred by phylogenetic analyses using SSU rDNA and gGAPDH genes. Parasitology. 2009;136:55–65. doi: 10.1017/S003118200800512X. [DOI] [PubMed] [Google Scholar]
- Yule G.U. A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. Philos. Trans. Roy. Soc. B – Biol. Sci. 1925;213:21–87. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


