Abstract
Rapid detection of severe fever with thrombocytopenia syndrome virus (SFTSV) is crucial for its control and surveillance. In this study, a rapid isothermal real-time reverse-transcription recombinase polymerase amplification (RT-RPA) assay was developed for the detection of SFTSV. The detection limit at 95% probability was 241 copies per reaction. A test of 120 serum samples of suspected severe fever with thrombocytopenia syndrome (SFTS) patients revealed that the sensitivity and specificity of the RT-RPA assay was approximately 96.00% (95%CI: 80.46%–99.79%) and 98.95% (95% CI: 94.28%–99.95%), respectively; the kappa value was 0.9495 (P<0.001). The Bland-Altman analysis showed that 87.50% of the different data points were located within the 95% limits of agreement, indicating a good correlation between the results from RT-RPA assays and those of RT-qPCR assays. In conclusion, the rapid and efficient RT-RPA assay can be a promising candidate for point-of-care detection method of SFTSV.
Keywords: Severe fever with thrombocytopenia syndrome virus, Recombinase polymerase amplification, Detection assay
Highlights
-
•
A RT-RPA assay was developed to detect SFTSV RNA isothermally.
-
•
The assay can rapidly produce a result in 15 min at 39 °C.
-
•
The detection limit of the assay is 241 RNA sequences.
-
•
The results of RT-RPA compare well with RT-qPCR.
-
•
The RT-RPA assay may be used for field detection of SFTSV in resource-limited settings.
Table of abbreviations
- dT-FAM
Thymidine nucleotide carrying FAM fluorophore
- dT-BHQ1
Thymidine nucleotide carrying BHQ-1 quencher
- HHV
Human herpesvirus
- LAMP
Loop-mediated isothermal amplification
- LOD
Limit of detection
- PIV
Parainfluenza virus
- RPA
Recombinase polymerase amplification
- RT-LAMP
Reverse-transcription loop-mediated isothermal amplification
- RT-PCR
Reverse-transcription polymerase chain reaction
- RT-qPCR
Real-time reverse-transcription polymerase chain reaction
- RT-RAA
Reverse-transcription recombinase-aided amplification
- RT-RPA
Real-time reverse-transcription recombinase polymerase amplification
- SFTS
Severe fever with thrombocytopenia syndrome
- SFTSV
Severe fever with thrombocytopenia syndrome virus
1. Introduction
Severe fever with thrombocytopenia syndrome virus (SFTSV), belonging to the family Phenuiviridae, the genus Banyangvirus, the species Huaiyangshan banyangvirus, was first identified as the causative pathogen of the outbreak of severe fever with thrombocytopenia syndrome (SFTS) in the central and northeast regions of China in 2009 [1]. During the following decade, this emerging endemic tick-borne disease has been reported in many countries worldwide, including South Korea, Japan, United Arab Emirates, and the United States [[2], [3], [4], [5]]. Delayed admission due to nonspecific clinical manifestations, along with rapid progression to multiple organ dysfunction in severe cases led to a high case fatality rate of 6.40–30% in SFTS patients [6,7].
Currently, detection methods of SFTSV comprise nucleic acid testing, serological detection, and virus isolation [1,8,9]. Due to the long turnaround time of serological detection and virus isolation, nucleic acid testing, such as real-time reverse-transcription PCR (RT-qPCR) and reverse-transcription loop-mediated isothermal amplification (RT-LAMP), are more widely used for early diagnosis of this acute febrile disease [[10], [11], [12]]. Nevertheless, most PCR-based detection methods require sophisticated equipment and skilled personnel. Besides, LAMP faced problems of high false-positive rates and contamination, and the requirement for six primers adds up to the difficulty of primer design. Such limitations hampered the application of these nucleic acid detection methods as point-of-care diagnostic methods since the endemic areas of SFTS in China are usually rural mountain areas with limited medical resources [7].
Recombinase polymerase amplification (RPA) is a novel isothermal amplification technique first unveiled in 2006 [13], which employs three major proteins for DNA molecule amplification, including a T4 uvsX recombinase, a single-stranded DNA binding protein, and a polymerase. These enzymes show high activity at a constant temperature ranging from 37 to 42 °C. With the addition of reverse transcriptase and a fluorescence-modified probe, the RPA technique can also be used for real-time detection of RNA molecules. This user-friendly technique with short turnaround time can be a promising candidate for point-of-care detection assays in resource-limited SFTS endemic areas. In this study, a real-time reverse-transcription RPA (RT-RPA) assay for the rapid detection of SFTSV was established and the performance of this assay was evaluated with clinical specimens of suspected SFTS patients.
2. Materials and methods
2.1. Clinical samples and viruses
A total of 120 blood samples were collected from patients with suspected cases of SFTS admitted to the Zhoushan Hospital, China between January 2011 and December 2014. A 2 ml whole blood sample was obtained from each patient and the serum were stored at −80 °C. After tested by nested-PCR assays, 26 of these patients were eventually confirmed being infected with SFTSV.
Nucleic acids preparations from 15 SFTSV-negative clinical samples were kindly provided by Fudan University, China and Zhoushan Hospital, China. These samples were confirmed positive for either of the following non-SFTSV pathogens, including parainfluenza virus-1 (PIV-1), PIV-2, measles virus, mumps virus, rubella virus, coronavirus, respiratory syncytial virus, rotavirus, human adenovirus, human herpesvirus-1 (HHV-1), HHV-2, HHV-3, HHV-5, HHV-6, and dengue virus.
Written informed consents were obtained from all patients. This study was conducted in accordance with the national ethical regulations of China and approved by the Ethics Committee of Zhoushan Hospital, Zhoushan, China [(2016) approval No. 28].
2.2. RNA extraction
Total RNA was extracted from 200 μL of each serum specimen using the QIAamp Viral RNA Mini Kit (Qiagen, Shanghai, China) following the manufacturer's instruction. The extracted RNA was diluted using 50 μL nuclease-free water and stored at −20 °C until further analysis.
2.3. Design of primers and probes
Primers and probes used in this study were designed based on the conservative L-segment of the SFTSV genome. Highly-conserved sequence regions of the L-segment, which codes for the RNA dependent RNA polymerase of SFTSV, was identified using the CD-Search tool provided by the National Center for Biotechnology Information. The primers and exo-probes for RT-RPA assays were designed manually per the instructions of the reverse-transcription recombinase-aided amplification (RT-RAA) exo kit manufacturer (Qitian Bio-Tech Co. Ltd., Jiangsu, China). All the oligonucleotides, including RT-qPCR and RT-PCR primers, were designed using the Oligo 7 Primer Analysis software (Molecular Biology Insights, Colorado, U.S.A), and the specificity of each primer and probe was verified by BLAST analysis. The primers and probes were synthesized by Invitrogen (Shanghai, China) and Sangon Biotech Co., Ltd. (Shanghai, China), respectively. The sensitivity and specificity of these primers and probes was evaluated using RT-RAA basic kits and RT-RAA exo kits (Qitian Bio-Tech Co. Ltd., Jiangsu, China) to find out an optimal combination. Sequences and details of the optimal combination of oligonucleotides are listed in Table 1 .
Table 1.
Sequences and details of the primers and probe used for SFTSV RT-RPA assays, RT-qPCR assays and RT-PCR assays.
| Assay | Primer/Probe | Sequence (5′-3′) | Number of base pairs |
|---|---|---|---|
| RT-RPA | RPA-L-F4a | ATCACAATCCAGCTCTCTGAAGCGTATAAG | 128 |
| RPA-L-R4b | CATGTTGGACAGAACTCCTCCTGACGACACTAC | ||
| RPA-L-P2c | AGGCAGCATACAGGACAAAGATAGAAAAG[dT-FAM] [dSpacer][dT-BHQ1]AGGGACCCAATCTCAA-[C3 Spacer] | ||
| RT-qPCR | qPCR-Fa | CTTCTTTGGGGTTATTGTAGTGT | 131 |
| qPCR-Rb | CCATAGACCTAGCCTTAGTGT | ||
| RT-PCR | st-Fa | TGCCAGACCTAGATGTGACTG | 569 |
| st-Rb | CTCAAGCTCTTCTTCGCTCT |
F indicates forward primer.
R indicates reverse primer.
P indicates exo-probe. For probe modifications: dT-FAM, thymidine nucleotide carrying FAM fluorophore; dSpacer, a tetrahydrofuran residue; dT-BHQ1, thymidine nucleotide carrying BHQ-1 quencher; C3 Spacer, 3′-block.
2.4. Generation of RNA standard
A 569-bp (nt114-682, GenBank accession no. KR017845.1) fragment of the L-segment of SFTSV was amplified using a PrimeScript™ RT-PCR kit (TaKaRa, Shanghai, China) according to the manufacturer's protocol. Primer pair st-F/st-R (Table 1) and extracted RNA from a SFTSV-positive clinical sample was used to perform RT-PCR. Target amplicons were ligated using a pMD19-T vector cloning kit (TaKaRa, Shanghai, China), and ligations were transformed into Escherichia coli. The recombinant plasmids were verified by sequencing and then linearized. In vitro transcription of L-segment RNA standard was performed using T7 RNA polymerase (Thermo Scientific™, Shanghai, China) according to the manufacturer's instructions. Quantification of RNA standards was carried out using an ND-2000c spectrophotometer (NanoDrop, Wilmington, U.S.A).
2.5. Development of SFTSV RT-RPA assays
RT-RPA assay with exo-probe to detect SFTSV was performed in a 20 μL reaction system using RT-RAA exo kits (Qitian Bio-Tech Co.Ltd., Jiangsu, China). Details of primer set and probe used for RT-RPA assays are shown in Table 1. The reaction mixture contained 25 μL of rehydration buffer, 2.1 μL of each primer (10 μM, final concentration: 420 nM), 0.3 μL of exo-probe (10 μM, final concentration: 60 nM), and 15.5 μL of nuclease-free water. Lyophilized pellets were rehydrated with 45 μL of the reaction mixture. 18 μL aliquot of the resulting mixture was then transferred to each reaction tube. Then, 1 μL of the RNA template was added to the aliquot and 1 μL of 280 nM magnesium acetate was pipetted into the lid of each reaction tube. Lid was closed carefully, and the magnesium acetate was spun down into the mixture to catalyze the reaction by a mini centrifuge. After gentle inversion and quick spin down by a mini centrifuge, the reaction tubes were placed immediately into the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) and incubated at 39 °C for 30 min. Optimal reaction conditions were defined after testing different incubation temperatures (37 °C, 38 °C, 39 °C, 40 °C, 41 °C), as well as different final concentrations of primer (150–600 nM) and exo-probe (50–150 nM).
The fluorescence signal was collected in the FAM-Channel (excitation 488 nm and emission 520 nm) once every 30 s. The threshold time of each sample, representing the time a reaction needed to generate fluorescence signals that exceeded the threshold, was recorded. The threshold was defined as the mean value of negative control (nuclease-free water) plus three times its standard deviation.
2.6. RT-qPCR assays
The RT-qPCR assay was established as the reference method and performed using SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase (Invitrogen, Shanghai, China). A 12.5 μL reaction system was prepared according to the manufacturer's instructions, with the addition of a primer set as shown in Table 1. The 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) was used and programmed as follows: reverse transcription for 15 min at 50 °C; polymerase activation for 2 min at 95 °C; followed by 45 cycles of 95 °C for 5 s, 56 °C for 30 s, and 68 °C for 30 s. The cycle threshold value of each sample was recorded.
2.7. Analysis of sensitivity and specificity
Ten-fold serial dilutions of the SFTSV RNA standard from 106 to 10 copies/μL were prepared and tested in three replicates for two independent runs to determine the sensitivity of the RT-RPA assay. The threshold time was plotted against molecules detected and a semi-logarithmic regression was calculated using Prism 7.0 software (Graphpad Software Inc., California, U.S.A.). Also, a probit regression was performed to determine the limit of detection (LOD) of the RT-RPA assay at 95% probability using SPSS Statistics software (IBM Corporation, New York, U.S.A).
The specificity of the RT-RPA assay was evaluated using nucleic acids of fifteen non-SFTSV viruses (listed above) that could cause similar clinical symptoms. RNA extraction from one SFTSV-positive clinical sample was tested concurrently as a positive control.
2.8. Detection of clinical samples
All the clinical samples were tested to evaluate the sensitivity and specificity of SFTSV RT-RPA assays. Alongside all the samples were tested by RT-qPCR assays. Results of RT-RPA assays were compared with those of RT-qPCR assays. To assess the agreement of RT-RPA and RT-qPCR, the kappa value was calculated, and the Bland-Altman analysis was performed using results from both assays with SPSS Statistics software (IBM Corporation, New York, U.S.A.).
3. Result
3.1. Optimization of SFTSV RT-RPA assays
The candidate primer pairs were tested for detection of the SFTSV RNA standard using RT-RAA basic kits and the amplicons were analyzed with 0.8% agarose gel. The primer pair with the best amplification efficiency and specificity was used for the screening of exo-probe with RT-RAA exo kits. Of the primer pairs, the RPA-L-F4/RPA-L-R4/RPA-L-P2 set was identified as the ideal combination that produces maximal fluorescence signals in the least time and was incorporated in further assays (listed in Table 1). Besides, different incubation temperatures and concentrations of primers and exo-probe were tested for the specificity and rapidity of the assay. The optimized performance was achieved when incubated at 39 °C with a primer/probe final concentration settled to 420 nM/60 nM.
3.2. Sensitivity and specificity of SFTSV RT-RPA assay
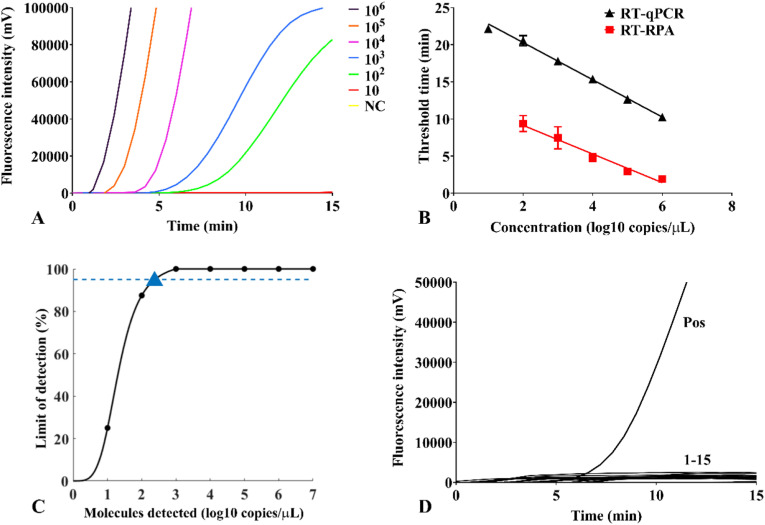
The sensitivity of the SFTSV RT-RPA assay was determined by testing with a serial dilution of RNA standard from 106 to 10 copies/μL. Robust fluorescence signals of the 106 copies/μL standards were rapidly observed within 1.85–1.92 min. Positive signals of 105–102 copies/μL dilutions were successfully detected within 2.73–9.41 min (Fig. 1 A). Results from semi-logarithmic regression showed a good correlation between molecule concentrations and detection time of real-time RPA assay, with the R2 value of 0.967 (Fig. 1B). Calibration curves of RT-RPA and RT-qPCR assay also illustrated that the detection time of RT-RPA assay was shorter than that of RT-qPCR assay by an average of 10 min.
Fig. 1.
Analysis of sensitivity and specificity of SFTSV RT-RPA assays. (A) Representative fluorescence amplification curves of SFTSV RT-RPA assays were developed with the 106–10 copies/μL RNA standard dilutions. NC indicated negative control. (B) Calibration curves of SFTSV RT-RPA assays (red square, R2 = 0.967) and RT-qPCR assays (black triangle, R2 = 0.996). Each dilution was tested in three replicates by both assays. (C) Probit regression analysis of SFTSV RT-RPA assays predicted LOD at 95% probability (triangle). (D) No cross-reactivity was observed. Nucleic acids of fifteen non-SFTSV pathogens were tested, including PIV-1, PIV-2, measles virus, mumps virus, rubella virus, coronavirus, respiratory syncytial virus, rotavirus, human adenovirus, HHV-1, HHV-2, HHV-3, HHV-5, HHV-6, and dengue virus. Pos indicated positive control with the SFTSV RNA. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The detection limit was further determined by applying probit regression analysis using a data set of six RT-RPA runs on the RNA standard, and the predicted LOD was 241 copies per reaction at 95% probability (Fig. 1C).
The specificity of the SFTSV RT-RPA assay was determined using nucleic acid extracted from a panel of viruses that can lead to clinical manifestations similar to SFTS. No unspecific amplification was observed with nucleic acids of fifteen non-SFTSV pathogens. Only the positive control of RNA extraction from a SFTSV-positive clinical sample produced robust fluorescence signals (Fig. 1D).
3.3. Evaluation of SFTSV RT-RPA assay using clinical samples
A total of 120 extracted nucleic acids from serum samples of suspected SFTS patients were tested to evaluate the clinical applicability of SFTSV RT-RPA assays, and the results were compared with those of the RT-qPCR assays (Table 2 ). 25 of the 120 samples were tested positive by RT-qPCR assays. RT-RPA assays successfully detected 24 (96.00%) of these RT-qPCR-positive samples. Among the 95 samples tested negative by RT-qPCR assays, 94 (98.95%) samples were also determined negative by RT-RPA assays.
Table 2.
Detection of SFTSV nucleic acids in clinical samples using SFTSV RT-RPA assays and RT-qPCR assays.
| Method | RT-qPCR |
Total | Kappa | P-value of Kappa | ||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| RT-RPA | Positive | 24 | 1 | 25 | 0.9495 | <0.001 |
| Negative | 1 | 94 | 95 | |||
| Total | 25 | 95 | 120 | |||
When RT-qPCR was used as the reference method, the sensitivity and specificity of RT-RPA assay were estimated to be 96.00% (95%CI: 80.46%–99.79%) and 98.95% (95% CI: 94.28%–99.95%), respectively. The agreement of these methods was 98.39%. A kappa value of 0.9495 (z = 10.40, P<0.001) indicated that the results of RT-RPA were consistent with those of the conventional RT-qPCR. The agreement of the RT-RPA and RT-qPCR assay was further assessed using the Bland-Altman analysis (Fig. 2 ). A total of 21 (87.50%) different data points were located within the 95% limits of agreement, which indicate a good correlation between the results of these assays. Only two samples failed to produce consistent results. The RT-RPA-positive but RT-qPCR-negative sample was confirmed as positive by nested-PCR. The RT-RPA-negative but RT-qPCR-positive sample was also scored positive by nested-PCR and sent for further sequencing. The sequencing result revealed that there were two mismatches of the probe (442C/T and 460G/A), one of which was located next to the BHQ-coupled nucleotide, and two mismatches of the reverse primer (562G/C and 544A/G).
Fig. 2.
Bland-Altman analysis of SFTSV RT-RPA assays and RT-qPCR assays. Mean of difference between results from RT-RPA assays and RT-qPCR assays is indicated with the red line. The 95% limit of agreement was indicated with the blue lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
SFTS is an acute infectious disease with a high case fatality rate [6]. The most commonly seen clinical manifestations of SFTSV infection, including fever, thrombocytopenia and lymphocytopenia, are often unspecific and need to be differentiated from other infectious diseases, such as haemorrhagic fever with renal syndrome, human anaplasmosis [6,14]. A recent study showed an increasing incidence of SFTSV-specific antibody seropositivity among healthy people in high endemic areas of China, suggesting the existence of asymptomatic SFTSV infections [15]. Human-to-human transmission through blood and/or body fluid contact or aerosols and resulting multiple cluster cases have also made SFTS a threat to public health [16,17]. Therefore, a rapid detection method is in urgent need for early diagnosis of SFTSV infection to improve clinical outcome and prevent transmission. Currently, the diagnosis of SFTS is based on serological methods and PCR-based viral nucleic acid testing [14]. However, specific antibodies to SFTSV are detectable after about 7 days from disease onset [18], and PCR-based nucleic acid tests usually require a centralized laboratory with high-end instrumentation and skilled personnel to perform. The novel RT-RPA technique might be an ideal candidate for point-of-care detection of SFTSV.
This study established a highly sensitive and specific diagnostic approach for the detection of SFTSV nucleic acids in clinical serum specimens. The detection limit of SFTSV RT-RPA assay was statistically estimated as 241 copies of target RNA per reaction, which is consistent with other published literature [[19], [20], [21]]. While the sensitivity of this RT-RPA assay is lower than conventional RT-qPCR for SFTSV, the detection limit is still far below the viremia level in most clinical SFTS patients [18]. With the application of RT-RPA, the detection time was reduced to <30 min, whereas RT-qPCR generally takes >1.5 h. In addition to the reduced detection time, the constant reaction temperature of 39 °C allowed RT-RPA assays to be performed using simple equipment such as a water or metal bath, while RT-qPCR requires sophisticated thermocyclers. The short detection time and isothermal reaction condition therefore provided the RT-RPA assay with advantages over RT-qPCR as a point-of-care detection assay in resource-limited areas.
The clinical performance of this RT-RPA assay was also proved to be satisfactory using a series of clinical serum samples. The sensitivity and specificity are comparable with other established detection methods [10,11,[22], [23], [24], [25]]. One RPA-negative sample was positive by nest-PCR. The sequencing result of the RPA-negative sample revealed that there were several mismatches within the binding sites of the probe and the reverse primer. Previous study, regarding foot-and-mouth disease virus and human immunodeficiency virus, indicated that perhaps as a result of the length of primer and probe compensating for mismatches in target sequence, RPA can tolerate up to nine mismatches across the primer and probe binding sites [26,27]. However, in our study, one mismatch is located next to the fluorophore-modified nucleotide of the probe. Due to its location, it might hinder recognition of the tetrahydrofuran spacer by the double-strand-specific endonuclease IV and subsequent separation of the fluorophore/quencher complex, thereby, leading to significant reduction of fluorescence signal in the RT-RPA assay. Several studies also observed that mismatches covering the center and both extremities of the primer sequence would hamper RPA reaction [28]. Mismatches of probe show greater negative impact on the efficiency of RPA reaction than the primer, and RPA reaction efficiency decreases as the number of mismatches increases on the probe binding site [29]. Our SFTSV-specific probe and primers successfully distinguished target pathogen from fifteen other pathogens, exhibiting high specificity for SFTSV. Therefore, the RPA technique can probably be integrated into the detection of single nucleotide polymorphisms and pathogen-differentiating diagnostic methods, like the SHERLOCK detection platform, combining RPA with CRISPR-Cas13a, does in detecting Dengue or Zika virus ssRNA as well as mutations [30].
However, the RT-RPA assay we developed has certain limitations and should be improved in further studies. The RNA extraction method used in this study to process serum sample still require much effort and heavy centrifuges, which might hamper the practicality of this assay in point-of-care settings. Magnetic bead-based methods [21] or one-step RPA microfluidics [31] could be employed to simplify the sample preparation process. In detection of clinical samples, the RT-RPA assay has a certain probability to produce false-negative results [32]. To improve the clinical sensitivity of RT-RPA, investigation into the influence of the amount and distribution of mismatches on RPA and the employment of degenerate primers [33] is in need. Besides, with better fluorescence detection equipment, this technique can be integrated into a phone-size portable device and be easily used for field detection in SFTS-prevalent areas.
In conclusion, as shown in this study, we developed a rapid isothermal detection method for SFTSV with high sensitivity and specificity. The RT-RPA assay can be a suitable choice as a point-of-care detection assay for SFTSV.
Funding sources
This work was supported by the National Key Research and Development Program of China (No. 2016YFD0500604, No. 2016YFD0500603); National Natural Science Foundation of China (No. 81201306); the Thirteenth Five-Year National Science and Technology Major Project for Infectious Diseases (No. 2017ZX10305501-002); and Municipal Science and Technology Project of Zhoushan (No. 2016C11001, No. 2016C31038).
Ethical approval
All aspects of this study were performed in accordance with national ethic regulations and approved by the ethics committee of Zhoushan Hospital [(2016) approval No. 28].
CRediT authorship contribution statement
Jingyu Zhou: Conceptualization, Methodology, Data curation, Writing - original draft. Qiujing Wang: Resources, Methodology. Lijun Zhu: Methodology. Shibo Li: Resources. Wei Li: Formal analysis. Yongfeng Fu: Supervision, Conceptualization, Writing - review & editing. Xunjia Cheng: Project administration.
Declarations of competing interest
None.
Acknowledgements
We would like to thank Prof. Bo Dai (University of Shanghai for Science and Technology, Shanghai) for providing technical help, as well as Prof. Lingyun Shao (Huashan hospital, Shanghai) for proofreading the manuscript.
Contributor Information
Yongfeng Fu, Email: yffu@fudan.edu.cn.
Xunjia Cheng, Email: xjcheng@shmu.edu.cn.
References
- 1.Yu X.J., Liang M.F., Zhang S.Y., Liu Y., Li J.D., Sun Y.L., Zhang L., Zhang Q.F., Popov V.L., Li C., Qu J., Li Q., Zhang Y.P., Hai R., Wu W., Wang Q., Zhan F.X., Wang X.J., Kan B., Wang S.W., Wan K.L., Jing H.Q., Lu J.X., Yin W.W., Zhou H., Guan X.H., Liu J.F., Bi Z.Q., Liu G.H., Ren J., Wang H., Zhao Z., Song J.D., He J.R., Wan T., Zhang J.S., Fu X.P., Sun L.N., Dong X.P., Feng Z.J., Yang W.Z., Hong T., Zhang Y., Walker D.H., Wang Y., Li D.X. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 2011;364(16):1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denic S., Janbeih J., Nair S., Conca W., Tariq W.U., Al-Salam S. Acute thrombocytopenia, leucopenia, and multiorgan dysfunction: the first case of SFTS bunyavirus outside China? Case Rep Infect Dis. 2011:204056. doi: 10.1155/2011/204056. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMullan L.K., Folk S.M., Kelly A.J., MacNeil A., Goldsmith C.S., Metcalfe M.G., Batten B.C., Albarino C.G., Zaki S.R., Rollin P.E., Nicholson W.L., Nichol S.T. A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 2012;367(9):834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi T., Maeda K., Suzuki T., Ishido A., Shigeoka T., Tominaga T., Kamei T., Honda M., Ninomiya D., Sakai T., Senba T., Kaneyuki S., Sakaguchi S., Satoh A., Hosokawa T., Kawabe Y., Kurihara S., Izumikawa K., Kohno S., Azuma T., Suemori K., Yasukawa M., Mizutani T., Omatsu T., Katayama Y., Miyahara M., Ijuin M., Doi K., Okuda M., Umeki K., Saito T., Fukushima K., Nakajima K., Yoshikawa T., Tani H., Fukushi S., Fukuma A., Ogata M., Shimojima M., Nakajima N., Nagata N., Katano H., Fukumoto H., Sato Y., Hasegawa H., Yamagishi T., Oishi K., Kurane I., Morikawa S., Saijo M. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J. Infect. Dis. 2014;209(6):816–827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin J., Kwon D., Youn S.K., Park J.H. Characteristics and factors associated with death among patients hospitalized for severe fever with thrombocytopenia syndrome, South Korea, 2013. Emerg. Infect. Dis. 2015;21(10):1704–1710. doi: 10.3201/eid2110.141928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo C.T., Lu Q.B., Ding S.J., Hu C.Y., Hu J.G., Wo Y., Fan Y.D., Wang X.J., Qin S.L., Cui N., Yang Z.D., Zhang X.A., Liu W., Cao W.C. Epidemiological and clinical characteristics of severe fever with thrombocytopenia syndrome (SFTS) in China: an integrated data analysis. Epidemiol. Infect. 2016;144(6):1345–1354. doi: 10.1017/S0950268815002678. [DOI] [PubMed] [Google Scholar]
- 7.Sun J., Lu L., Wu H., Yang J., Ren J., Liu Q. The changing epidemiological characteristics of severe fever with thrombocytopenia syndrome in China, 2011-2016. Sci. Rep. 2017;7(1):9236. doi: 10.1038/s41598-017-08042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiao Y., Zeng X., Guo X., Qi X., Zhang X., Shi Z., Zhou M., Bao C., Zhang W., Xu Y., Wang H. Preparation and evaluation of recombinant severe fever with thrombocytopenia syndrome virus nucleocapsid protein for detection of total antibodies in human and animal sera by double-antigen sandwich enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2012;50(2):372–377. doi: 10.1128/JCM.01319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo X., Zhang L., Zhang W., Chi Y., Zeng X., Li X., Qi X., Jin Q., Zhang X., Huang M., Wang H., Chen Y., Bao C., Hu J., Liang S., Bao L., Wu T., Zhou M., Jiao Y. Human antibody neutralizes severe fever with thrombocytopenia syndrome virus, an emerging hemorrhagic fever virus. Clin. Vaccine Immunol. 2013;20(9):1426–1432. doi: 10.1128/CVI.00222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y., Liang M., Qu J., Jin C., Zhang Q., Li J., Jiang X., Wang Q., Lu J., Gu W., Zhang S., Li C., Wang X., Zhan F., Yao W., Bi Z., Wang S., Li D. Early diagnosis of novel SFTS bunyavirus infection by quantitative real-time RT-PCR assay. J. Clin. Virol. 2012;53(1):48–53. doi: 10.1016/j.jcv.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Yang G., Li B., Liu L., Huang W., Zhang W., Liu Y. Development and evaluation of a reverse transcription loop-mediated isothermal amplification assay for rapid detection of a new SFTS bunyavirus. Arch. Virol. 2012;157(9):1779–1783. doi: 10.1007/s00705-012-1348-1. [DOI] [PubMed] [Google Scholar]
- 12.Liu S., Chai C., Wang C., Amer S., Lv H., He H., Sun J., Lin J. Systematic review of severe fever with thrombocytopenia syndrome: virology, epidemiology, and clinical characteristics. Rev. Med. Virol. 2014;24(2):90–102. doi: 10.1002/rmv.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4(7) doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.China National Health and Family Planning Commission Guideline for prevention and treatment of sever fever with thrombocytopenia syndrome (2010 version) Chin J Clin Infect Dis. 2011;(4):193–194. 04. [Google Scholar]
- 15.Du Y.H., Cheng N.N., Li Y., Wang H.F., You A.G., Su J., Nie Y.F., Ma H.X., Xu B.L., Huang X.Y. Seroprevalance of antibodies specific for severe fever with thrombocytopenia syndrome virus and the discovery of asymptomatic infections in Henan province, China. PLoS Neglected Trop. Dis. 2019;13(11):10. doi: 10.1371/journal.pntd.0007242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong L., Song D.D., Wu J.B., Cao M.H., Su B., Sun Y., Lyu Y., Zhang L., Wang F., He Y.X., Wang J.S. Human-to-human transmissions of severe fever with thrombocytopenia syndrome virus in Anhui province, 2010-2017. Clin. Microbiol. Infect. 2018;24(8):920–922. doi: 10.1016/j.cmi.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Moon J., Lee H., Jeon J.H., Kwon Y., Kim H., Wang E.B., Seo C.W., Sung S.A., Kim S.H., Seok H., Choi W.S., Choi W., Park D.W. Aerosol transmission of severe fever with thrombocytopenia syndrome virus during resuscitation. Infect. Control Hosp. Epidemiol. 2019;40(2):238–241. doi: 10.1017/ice.2018.330. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q., He B., Huang S.Y., Wei F., Zhu X.Q. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect. Dis. 2014;14(8):763–772. doi: 10.1016/S1473-3099(14)70718-2. [DOI] [PubMed] [Google Scholar]
- 19.Patel P., Abd El Wahed A., Faye O., Pruger P., Kaiser M., Thaloengsok S., Ubol S., Sakuntabhai A., Leparc-Goffart I., Hufert F.T., Sall A.A., Weidmann M., Niedrig M. A field-deployable reverse transcription recombinase polymerase amplification assay for rapid detection of the chikungunya virus. PLoS Neglected Trop. Dis. 2016;10(9):15. doi: 10.1371/journal.pntd.0004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Wu Y., Yin D., Tang S., Hu G., He Y. Development and evaluation of a rapid recombinase polymerase amplification assay for detection of coxsackievirus A6. Arch. Virol. 2017;162(1):287–290. doi: 10.1007/s00705-016-3100-8. [DOI] [PubMed] [Google Scholar]
- 21.Xi Y., Xu C.Z., Xie Z.Z., Zhu D.L., Dong J.M., Xiao G. Development of a reverse transcription recombinase polymerase amplification assay for rapid detection of human respiratory syncytial virus. Mol. Cell. Probes. 2019;45:8–13. doi: 10.1016/j.mcp.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Cui L., Ge Y., Qi X., Xu G., Li H., Zhao K., Wu B., Shi Z., Guo X., Hu L., You Q., Zhang L.H., Freiberg A.N., Yu X., Wang H., Zhou M., Tang Y.W. Detection of severe fever with thrombocytopenia syndrome virus by reverse transcription-cross-priming amplification coupled with vertical flow visualization. J. Clin. Microbiol. 2012;50(12):3881–3885. doi: 10.1128/JCM.01931-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z., Cui L., Zhou M., Qi X., Bao C., Hu J., Shan J., Wu B., Wang S., Guo X., Jiao Y., Tang F., Wang H. Development and application of a one-step real-time RT-PCR using a minor-groove-binding probe for the detection of a novel bunyavirus in clinical specimens. J. Med. Virol. 2013;85(2):370–377. doi: 10.1002/jmv.23415. [DOI] [PubMed] [Google Scholar]
- 24.Li Z., Qi X., Zhou M., Bao C., Hu J., Wu B., Wang S., Tan Z., Fu J., Shan J., Zhu Y., Tang F. A two-tube multiplex real-time RT-PCR assay for the detection of four hemorrhagic fever viruses: severe fever with thrombocytopenia syndrome virus, Hantaan virus, Seoul virus, and dengue virus. Arch. Virol. 2013;158(9):1857–1863. doi: 10.1007/s00705-013-1677-8. [DOI] [PubMed] [Google Scholar]
- 25.Xu H., Zhang L., Shen G., Feng C., Wang X., Yan J., Zhang Y. Establishment of a novel one-step reverse transcription loop-mediated isothermal amplification assay for rapid identification of RNA from the severe fever with thrombocytopenia syndrome virus. J. Virol Methods. 2013;194(1–2):21–25. doi: 10.1016/j.jviromet.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 26.Abd El Wahed A., El-Deeb A., El-Tholoth M., Abd El Kader H., Ahmed A., Hassan S., Hoffmann B., Haas B., Shalaby M.A., Hufert F.T., Weidmann M. A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0071642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle D.S., Lehman D.A., Lillis L., Peterson D., Singhal M., Armes N., Parker M., Piepenburg O., Overbaugh J. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio. 2013;4(2) doi: 10.1128/mBio.00135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daher R.K., Stewart G., Boissinot M., Boudreau D.K., Bergeron M.G. Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol. Cell. Probes. 2015;29(2):116–121. doi: 10.1016/j.mcp.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Liu X., Yan Q., Huang J., Chen J., Guo Z., Liu Z., Cai L., Li R., Wang Y., Yang G., Lan Q. Influence of design probe and sequence mismatches on the efficiency of fluorescent RPA. World J. Microbiol. Biotechnol. 2019;35(6):95. doi: 10.1007/s11274-019-2620-2. [DOI] [PubMed] [Google Scholar]
- 30.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi G., Jung J.H., Park B.H., Oh S.J., Seo J.H., Choi J.S., Kim do H., Seo T.S. A centrifugal direct recombinase polymerase amplification (direct-RPA) microdevice for multiplex and real-time identification of food poisoning bacteria. Lab Chip. 2016;16(12):2309–2316. doi: 10.1039/c6lc00329j. [DOI] [PubMed] [Google Scholar]
- 32.Li J., Macdonald J., von Stetten F. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. 2019;144(1):31–67. doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- 33.Coertse J., Weyer J., Nel L.H., Markotter W. Reverse transcription recombinase polymerase amplification assay for rapid detection of canine associated rabies virus in Africa. PloS One. 2019;14(7):15. doi: 10.1371/journal.pone.0219292. [DOI] [PMC free article] [PubMed] [Google Scholar]