Abstract
Internal ribosome entry site (IRES) elements consist of cis-acting regions that recruit the translation machinery to an internal position in the mRNA. The biological relevance of RNA structure-mediated mechanisms involved in internal ribosome recruitment is now emerging from the structural and functional analysis of viral IRES elements. However, because IRES elements found in genetically distant mRNAs seem to be organized in different RNA structures, the definition of the structural requirements for IRES activity is challenging and demands multidisciplinary approaches. This review discusses the latest reports that establish a relationship between RNA structure and IRES function in picornavirus genomes, the first RNAs described to contain these specialized regulatory elements.
Internal initiation of protein synthesis in picornavirus
Picornaviruses belong to an important group of animal pathogens that have evolved a specialized mechanism to promote translation initiation of their mRNAs internally [1]. This mechanism of translation initiation enables the preferential translation of their genome while avoiding the modification of translation initiation factors (eIFs) and other host factors that occur in infected cells 2, 3, 4 and induce the inactivation of host protein synthesis. The process of internal initiation of translation in eukaryotic mRNAs is dependent on a cis-acting element known as the internal ribosome entry site (IRES) that uses an internal codon in the mRNA to start protein synthesis. IRES elements were originally found in the genomes of picornaviruses, which are a family of positive strand RNA viruses 5, 6. IRES elements have also been found in a growing number of genetically distant viral RNAs, including flaviviruses, retroviruses, coronaviruses, dicistroviruses and plant RNA viruses, in addition to several cellular mRNAs that are translated under conditions when cap-dependent initiation is inhibited 7, 8, 9, 10, 11, 12, 13, 14.
Most eukaryotic mRNAs use a cap structure (m7Gppp) (see Glossary) at the 5′ end to initiate translation via the translation initiation factor eIF4F [15]. The process of protein synthesis is initiated by the interaction of the 40S ribosomal subunit (bound to eIF3–eIF2–methionyl-tRNA) with the eIF4F complex (eIF4G–eIF4A–eIF4E), which in turn is bound to the 5′ cap of the mRNA. This macromolecular complex then scans the 5′ untranslated region (UTR) to reach an AUG triplet in an appropriate context [16]. By contrast, IRES-dependent translation initiation recruits the translational machinery to an internal position in the mRNA, regardless of the presence of upstream AUG codons, stable RNA structure and protein-binding sites 1, 17.
The picornavirus genome consists of a positive sense RNA of ∼7400–8500 nucleotides, in which 5′ and 3′ UTRs flank the single open reading frame (Figure 1 ). The 5′ end of the viral RNA is covalently linked to viral protein g (VPg). All members of the different genera belonging to the picornavirus family initiate translation via an IRES element [17]. The success of the picornavirus replication cycle, and thus the effectiveness of infection, is dependent on the correct function of the IRES; the IRES region is therefore a target for antiviral drugs aiming to inactivate the IRES [18], and is a determinant of viral pathogenesis and virulence 19, 20, 21.
Figure 1.

Diagram of protein interactions with a picornavirus IRES. (a) Upstream of the IRES element in the 5′UTR of the aphthovirus genome (top), several structural elements are arranged consecutively: the S region, the poly(C) tract, the pseudoknotted region (PK), the cis-replicative element (cre). The poly(A) tail is located at the 3′UTR. The grey circle at the 5′ end depicts the viral protein VPg. Approximate nucleotide content of each region of the viral genome is indicated. The IRES region is enlarged from the aphthovirus genome (b). Position of IRES domains [2(H) to 5 (L)] is indicated at the bottom of the corresponding stem-loops. Interaction of specific sequences within domain 4 with initiation factor eIF4G is essential for IRES activity. eIF3, which is also required for IRES activity, shows preferential binding in domain 5. eIF4B-binding is restricted to the hairpin of domain 5 and moderately enhances IRES activity. The auxiliary protein PTB, which contributes to enhanced IRES activity, recognizes two separate polypyrimidine tracts, one in domain 2 and another in domain 5. The host factor PCBP interacts with the C-rich bulge in the central domain; its interaction is needed for entero- and rhinovirus IRES, but not for cardio- and aphthovirus IRES activity.
Picornavirus IRES-driven translation initiation depends on the recognition of IRES by specific cellular proteins [22]. Translation initiation factors such as eIF4G are essential for picornavirus IRES activity 23, 24, 25. In addition, RNA-binding proteins such as the polypyrimidine tract-binding protein (PTB), the poly(rC)-binding protein (PCBP), the SRp20 splicing factor, the proliferation-associated IRES transacting factor (ITAF)45 or the upstream-of-Nras protein (Unr) interact with IRES elements 26, 27, 28, 29, 30, 31 presumably facilitating their structural organization by acting as RNA chaperones.
In this review, we discuss structural aspects of picornavirus IRES elements that are crucial for internal initiation activity. We suggest that the acquisition of specific structural organization by the IRES has a dual function; it is necessary for recognition by RNA-binding proteins and for RNA-mediated regulation of internal translation initiation.
Functional link between the 5′ and 3′ ends of the viral RNA
Polyadenylation is a conserved feature in picornavirus RNAs and cellular mRNAs. The poly(A) tail present in the majority of eukaryotic mRNAs improves the efficiency of translation initiation through recruitment of poly(A)-binding protein (PABP), enabling its interaction with eIF4F located at the mRNA 5′ end. In picornavirus RNAs the poly(A) tail together with the sequence of the 3′ UTR, which is specific for each genus, constitute essential determinants of the virus multiplication cycle 32, 33, 34. The efficiency of viral RNA replication is enhanced by protein bridges that mediate RNA circularization by interacting with both ends of the viral RNA. In poliovirus RNAs the cloverleaf (CL) structure and the adjacent C-rich spacer interact with the host protein PCBP2 and the viral protein 3CD [35] generating a ternary complex that is essential for viral RNA replication, and that possibly regulates the switch from translation to replication [36]. Viral proteins 3A and 3C also contribute to the formation of a functional complex with the IRES and 3′ UTR, which is important for viral propagation [37].
Poliovirus IRES activity in neuronal cells was stimulated by the presence of specific 3′ UTR structural elements [38] suggesting a functional role for the 3′ end–IRES interaction in viral pathogenesis. Similarly, a functional link between the aphthovirus IRES and the 3′ end of the viral RNA was suggested by the specific stimulation of IRES activity by the foot-and-mouth disease virus (FMDV) 3′ UTR [39]. In agreement with data reported in other RNA viruses [40], the 3′ end of the FMDV genome establishes strand-specific long range RNA–RNA interactions, but in this case there are two different interactions, one with the IRES element and another with the S region at the 5′ end [41] (Figure 1). Different RNA motifs seem to be involved in these interactions because a high-order structure adopted by the entire IRES and the 3′ UTR was essential for RNA interaction, whereas the S region interacted with each of the stem-loops at the genome 3′ end. The 3D structure of the FMDV 3′ UTR is unknown; however in enteroviruses the 3′ UTR is organized as two stem-loops that adopt a quasi globular organization [42].
Thus, bridging of 5′ and 3′ ends in the picornavirus genome involves direct RNA–RNA contacts and RNA–protein interactions. It is noteworthy that the host factor PCBP2, which is required for translation initiation and viral RNA replication, is cleaved during poliovirus infection [2] resulting in a truncated protein that is unable to function in translation but that maintains its activity in viral RNA replication. This event might promote the switch from viral translation to RNA replication, a key step in the picornavirus infection cycle that needs to be tightly regulated.
Relevance of RNA structure in IRES function
Understanding IRES biology is crucial for increasing our knowledge of translational control in viral RNAs. Several studies have shown a strong relationship between RNA structure and IRES function, but the basic mechanisms through which IRES elements recruit the ribosome have only begun to emerge recently. This is due, at least in part, to the lack of a conserved core either in the primary sequence or in the predicted RNA structure of genetically distant IRES elements.
From a functional and structural point of view, the picornavirus IRES together with IRES elements present in the viral RNA of hepatitis C virus (HCV) [7] and the dicistrovirus intergenic region (IGR) [14] provide good model systems for addressing the relevance of RNA structure in IRES function. The IRES elements present in these three positive strand RNA viruses belonging to different families possibly encounter the largest variation in RNA organization and eIF requirements. The dicistrovirus IGR is organized in a pseudoknotted RNA structure and it does not require eIFs for assembly of 48S initiation complexes 43, 44. The HCV IRES adopts a different folded RNA structure (distributed in three domains, II, III and IV, with IV including a pseudoknot) and requires eIF3 and eIF2, but no eIF4G, to assemble initiation complexes 45, 46, 47. In this scale of growing complexity, picornavirus IRES represent the most complex group in terms of sequence length and factor requirements 23, 25.
In the genome of all picornaviruses, the IRES element is surrounded by RNA structural elements. This is particularly evident in cardiovirus and aphthovirus, which have long 5′ UTRs burdened with different structural elements required for viral infectivity [41] (Figure 1). The IRES region spans approximately 450 nucleotides of the 5′ viral RNA, and on average <50% of the primary nucleotide sequence is conserved between different picornaviruses. However, the secondary structure is conserved between different members of this family [48]. According to secondary structure prediction, representative members such as poliovirus (PV), coxsackievirus B3 (CVB3) or human rhinovirus (HRV) belong to group I, whereas encephalomyocarditis virus (EMCV) or FMDV belong to group II. A novel group that shares RNA structural similarities with the IRES element of HCV has been described recently 49, 50.
All picornavirus IRES elements are organized in stable domains termed II to VII in enteroviruses and G to L or 2 to 5 in cardio- and aphthovirus IRES 1, 17, 48. Structural analysis of type II IRES encompassing either single domains or the entire IRES have shown the presence of self-folding structural elements that acquire a similar conformation 51, 52. Consistent with the lack of effect of upstream sequences, the IRES element occupies an internal position in the viral RNA where the different 5′ UTR upstream elements perform their specific roles during the replication cycle without interfering with IRES activity. Consequently, the picornavirus IRES elements promote efficient protein synthesis in polycistronic vectors [53].
Biochemical and functional analyses have shown that distal domains within the IRES structure are involved in interactions with host factors [17]. Recognition of the different picornavirus IRES elements by host transacting factors is mainly dependent on the structural organization of specific domains, as demonstrated by mutational analysis of the binding site of proteins such as eIF4G, eIF4B or PTB (Figure 1; reviewed in [54]). However, the role of the central domain of picornavirus IRES that occupies a significant portion of the entire sequence remains elusive. Few interactions with host factors have been found in this region; a conserved C-rich motif that interacts with PCBP2 is essential for IRES activity in the viral RNA of entero- and rhinovirus, but not in cardio- and aphthovirus [27].
A distinctive feature of the central domain (termed 3 or I in cardio- and aphthovirus; Figure 2 ) is the presence of a cruciform structure at the most apical region [51]. This region of domain 3 contains two conserved purine-rich motifs, GNRA and RAAA (N, any nucleotide, R, purine), located in distal loops that do not tolerate nucleotide substitutions, deletions or insertions 55, 56. The proximal region of this domain is organized as a base-paired structure interrupted with bulges that includes several non-canonical base pairings and a helical structure at its proximal region that is crucial for IRES activity [57]. These results suggest that the central domain could have a regulatory role during internal initiation, in which the proximal region of the central domain contributes to IRES activity by serving as a platform that holds the apical region in a conformation appropriate for its recognition by the translation machinery.
Figure 2.
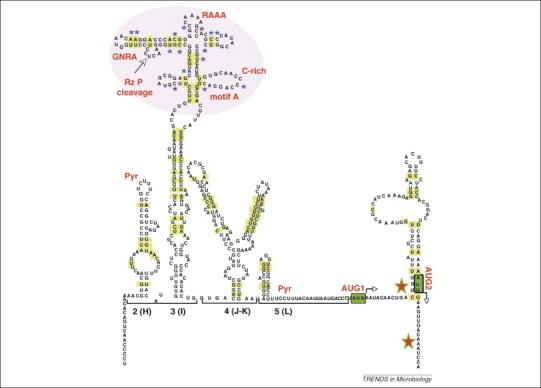
Schematic representation of the secondary structure of a picornavirus IRES. Nucleotide covariation is indicated by yellow rectangles; Pyr denotes the polypyrimidine tracts; GNRA, RAAA, C-rich and motif A denote picornavirus IRES conserved motifs; Rz P cleavage denotes the cleavage site of the FMDV IRES transcript in vitro by the cyanobacteria RNase P ribozyme. The apical region of domain 3 (I) is highlighted by a purple circle that contains the RNase P recognition motif overlapping with the GNRA stem-loop. In this region the nucleotides marked with blue asterisks showed a differential accessibility in the cellular cytoplasm. The positions of the functional AUG codons (arrows on green boxes) as well as the toe-prints (green and red stars) are depicted.
The GNRA motifs of different picornavirus IRES (FMDV, PV, CVB3 and EMCV) adopt a tetraloop conformation at the tip of a stem-loop 51, 58, 59, 60. GNRA tetraloops are often involved in RNA folding, generating RNA tertiary contacts [61]. Sequence variability of the IRES region in FMDV field isolates shows substitutions in the GUAA sequence yielding GCAA or GCGA, always compatible with the GNRA consensus motif. Conversely, as shown by RNA probing, IRES residues involved in base pairing often show nucleotide covariation (Figure 2), strongly supporting the need for preserving IRES structure for internal initiation [54]. Site-directed nucleotide substitutions disrupting the FMDV GNRA motif led to the reorganization of the apical region of the central domain, demonstrating the significant role of this conserved motif in dictating the stability of the cruciform structure. Subsequent structural probing of GNRA mutants enabled the identification of a distant region (motif A) that became more susceptible to RNase attack [51]. Functional analysis of mutants in this region indicated a reduction in IRES activity similar to that reported for GUAG mutants. Conversely, RNA probing of the second-site substitutions showed that motif A mutants contained a reorganization that affected the mutated region and also the distant GNRA motif [62]. Based on the reciprocity of RNA structural changes this short region behaves as a receptor of the GNRA motif. Lack of genetic variability in this region in approximately one hundred FMDV sequences [63] showed a strong selective pressure for keeping the primary sequence. This feature can be explained either because of its involvement in distant contacts with other RNA motifs, or by recognition by auxiliary factors involved in translation initiation that remain to be identified.
The organization of the RAAA stem-loop was also dependent on the local RNA structure determined by GNRA-dependent interactions [62]. Thus, multiple contacts within the apical region of the central domain presumably contribute to formation of a tight RNA structure that is essential for IRES activity, as also suggested by the finding of Mg2+-dependent RNA–RNA interactions mediated by this IRES region [64]. Along this line, RNA probing of the entire CVB3 5′ NTR has shown that the previously known domains II to VII, defined by mutational and bioinformatic approaches, comprises a long-range tertiary interaction involving bases in domain II and domain V [60] that connects the 5′ end (domain I, which folds in a CL structure) with the IRES element. This physical association opens new perspectives for crosstalk between the IRES element and the CL regarding translation and replication of viral RNA.
Structural studies performed in other viral IRES showed that the tertiary structure of IRES elements contributes to regulation of translation efficiency 46, 43, 47. The IRES elements of cricket paralysis virus (CrPV) and HCV have different RNA structures and distinct binding sites in the ribosomal subunit, yet they induce similar conformational changes in the 40S ribosomal subunit [65]. This observation suggested that IRES elements could share the property of having unique structural elements that mediate direct interaction with the 40S subunit. Accordingly, it has been speculated that the central domain of the aphthovirus and its structurally related cardiovirus IRES could have a fundamental role in dictating the formation of a stable tertiary structure, thereby providing the correct orientation to recruit the ribosome subunits to the initiation sites [62].
As mentioned previously, the IRES region constitutes a structural entity within the viral genome; this model is supported by the ability of IRES to fold in essentially the same manner, regardless of the downstream viral sequences that encompass the initiator codons, and fold as a stem-loop [66] (Figure 2). In the aphthovirus genome, two in-frame AUG triplets separated by 84 nucleotides are used as translation initiator codons of the viral polyprotein. The second functional triplet (AUG2) is used to initiate protein synthesis more frequently (80%) than AUG1 in infected cells, and also in cells transfected with chimeric RNAs 67, 68. A conserved A-rich sequence precedes AUG2 (Figure 2) and is engaged in base pair formation within a stem-loop phylogenetically conserved in FMDV field isolates. By contrast, the first functional start codon (AUG1) is located in a single-stranded region, as judged by its accessibility to chemicals and enzymes [66]. Initiation of protein synthesis from both AUGs is IRES-dependent but the mechanism operating in codon selection remains poorly understood. It has been proposed that following entry of the ribosome the initiation complex scans along this viral region until AUG2 is reached [68], or that the initiation complex is transferred to the vicinity of AUG2 [67]. Recently it was shown that translation initiation at AUG2 was barely abrogated by the presence of a modified, more stable RNA structure in the spacer sequence [66], consistent with previous observations of lack of effect of antisense molecules bound to AUG1 [67]. However, the factors required for reconstituting 48S initiation complexes at each initiation codon in vitro are different. In addition to eIF3, eIF4G and eIF4A, recognition of AUG1 is dependent on eIF1A, whereas initiation at AUG2 is dependent on eIF1 [66], suggesting that the mechanisms operating for selection of the initiation codon in FMDV depend on the sequence preceding the initiation triplet. This conclusion is in agreement with the observation that non-viral sequences adopting a stable hairpin structure placed in a spacer region between the IRES and the AUG of a reporter coding region abrogated translation initiation [69], but did not do so in the viral RNA 66, 67.
Structural organization of picornavirus IRES elements in living cells
The basis of RNA structure-mediated mechanisms involved in ribosome recruitment during internal initiation is currently under active investigation. In the past few years, determination of the RNA structure of several viral IRES elements using in vitro approaches has represented a major challenge in this area of research 44, 70. IRES elements found in unrelated mRNAs do not share primary sequences and they seem to be organized in different RNA structures 7, 54. Thus, the structural requirements for IRES activity remain to be defined.
The picornavirus IRES have been found to interact specifically with some eIFs in vitro, using reconstituted initiation complexes [1], or in functional analysis performed in transfected cells 24, 71. Therefore, it is conceivable that in the competitive environment of the cellular cytoplasm IRES function depends on the availability of eIFs and other ITAFs and their coordinated interaction with the IRES RNA. These functional interactions might be compromised owing to their preferential use in cap-dependent initiation as well as in other processes related to RNA biology [17].
Despite multiple efforts to understand IRES biology, information on the organization of IRES elements in living cells remains limited. Recent studies have taken advantage of reagents permeable to the cellular membrane that recognize RNA molecules in a structure-dependent manner to elucidate the organization of picornavirus IRES elements in living cells [52]. Notably, specific bases of the central domain in the FMDV IRES were differentially reactive towards dimethyl sulfate (DMS) in vivo than in vitro (Figure 2), demonstrating that the pattern of modification found in the cytoplasm of living cells differed from that obtained in vitro. In addition, new DMS-reactive bases specifically detected in vivo affected nucleotides involved in the formation of G:C base pairs in the secondary structure model generated in vitro. This result again suggests that the central region of the IRES adopts a different local RNA conformation in the cellular cytoplasm. Decreased attack of residues in the C-rich loop in vivo suggested a potential RNA–protein interaction; this conserved motif is a candidate for interaction with PCBP [27]. Conversely, strong protection in the region upstream of the initiator codon suggested the formation of RNA–protein complexes, which overlap the region described to interact with translation initiation factors in tissue culture cells 24, 71.
Complementary studies on the organization of FMDV IRES in the cell cytoplasm using the photoreactive reagent amino-methyl psoralen revealed a hot-spot of crosslinked pyrimidines in the central domain [34], consistent with the formation of inter-strand crosslinks in the secondary structure. The comparison of the UV-psoralen crosslink pattern performed in vitro to that seen in vivo showed changes compatible with a local reorganization of RNA structure within the apical region of the central domain.
Overall, the results derived from different approaches are consistent with an active role of the RNA structure in this region during translation initiation. The modification of RNA accessibility detected in the central domain in vivo could be because of different scenarios. The interaction with host transacting factors in the cellular cytoplasm could be responsible for this reorganization, as observed in vitro for other IRES elements [72]. Another possibility is the acquisition of a structural conformation that might facilitate a direct involvement of the apical region of the central domain with the ribosomal components. In support of this hypothesis, a tRNA-like structural motif that serves as substrate for RNase P in vitro resides in this IRES region [73]. These two possibilities are not mutually exclusive, and both need further investigation because no 48S initiation complexes have been detected in the absence of eIFs and the corresponding IRES-dependent toe-prints are located downstream of the respective initiation codon [66] (Figure 2).
Functional and evolutionary implications of IRES-related structural elements in viral RNAs
At present, the question of how the IRES elements appeared during evolution is unresolved. Although the primary sequence is remarkably divergent, conservation of specific motifs in IRES structural organization could provide some hints on their evolutionary history. As an example, the conservation of the pseudoknot motif – a characteristic feature of the HCV IRES – supports the possibility that the differences in RNA structure found among picornavirus IRES might have come about by horizontal transmission between different ancestors [50]. Another conserved feature of viral IRES is the presence of one or more polypyrimidine tracts 1, 48. However, a search of IRES elements on the basis of short conserved primary sequences (e.g. the polypyrimidine tract or the C-rich motifs that provide PTB or PCBP-binding sites, respectively) does not guarantee the presence of a functional element. The data available are theoretically compatible with a high-order structure generated by a combination of several structural elements that differ between divergent IRES elements; how many different combinations can generate a functionally active element remains an unresolved question.
Structural motifs in HCV RNA that serve as substrate for the human RNase P have been discovered recently, one in the IRES and another in the coding region [74]. On the basis of the recognition by RNase P it was inferred that the HCV and pestivirus IRES contained a structural element that mimics the tRNA-like structure [75]. RNase P is a structure-dependent endonuclease involved in the processing of the tRNA precursor [76], which also recognizes RNA viruses containing tRNA-like structures at the 3′ end of the viral genome as substrate [77]. The aphthovirus IRES contains a structural motif that acts as substrate for the RNase P ribozyme in vitro (Figure 2). The core structural element recognized by the ribozyme resides in the central domain of the FMDV IRES [73], overlapping with a self-folding region that contains the conserved motif GNRA, which is essential for IRES activity. As mentioned previously, the GNRA motif mediates the local RNA structure [51], presumably with the involvement of tertiary contacts. In support of the recognition of a structural element, defective FMDV IRES mutants with modified RNA structure responded differentially to ribozyme cleavage leading to an enhanced recognition that was accompanied by cleavage sites in nearby residues [73] as it also occurs in a variant RNA molecule of HCV [78]. As the viral cycle of picornaviruses and flaviviruses occurs in the cell cytoplasm, the viral RNA has no access to RNase P, which resides in the nucleus of eukaryotic cells [76].
The ribozyme recognition motif in FMDV is located in a different position relative to the HCV or the pestivirus IRES, which have this motif at the 3′ end of the IRES region [74]. From an evolutionary point of view, this observation can be interpreted in several ways. First, the differences in the relative position within the IRES and also within the genome of other distantly related viral RNAs 74, 77 could be the remnants of an ancestor module without function in the viral genome. Second, because overlapping information in the small genome of RNA viruses occurs often, it is also possible that this motif is involved in a new function still to be determined. Third, regarding the possibility of an active role of this motif during IRES activity, the differences found between IRES elements could be indicative of the distinct strategies used by the genome of RNA viruses belonging to different families to interact with the translational machinery.
In this regard, structural studies performed on HCV and the IGR of dicistroviridae members have shown that these IRES elements are located in the interface of the ribosomal subunits 79, 70, 44. These results suggested that specialized motifs of the IRES region mimic the initiator tRNA (tRNAi) during initiation of protein synthesis and have recently been confirmed in domain 3 of the CrPV IGR [80]. It is proposed that the IGR structure undergoes subtle structural changes during the process of translation initiation, suggesting a direct involvement of the RNA structure in the ribosome assembly and translocation process. It can be envisaged that a tRNA-like motif within the picornavirus IRES could contribute to efficient recognition of the viral RNA by the translational machinery because it seems to occur in the dicistrovirus IGR 14, 80. However, to date there is no evidence of the formation of binary complexes mediated by the picornavirus IRES and 40S subunits in the absence of eIFs. Thus, a functional connection between RNA organization detected by structural analysis and a putative tRNA-like structure detected by ribozyme recognition in IRES elements requires further investigation.
Concluding remarks and future perspectives
Understanding how RNA structure-mediated mechanisms operate in genetically divergent IRES elements is necessary for generating a model of the internal initiation process occurring in eukaryotic cells. Picornavirus IRES elements seem to work as a ribonucleoprotein engine that assembles a large number of factors into a specialized, appropriately folded RNA structure that can be recognized by the ribosomal subunits. Other viral IRES elements seem to work as docking RNA structures that can gain access to specific sites in the ribosomal subunit without the help of additional factors. In between these divergent types of IRES elements, large variations in the mode of action exist that extend from picornavirus-like to the poorly understood cellular IRES elements. The idea that a universal IRES motif could consist of a structural element that mediates the interaction with the ribosomal machinery has gained support from studies using different approaches. In particular, structural and functional studies of the relatively small dicistrovirus IRES models have led to the idea that the IRES element itself mimics the initiator tRNA during internal initiation, in a process in which flexibility of the RNA molecule is crucial. Evaluating this model in the more complex, but also more efficient, picornavirus IRES elements represents a major challenge for the near future.
Acknowledgements
I am grateful to C. Gutierrez for helpful suggestions. This work was supported by grant BFU-2005–00948 and by an Institutional grant from Fundación Ramón Areces.
Glossary
- Cap structure
m7Gppp residue present in the 5′ end of eukaryotic mRNAs that is recognized by the initiation factor eIF4E.
- Dimethyl sulfate (DMS)
A reagent that reacts with unpaired bases in the RNA structure, used to measure accessibility to C and A bases.
- GNRA
A motif in the RNA structure that consist of 4 bases, G, N (any nucleotide), R (purine) and A.
- Intergenic region (IGR)
The region in the genome of dicistroviruses that separates two open reading frames and enables internal initiation of translation of the second cistron.
- Internal ribosome entry site (IRES)
A region in eukaryotic mRNA that enables translation initiation independently of the 5′ end.
- ITAFs
Trans-acting factors that interact with IRES regions.
- Picornaviruses
A group of unsegmented and non-enveloped positive-strand RNA viruses, including enteroviruses, rhinoviruses, cardioviruses, aphthoviruses, hepatoviruses, erboviruses, teschoviruses, parechoviruses and kobuviruses.
- Toe-print
A technique that determines the position of assembled 48S initiation complexes by measuring the inhibition of reverse transcriptase elongation of antisense primers.
- Untranslated regions (UTR)
The sequences on either end of the mRNA, flanking the coding region.
References
- 1.Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 2.Perera R. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez Pulido M. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology. 2007;364:466–474. doi: 10.1016/j.virol.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 6.Jang S.K. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C.S., Doudna J.A. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 8.O’Connor J.B., Brian D.A. Downstream ribosomal entry for translation of coronavirus TGEV gene 3b. Virology. 2000;269:172–182. doi: 10.1006/viro.2000.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baird S.D. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balvay L. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karetnikov A., Lehto K. The RNA2 5′ leader of blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J. Gen. Virol. 2007;88:286–297. doi: 10.1099/vir.0.82307-0. [DOI] [PubMed] [Google Scholar]
- 12.Dinkova T.D. Cap-independent translation of maize Hsp101. Plant J. 2005;41:722–731. doi: 10.1111/j.1365-313X.2005.02333.x. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert W.V. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 14.Wilson J.E. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 15.Merrick W.C. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Salas E. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 2001;82:973–984. doi: 10.1099/0022-1317-82-5-973. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez A. Specific inhibition of aphthovirus infection by RNAs transcribed from both the 5′ and the 3′ noncoding regions. J. Virol. 1994;68:7426–7432. doi: 10.1128/jvi.68.11.7426-7432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilipenko E.V. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 2001;20:6899–6908. doi: 10.1093/emboj/20.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malnou C.E. Poliovirus internal ribosome entry segment structure alterations that specifically affect function in neuronal cells: molecular genetic analysis. J. Virol. 2002;76:10617–10626. doi: 10.1128/JVI.76.21.10617-10626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn J.J. The stem loop II within the 5′ nontranslated region of clinical coxsackievirus B3 genomes determines cardiovirulence phenotype in a murine model. J. Infect. Dis. 2003;187:1552–1561. doi: 10.1086/374877. [DOI] [PubMed] [Google Scholar]
- 22.Belsham G.J., Sonenberg N. Picornavirus RNA translation: roles for cellular proteins. Trends Microbiol. 2000;8:330–335. doi: 10.1016/s0966-842x(00)01788-1. [DOI] [PubMed] [Google Scholar]
- 23.Kolupaeva V.G. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 24.Lopez de Quinto S., Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez de Quinto S. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monie T.P. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter B.L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luz N., Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1991;65:6486–6494. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boussadia O. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003;77:3353–3359. doi: 10.1128/JVI.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilipenko E.V. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 31.Bedard K.M. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svitkin Y.V. Stimulation of picornavirus replication by the poly(A) tail in a cell-free extract is largely independent of the poly(A) binding protein (PABP) RNA. 2007;13:2330–2340. doi: 10.1261/rna.606407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobrikova E.Y. Competitive translation efficiency at the picornavirus type 1 internal ribosome entry site facilitated by viral cis and trans factors. J. Virol. 2006;80:3310–3321. doi: 10.1128/JVI.80.7.3310-3321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saiz M. Deletion or substitution of the aphthovirus 3′ NCR abrogates infectivity and virus replication. J. Gen. Virol. 2001;82:93–101. doi: 10.1099/0022-1317-82-1-93. [DOI] [PubMed] [Google Scholar]
- 35.Toyoda H. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J. Virol. 2007;81:10017–10028. doi: 10.1128/JVI.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Florez de Sessions P. Genetic adaptation to untranslated region-mediated enterovirus growth deficits by mutations in the nonstructural proteins 3AB and 3CD. J. Virol. 2007;81:8396–8405. doi: 10.1128/JVI.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobrikova E. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez de Quinto S. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filomatori C.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrano P. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA–RNA interactions with the 5′ end region. J. Gen. Virol. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- 42.van Ooij M.J. Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative- and positive-strand RNA synthesis. J. Gen. Virol. 2006;87:103–113. doi: 10.1099/vir.0.81297-0. [DOI] [PubMed] [Google Scholar]
- 43.Jan E., Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 44.Pfingsten J.S. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pestova T.V. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kieft J.S. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 47.Locker N. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson R.J., Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 49.Pisarev A.V. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hellen C.U., de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez-Miragall O., Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez-Miragall O., Martinez-Salas E. In vivo footprint of a picornavirus internal ribosome entry site reveals differences in accessibility to specific RNA structural elements. J. Gen. Virol. 2007;88:3053–3062. doi: 10.1099/vir.0.83218-0. [DOI] [PubMed] [Google Scholar]
- 53.de Felipe P. Polycistronic viral vectors. Curr. Gene Ther. 2002;2:355–378. doi: 10.2174/1566523023347742. [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Salas E., Fernandez-Miragall O. Picornavirus IRES: structure function relationship. Curr. Pharm. Des. 2004;10:3757–3767. doi: 10.2174/1381612043382657. [DOI] [PubMed] [Google Scholar]
- 55.Lopez de Quinto S., Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson M.E. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez-Salas E. Identification of an essential region for internal initiation of translation in the aphthovirus internal ribosome entry site and implications for viral evolution. J. Virol. 1996;70:992–998. doi: 10.1128/jvi.70.2.992-998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phelan M. NMR studies of the structure and Mg2+ binding properties of a conserved RNA motif of EMCV picornavirus IRES element. Nucleic Acids Res. 2004;32:4715–4724. doi: 10.1093/nar/gkh805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du Z. NMR structures of loop B RNAs from the stem-loop IV domain of the enterovirus internal ribosome entry site: a single C to U substitution drastically changes the shape and flexibility of RNA. Biochemistry. 2004;43:5757–5771. doi: 10.1021/bi0363228. [DOI] [PubMed] [Google Scholar]
- 60.Bailey J.M., Tapprich W.E. Structure of the 5′ nontranslated region of the coxsackievirus b3 genome: chemical modification and comparative sequence analysis. J. Virol. 2007;81:650–668. doi: 10.1128/JVI.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Correll C.C., Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 A resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernandez-Miragall O. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carrillo C. Comparative genomics of foot-and-mouth disease virus. J. Virol. 2005;79:6487–6504. doi: 10.1128/JVI.79.10.6487-6504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramos R., Martinez-Salas E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES) RNA. 1999;5:1374–1383. doi: 10.1017/s1355838299991240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spahn C.M. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 66.Andreev D.E. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez de Quinto S., Martinez-Salas E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255:324–336. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- 68.Belsham G.J. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1105–1110. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez de Quinto S., Martinez-Salas E. Parameters influencing translational efficiency in aphthovirus IRES-based bicistronic expression vectors. Gene. 1998;217:51–56. doi: 10.1016/s0378-1119(98)00379-5. [DOI] [PubMed] [Google Scholar]
- 70.Schuler M. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 71.Clark A.T. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J. Virol. 2003;77:12441–12449. doi: 10.1128/JVI.77.23.12441-12449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolupaeva V.G. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Serrano P. Characterization of a cyanobacterial RNase P ribozyme recognition motif in the IRES of foot-and-mouth disease virus reveals a unique structural element. RNA. 2007;13:849–859. doi: 10.1261/rna.506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nadal A. Specific cleavage of hepatitis C virus RNA genome by human RNase P. J. Biol. Chem. 2002;277:30606–30613. doi: 10.1074/jbc.M203595200. [DOI] [PubMed] [Google Scholar]
- 75.Lyons A.J., Robertson H.D. Detection of tRNA-like structure through RNase P cleavage of viral internal ribosome entry site RNAs near the AUG start triplet. J. Biol. Chem. 2003;278:26844–26850. doi: 10.1074/jbc.M304052200. [DOI] [PubMed] [Google Scholar]
- 76.Evans D. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 77.Guerrier-Takada C. Novel reactions of RNAase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988;53:267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- 78.Piron M. Characterizing the function and structural organization of the 5′ tRNA-like motif within the hepatitis C virus quasispecies. Nucleic Acids Res. 2005;33:1487–1502. doi: 10.1093/nar/gki290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boehringer D. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 80.Costantino D.A. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]


