Abstract
Bacterial infections and antibiotic resistant bacteria have become a growing problem over the past decade. As a result, the Centers for Disease Control predict more deaths resulting from microorganisms than all cancers combined by 2050. Currently, many traditional models used to study bacterial infections fail to precisely replicate the in vivo bacterial environment. These models often fail to incorporate fluid flow, bio-mechanical cues, intercellular interactions, host-bacteria interactions, and even the simple inclusion of relevant physiological proteins in culture media. As a result of these inadequate models, there is often a poor correlation between in vitro and in vivo assays, limiting therapeutic potential. Thus, the urgency to establish in vitro and ex vivo systems to investigate the mechanisms underlying bacterial infections and to discover new-age therapeutics against bacterial infections is dire. In this review, we present an update of current in vitro and ex vivo models that are comprehensively changing the landscape of traditional microbiology assays. Further, we provide a comparative analysis of previous research on various established organ-disease models. Lastly, we provide insight on future techniques that may more accurately test new formulations to meet the growing demand of antibiotic resistant bacterial infections.
Keywords: Infection models, In vitro, Ex vivo, Nanotechnology, Organ-on-a-chip
1. Background
1.1. Rising concerns of bacterial infections
The rising number of bacterial infections, especially from antibiotic resistant bacteria, has become a global threat to public health over recent decades. According to the 2013 Center for Disease Control and Prevention (CDC) report on “antibiotic resistance threats in the U.S.”, more than 2 million people become infected with antibiotic resistant bacteria in the United States alone every year [1]. This directly results in a death toll of over 23,000 people each year, with many more dying from infection complications. Furthermore, infections caused by drug resistant bacteria often require an extended hospital stay and expensive treatments, which add a considerable economic burden to the healthcare system estimated to be as high as $20 billion [2]. In addition to the emergence of antibiotic resistance, concerns have also been raised for infections associated with a variety of medical devices including joint prostheses, heart valves, pacemakers and catheters [3]. Device-associated infections are typically caused by microorganisms that grow in biofilms, and are introduced via surgery or implants [4,5]. Treatment of these infections are generally more challenging and require prolonged antibiotic therapy and even revision surgeries, which is inevitably associated with increased patient suffering and high costs [6]. Finally, biofilms also dominate numerous chronic bacterial infections, such as pneumonia in cystic fibrosis patients and chronic wounds. Treatments for such chronic conditions remain a significant challenge to healthcare systems worldwide [7]. Collectively, the total annual cost associated with biofilm infections was estimated to be in excess of $94 billion in the United States alone, resulting in more than half a million deaths [8]. To efficiently diagnose, treat and prevent such devastating bacterial infections, a better understanding of the mechanisms involved in their formation, virulence, and persistence using physiologically relevant models is critically needed. It is now widely accepted that the models we have today, from in vitro to in vivo, do not provide an accurate environment to test new antibacterial approaches and have contributed to our poor understanding of how to limit bacteria adhesion, growth, and biofilm formation.
1.2. Lack of adequate model systems
Commonly, a variety of inconsistent, in vitro and in vivo models have been progressively utilized for antibiotic/drug development and pathophysiological studies. These systems range from simple in vitro models using microtiter plate assays and flow cells to more complex in vivo models involving rodents, rabbits or pigs [[9], [10], [11], [12]]. In vitro models are widely used for antimicrobial susceptibility screening as they are cheap, easy to set up, and amenable to high-throughput designs and automation. While in vivo models remain the best option available for safety and efficacy evaluation and are indispensable in connecting in vitro experiments and clinical trials. Antimicrobials of interest must demonstrate sufficient activity in vivo following in vitro evaluation to justify the initiation of clinical trials. In addition, in vivo systems have also been invaluable towards investigating disease pathogenesis and the complex interactions between the host and pathogens. For example, the causal agents of several infectious diseases, including Bacillus anthracis, Mycobacterium tuberculosis or the rabies virus was uncovered through the use of animal models [13]. From a pharmaceutical and device development perspective, both in vitro and in vivo models have been instrumental in screening effective strategies to fight bacterial infections, albeit sometimes with contradictory results. One notable example is the discovery of the first sulfonamide drug, Prontosil, that targets a broad spectrum of Gram-positive cocci. The drug was effective in a pneumococcal infection mice model even though no activity was identified using an in vitro model [14], which should cause us all to pause on the inappropriateness of commonly used in vitro bacteria models.
Nonetheless, mimicking bacterial infections in a physiologically relevant manner has proved to be a daunting task. In fact, there still lacks a consensus among the scientific community on what these assays should incorporate. Since most of the existing in vitro models fail to recapitulate the complex microenvironment and disease processes at the organ level, expensive and time-consuming animal tests are often implemented, despite low predictive results and high failure rates. In addition, in vivo models, although more physiologically relevant, suffer from interspecies differences that puts into question the amount and manner in which bacteria should be introduced (which, for example, remain significant clinic questions for hospital acquired infections). A recent review examining differences in the innate immune response revealed that, when compared to humans, sufficient differences in the organization of the murine immune system hinders the direct translation of murine experimental data to human pathological events, despite the efforts to bridge this gap by creating humanized mice [15]. This difference is further exacerbated in studying bacterial infections as the pathogenesis of infection is often a result of a lost balance between the host immune responses and bacterial overloads. Another important limitation in the use of in vivo models lies in the difference between the pharmacokinetic profiles in most animal models and those occurring in humans, which can dramatically affect drug efficacy. In addition to their inherent differences to humans, the increased use of animal models in biomedical research has also raised concerns over animal welfare and related ethical issues. This growing awareness of animal rights has provided the impetus for the recent ban on testing finished cosmetic products and cosmetic ingredients on animals in the European Union [16]. Consequently, no appropriate experimental animal models or 2D in vitro models are able to accurately predict the required drug doses and drug efficiencies for human use [17,18]. Further complicating the matter is the widespread use of biofilm formation and more specifically, what even qualifies as a biofilm, for many infections, chronic infections and device associated infections in particular.
1.3. Biofilm complicates disease modeling and antibacterial treatments
Beginning with the pioneering work by Robert Koch that started the field of medical bacteriology, for centuries, bacteria have been largely viewed as single and free-floating organisms, now referred to as the planktonic phenotype. The investigation of bacteria based on the single-species planktonic classification was enormously successful and led to the “Golden Age” of microbiology. During this period, antimicrobials against an abundancy of devastating human pathogens such as tuberculosis and diphtheria were discovered [19]. Initially, most acute bacterial infections, which are often dominated by planktonic bacteria, can be readily cured if the right treatment is promptly initiated [20]. It was not until the 1970s that the first observation of biofilms, aggregated bacteria enclosed within a matrix of extracellular materials, was reported in the lungs of cystic fibrosis (CF) patients [21,22], a genetic disorder that often causes repeated lung infections. After decades of research, we are now increasingly aware that pathogenic bacteria often grow in a structured consortium, known as a “biofilm”, attached to biotic or abiotic surfaces during chronic infections [23]. When bacteria succeed in colonizing and forming a matured biofilm within the human host, the infection becomes phenotypically and physiologically different from their planktonic counterparts and is extremely tolerant to both the innate immune system and antibiotic treatments. It has been well characterized that bacteria in biofilms can tolerate up to 10–1000 times higher concentrations of antibiotics than planktonic bacteria [24,25]. Consequently, many of these biofilm infections develop into a chronic state [26]. In the case of CF, a chronic biofilm infection can persist in the airways of patients for over 30 years [27]. In addition to their resistance and persistence, the incidence rate of biofilm-related infections is also extremely high. While it is difficult to precisely determine, it is generally accepted that 65%–80% of human bacterial infections are biofilm-related. These include chronic wounds, lung-related infections, and device-associated infections [[28], [29], [30]]. As an example of their prevalence and severity, a recent survey revealed that more than a quarter of healthcare-associated infections (HAIs) were device-associated [31] and involved microorganisms that form biofilms. In view of this, an improved understanding of the underlying causes for their resistance and persistence is critically needed to better manage infections involving biofilms.
Towards this end, a number of structural and biochemical characteristics of biofilms have been implicated in this increased tolerance in both in vitro and in vivo model systems. One of the primary barriers that protects the underlying bacteria against the host immune system and some antimicrobials is the sticky matrix of extracellular polymeric substances (EPS), which consists of a wide variety of proteins, glycoproteins, glycolipids and extracellular DNA, that encompass the bacteria [32]. It has been suggested that this matrix, among other functions, limits the access of antimicrobials to the embedded bacteria cells by either physically absorbing or inactivating the compound with the EPS components [33]. For example, ampicillin was not able to penetrate wild-type K. pneumoniae biofilms in vitro, which was attributed to the production of the ampicillin-degrading enzyme β-lactamase [34]. Interestingly, biofilms formed by β-lactamase-deficient mutant K. pneumoniae were also resistant even though they are readily penetrable by ampicillin, suggesting that other resistance mechanisms are involved. Indeed, recent evidence suggested that efflux pump systems also played an important role in the resistance of K. pneumonia [35]. In addition to targeting antibiotics, biofilms are also capable of compromising the host's innate immune response by suppressing the antimicrobial activity of polymorphonuclear leukocytes (PMNs) in vivo [[36], [37], [38]]. In the case of biofilms produced by the opportunistic pathogen Pseudomonas aeruginosa (P. aeruginosa), which is commonly observed in the lungs of CF patients, the suppression of PMNs has been suggested to be quorum-sensing (QS) dependent [36], a mechanism through which bacteria respond to fluctuations in cell density. The QS-mediated upregulation of virulence factors, including rhamnolipids, was able to eliminate incoming PNMs on contact, thereby creating a ‘shield’ around biofilm bacteria [37]. Blocking QS by either mutation or administration of QS inhibitory drugs sensitized biofilm bacteria. Furthermore, the presence of multiple bacterial species and phenotypically diverse subpopulations within biofilms has also been linked to their increased tolerance and chronicity. For example, it was discovered that the presence of Haemophilus influenza within polymicrobial biofilms promoted Moraxella catarrhalis resistance to both antibiotics and host clearance in otitis media. This increased resistance was mediated via an autoinducer-2 (AI-2) quorum signaling dependent pathway [39]. Similarly, the increased effectiveness of multi-bacteria biofilms is supported by the mutualistic partnership between Sreptococus oralis and Actinomyces naeslundii in forming dental plaque. In this scenario, co-aggregation of these bacteria formed a nutritionally beneficial environment that allowed each bacteria to grow, where neither grew in the absence of the other [40]. Finally, phenotypical diversification of bacteria in response to steep nutrient gradients and varying environmental stresses has also been suggested to drive infection persistence [25,26]. One classic example is the presence of persister cells in biofilms [41,42], which are a subpopulation of cells that are slow-growing or growth arrested, and in some cases, metabolically inactive [43]. Antibiotic-tolerant persisters play a major role in the recalcitrance and relapse of chronic infections [42]. In addition to the increased tolerance, matured biofilms within the human host provide bacterial inoculums with an opportunity to spread [44] and can serve as reservoirs for plasmids carrying antibiotic resistance genes [45].
Collectively, these distinct structural and biochemical characteristics of biofilms not only make them extremely persistent and difficult to treat, but also further complicates the disease modeling using either in vitro or in vivo model systems. In vitro models often lack host immune components and fail to recapitulate the complex physical and chemical environments bacteria may experience in vivo, while the establishment of chronic infection models in animals is equally challenging. To date, most pathogenic bacteria have been studied using acute infection animal models [46] which do not involve biofilms or reflect the chronic state of infections. Another significant challenge when establishing these models concerns the amount and manner in which bacteria should be introduced. A high infecting dose is sometimes lethal and are ethically challenging, while infections at a lower dose often resolve rapidly by host immune systems, resulting in great inconsistency [47]. Only a handful of animal models aim to model chronic bacterial infections, and most of which involve embedding bacteria in a biofilm-like matrix such as agar or alginate to prevent host clearance [48] or the use of a preformed biofilm on implants [49]. Nonetheless, embedding bacteria in a polymeric matrix produce an artificial environment that inaccurately represents the flow, oxygen or nutrient environments in vivo. Since preformed biofilms in vitro differ morphologically and physiologically from in vivo biofilms, which will be discussed in detail later, inaccurate mimicry of infectious human diseases is common. Finally, most in vivo models to date focus on monospecies infections [50], which do not accurately represent most infections under physiological conditions as these often result from colonization by more than one microbe. A recent study demonstrated that P. aeruginosa used peptidoglycan shed by Gram-positive bacteria to stimulate the production of multiple lytic factors against prokaryotic and eukaryotic cells in a coinfection model using both Drosophila and murine models, further suggesting the importance of polymicrobial interactions in infection resistance and persistence [51].
In light of this, it is necessary to develop new assays and models that can simply, yet more precisely, replicate in vivo microenvironments and reliably predict and record tissue activities under both physiological and pathological conditions. These model systems are essential, as they allow for a mechanistic understanding of the dynamic interactions among relevant factors. Moreover, they provide better predictive power when assessing the clinical and translational potential of novel antimicrobials or antimicrobial materials. Towards this end, multiple in vitro and ex vivo models have emerged in recent years. This review will highlight recent technological advances in improving model relevance as well as advantages and disadvantages of each model system (Table 1 ). A firm understanding of such advantages and limitations can hopefully guide us to select the most appropriate system for probing infection mechanisms in a more effective and efficient manner. From this, we aim to have a better understanding of human physiology and pathology at the cellular level and subsequently establish a better representation of intercellular and extracellular interactions during inflammatory responses. Ultimately, we believe through the use of more advanced in vitro and in vivo models, safe and effective therapeutic strategies can be developed to address antibiotic resistance crisis on a global scale.
Table 1.
Comparison of in vitro, organoids, organ-on-a-chip, ex vivo and in vivo models.
| Cell type | Advantages | Limitations | Potential Improvements | Ref | |
|---|---|---|---|---|---|
| In vitro (MTP- and flow-based systems) | Bacteria only | Inexpensive; High-throughput; Real-time visualization; Incorporation of flow conditions; Well defined experimental conditions | Lack of immune response; Use of abiotic surfaces; Lack of 3D structure of native substrates | Use of synthetic media to mimic native chemical environment | [27,52] |
| Organoid | PSCs and ASCsa | Near-physiological conditions; Specific stem cell propagation; Access to varieties of patient-derived organoids, sufficient tissue mass for analytical approaches | Lack of biomechanical forces and flow conditions; Unable to study interactions between environmental cues | Incorporate microfluidic techniques; establish co-culture systems | [53] [54] |
| Organ-On-a-Chip | Primary cells or cell lines | Enable cell-cell and cell-environment interactions; Introduce biomechanical forces and fluidic flow that mimic microenvironments in vivo; Real-time monitor and high-resolution imaging; Ability to model ADMET properties | Only partial tissue function is presented; Poly-dimethylsiloxane (PDMS) substrates are not ideal for mimicking extracellular matrices; Limited spaces and tissue mass | Improve fabrication techniques; Use ESCs or iPSCs to serve as cell sources | [55,56] |
| Ex vivo | Tissue explants | Native physiochemical environment; Relatively cheap and high-throughput; Real time monitoring; Relatively controlled experimental conditions; Less ethical concerns | Limited life span; Lack of immune response; Lack of standardization | Use of synthetic media to mimic native chemical environments; Use of standardized culture system, e.g. BoDrum® | [[57], [58], [59], [60]] |
| In vivo (Non-mammalian) | Native cell population | High-throughput; Presence of immune system, low cost, easy maintenance, easy genetic manipulation, less ethic constraints | Limited similarities to humans; Llimited lifespan; Difference in body temperature | Focus on elucidating conserved and universal immune mechanisms | [61,62] |
| In vivo (Mammalian) | Native cell population | Presence of host immune systems; Native physicochemical environment | Ethical and animal welfare constraints; High costs; interspecies differences; Limited experiment duration to mimic chronic infections | Repeated bacteria exposure to mimic chronic conditions | [13,[63], [64], [65]] |
PSCs = pluripotent stem cells; ASCs = adult stem cells.
2. Development of in vitro models to study infection and biofilms
The development of in vitro models of bacterial infection and biofilms began after the initial observation of sessile bacteria and the recognition of their role in human chronic infections. Many in vitro models have since emerged, with most of them designed to mimic biofilm formation using specific bacteria under controlled environments. Although often regarded as over-simplistic, in vitro models are still largely used today and have been indispensable in our mechanistic understanding of the biology of bacterial infections and biofilm formation. In addition to their roles in elucidating the underlying biology, they are also heavily relied upon as screening tools to interrogate libraries of antimicrobial agents under the current drug discovery paradigm due to a number of advantages they offer such as low cost, easy set-up and amenability to high-throughput designs and automation [66]. Nonetheless, there is an increasing awareness that these simplified in vitro models often fail to include important environmental parameters such as a host immune system and other mammalian cells, and therefore may lack effective predictive power. In fact, many promising antimicrobial drugs fail to translate from the bench to bedside, partly due to a lack of in vitro models that can effectively predict their long-term antimicrobial performance in vivo. In this regard, orthogonal assays must be included to rule out false positive or false negative conditions, and extreme caution should be taken when interpreting results obtained from these in vitro models. Here, the general setup, advantages, and limitations of commonly used in vitro model systems are introduced, hopefully serving as a valuable guide for model selection as well as data interpretation.
2.1. In vitro biofilm model systems
2.1.1. Microtiter plate (MTP)-based system
Microtiter plate (MTP)-based systems are among the most commonly used biofilm model systems and have been an important tools for studying the early stages in biofilm formation [52,67]. In these systems, biofilms are typically grown on either the bottom or the walls of a microtiter plate. When evaluating the ability of antimicrobials to eradicate biofilms on specific surfaces, or a material's propensity to resist biofilm formation, biofilms can also be grown on the surface of a coupon placed in the well plate. Monitoring changes of biofilms in these systems is also straightforward, as a biofilm can be quantified for changes in mass using stains like crystal violet, safranin and Congo red [66] or for changes in metabolic activity using an XTT viability assay [68]. For example, a MTP-based system was utilized to investigate the influence of DNase I, Ca2+ and extracellular DNA on biofilm formation and growth, where biofilm biomass was quantified using crystal violet as an indication of the enhancement or diminishment of biofilm growth [69]. Alternatively, the combination of live/dead staining and confocal laser scanning microscopy (CLSM) allows for the visualization of changes in bacterial viability and biofilm morphology, even in real time [70]. Our group showed that 20 μm thick Staphylococcus epidermidis (S. epidermidis) biofilms could form in a 96-well and MTP and was successfully applied as a model system to assess the ability of novel superparamagnetic iron oxide nanoparticle (SPION) encapsulating polymersomes to eradicate such biofilms [71]. The combined use of CLSM and live/dead staining revealed a dose-dependent bacteria death as a function of drug and SPION loading.
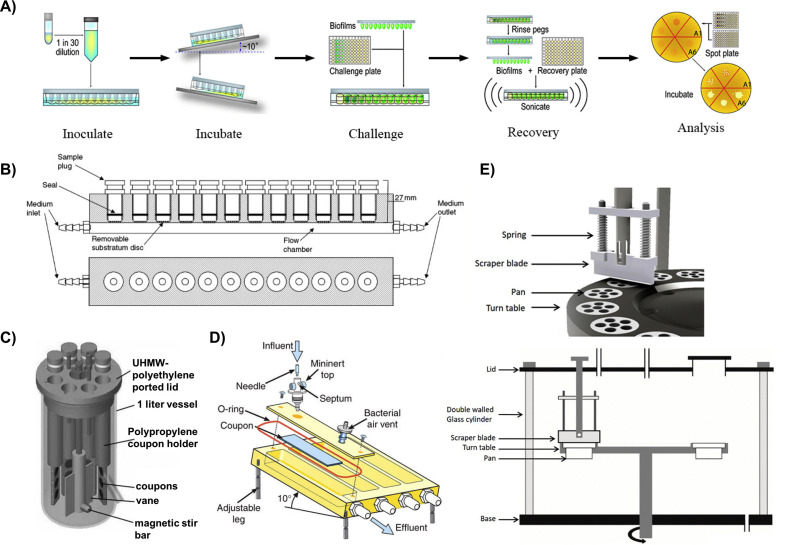
In these classic MTP-based systems, however, there are concerns that a portion of the accumulated biomass may not be a result of the biofilm forming process, but rather because of cell sedimentation and the subsequent entrapment of cell sediments within the EPS [72]. To address this concern, a variation of the MTP based system, called the “Calgary Biofilm Device (CBD)” (Fig. 1 A), was introduced, in which biofilms are formed on lids with pegs that fit into the wells of the microtiter plate containing bacteria [73]. The CBD has been successfully used as a rapid and reproducible assay to screen for biofilm susceptibility to antibiotics. For instance, both gram-negative and gram-positive pathogenic bacteria from various veterinary sources could readily form biofilms on the CBD and were deployed to determine the minimum inhibitory concentration (MIC) and minimum biofilm eradication concentration (MBEC) of a wide array of antibiotics [74]. More recently, biofilms formed by Escherichia coli (E.coli) and Staphylococcus aureus (S.aureus) on the CBD was exploited as a tool to investigate the role of carbon sources (such as glucose, mannitol, fructose, glycerol, etc.) in eradicating biofilms by aminoglycosides [75]. More importantly, screen results obtained from this CBD-based biofilm assay successfully translated in a mouse chronic urinary tract infection model. A combination of gentamicin and mannitol resulted in an almost 1.5 log reduction in biofilm viability and suppressed the spreading of bacterial infections to the kidneys. Quantification of viable cells in biofilms formed on these devices typically involves bacteria recovery using sonication. However, anywhere from 5% to 90% of the cells may disassociate during this procedure [76] depending on the protocols followed and equipment used, resulting in data discrepancies and subsequently hindering appropriate data interpretation. Visualization of biofilm architectures by microscopy methods (such as scanning electron microscopy (SEM) and CLSM) is also challenging as they require the detachment of pegs from the lids using pliers, making these methods labor-intensive and not amendable to high-throughput screening [77]. In addition, it has been argued that the physiological profiles of the detached population may not be representative of the whole population. This is because the outermost cells tend to detach first, and these cells may be phenotypically different from those that are attached to microtiter surfaces due to steep gradients of nutrients and gases [77].
Fig. 1.
In vitro biofilm model systems. A) A work flow of the Calgary Biofilm Device [106]; B) The modified Robbin device (MRD) [107]; C) A CDC biofilm reactor [108]; D) Schematic diagram of a drip flow reactor with its various components [109]; and a E) Schematic diagram of a constant depth film fermenter (CDFF) and a close-up view of the scraper blade sliding over the biofilms to maintain constant depth when biofilm thickness exceeds the thickness of the well [110].
To study the kinetics of early stage biofilm formation, particularly the initial adhesion of bacteria during early stages of biofilm formation, another modified MTP-based method, the ‘Biofilm Ring Test’, was developed based on the ability of bacteria to immobilize magnetic beads when forming biofilms [78]. The kinetics of biofilm formation can be determined by measuring the motility of magnetic beads over time using a plate reader. Applications of the Biofilm Ring Test range from the evaluation of antibiotic susceptibility of biofilms [79] to the assessment of biofilm forming potential [80] and kinetics [81] of clinical isolates. It has also been useful in studying the contribution of extracellular polymeric substances (EPS) [82] and molecular pathways [83] to the formation of biofilms. In comparison with standard crystal violet staining methods, the Biofilm Ring Test is much faster, more reproducible and allows for high-throughput screenings, as no washing or staining is involved. It is worth noting, however, that the Biofilm Ring test is limited to the investigation of early stage biofilm formation and does not provide information on matured biofilms.
The use of MTP-based assays offers a multitude of advantages. These assays are fairly cheap, as no specialized equipment is needed. They also provide the opportunity for multiplexing, as multiple organisms and treatments can be incorporated in a single run and, as such, are ideal for identifying potential anti-biofilm biomaterials and pinpointing genes that are essential for biofilm surface attachment [52]. In addition, culturing conditions such as temperature, oxygen and CO2 concentrations, and composition of growth media can be easily manipulated to investigate the effects of environmental factors on biofilm formation. The choice of which system to use really depends on what questions one wants to answer and often times a combination of different approaches is required to provide a complete answer. Nevertheless, these MTP-based systems are closed systems under static conditions. The environment including nutrient availability, signaling molecules, etc., in which biofilms are formed, changes with time and often does not recapitulate in vivo conditions. Recent transcriptomic analysis has suggested that biofilms formed under static conditions have different gene expression patterns from those formed under flow conditions [84]. This prompted the development of flow-based systems to mimic fluid flow present in many in vivo scenarios. Of course, one can make an argument that nothing is ever static in the human body, yet static in vitro bacteria cultures are commonplace.
2.1.2. Flow-based systems
In contrast to MTP-based systems, flow-based systems, such as the modified Robbins devices (MRD), flow cells, Centers for Disease Control biofilm reactors, drip flow reactors and rotating disc reactors, are open systems in which spent medium with metabolic byproducts and dead cells are removed and constantly replaced by fresh medium at a user defined speed and composition [24]. The use of flow-based systems allows for the formation of matured biofilms. In addition, by controlling the hydrodynamic conditions of flow, parameters such as shear forces can be modulated and are therefore suitable for investigating the physical resistance of biofilms. Similar to MTP-based systems, the structure and physiology of a live biofilm can be monitored non-invasively when coupled with CLSM [85].
One of the first flow systems was the modified Robbins device (MRD, Fig. 1B), which consisted of a pipe with several threaded holes where coupons are located [86]. Although it was originally designed to monitor biofilm formation under different flow speeds in a completely mixed tubular recycle reactor, it has since been adapted to study various aspects of biofilm formation. For example, it proved to be particularly useful in evaluating the effects of surface modifications on biofilm formation under controlled flow conditions [[87], [88], [89]]. As stated by Nava-Ortiz et al., after γ-ray pre-irradiation, glycidyl methacrylate (GMA) grafted polyethylene (PE) and polypropylene (PP) were further functionalized with cyclodextrins on their surfaces.
In the following studies, this surface functionalization strategy provided PE and PP an opportunity to incorporate the anti-fungal drug miconazole, which prevented the adhesion and biofilm formation of Candida albicans on medical devices made from these polymers. In addition, the MDR was effective in evaluating effects of antibiotic lock therapy in catheter-related biofilm infections, as it could accurately simulate fluid dynamics during biofilm formation [90,91]. Compared to the static microtiter plate systems, MRDs are advantageous as they can support continued biofilm growth and maturation for several weeks [76]. However, MRDs are not designed to allow for direct observation or quantification of biofilms. Coupons must be removed from the device for further analysis and therefore suffer from lower throughput.
To allow for direct inspection of biofilm development, several flow cell systems have been developed and are now commercially available [[92], [93], [94]]. The original flow cell systems consist of two chambers connected by a beam, which was later miniaturized to include multiple channels [92]. In combination with microscopy methods, these systems allow for a real-time, non-destructive recording of structural dynamics during biofilm development. This is helpful as it allows scientists to monitor the earliest stage of bacterial surface attachment and therefore gain a better understanding of how these processes can contribute to bacterial colonization and biofilm formation [95,96]. Recently, a multispecies biofilm model consisting of four oral bacterial species was also established in flow cells to study peri-implant infections [97]. As biofilms are polymicrobial in nature, this could be extremely helpful not only in building a more realistic biofilm model in vitro but also in elucidating the role of multispecies interaction in biofilm colonization. Similar to MRDs, however, these systems are relatively low-throughput compared to the MTP-based systems. In addition, the geometry of the flow cell systems has to be carefully designed because it critically affects the uniform flow regions, which can determine the validity of the system when evaluating bacterial adhesion and biofilm formation [93]. Finally, air bubbles frequently form in these systems because of changes in temperature or pressure, which can lead to the destruction of developing biofilms and consistent results. Learning from the renal dialysis process, a recent study proposed modifications to flow cell systems that could potentially minimize bubble formation [98].
The Centers for Disease Control (CDC) biofilm reactor (Fig. 1C) is another example of commercially available flow systems. It is a one-liter vessel with a polyethylene top that supports eight independent rods, which house a total of 24 removable coupons of customizable materials [99]. In this reactor, the magnetic stir bar in the middle of the device rotates and applies a constant high shear on the aforementioned coupons, which is adjustable by altering the rotational speed and is independent of feed rate. This allows for simultaneous investigation of the effects of shear stress and medium composition on biofilm formation. Compared to MDR and flow cells, another advantage of the CDC biofilm reactor is that biofilms can be grown reproducibly under much higher shear stress. As a result, it is recognized by ASTM International as a standard method for the quantification of P. aeruginosa biofilms grown with high shear and continuous flow [76]. Unfortunately, the CDC biofilm reactor shares a common disadvantage with MRD and flow cell systems: low throughput. Individual coupons have to be taken out at predetermined time points for analysis. Efforts are being made to create in/on-line monitoring system for rapid and accurate detection of bacterial attachment and growth in real time. For instance, a label-free interdigitated microelectrode biosensor was integrated with implantable devices [100]. Impedance characterization of S. epidermidis biofilm development on these devices allows for monitoring of bacterial growth as early as a few hours from the inoculation with high effectiveness. This same technique has since been extended to petri dishes [101] and 96-well plates [102]. Nonetheless, this method of detection does not provide any information on biofilm structure and physiology. Additionally, the large volume of the reactor makes it less appealing for screening antimicrobials, especially those that are not surface-tethered, as large amounts of materials are needed. Finally, the semi-open design of these reactors makes them prone to contamination.
Other specialized in vitro biofilm model systems have also been developed to investigate the effects of certain parameters on biofilm formation. For example, a drip flow reactor (Fig. 1D) is designed to investigate biofilm formation under low shear conditions at the air-liquid interface and to study a biofilms' vertical heterogeneity [103]. With the incorporation of microelectrode sensors, biofilm culture under drip flow conditions can also be useful for studying mass transport and chemical gradients within biofilms [104]. Another custom biofilm model, the constant depth film fermenter (CDFF, Fig. 1E), allows for the growth of a biofilm with well-defined thickness. Within this system, biofilms are grown on the bottom of wells with set depths and a scraper blade removes anything above the wells. This has been particularly useful when studying the effects of biofilm thickness on antimicrobial penetration [105].
While these systems incorporate varying flow conditions (e.g., high vs. low shear) and maintain a relatively stable nutrient concentration over time to allow for biofilm maturation, in vitro flow systems are less adapted to high-throughput analysis, more labor-intensive, and often require specialized equipment. As biofilms form on surfaces throughout these model systems, except in drip flow reactors, where there are isolated compartments, only a single bacteria species or a single combination can be tested per run. Furthermore, other aspects of the in vivo environment, such as the host's immune system, are still missing from these systems. In addition to their individual advantages and disadvantages, both MTP-based and flow-based systems share several common limitations. One pitfall in designing in vitro biofilm models to mimic in vivo environments lies in the use of irrelevant bacterial strains. The lack of knowledge in disease-related microbial consortia and the interactions between healthy and pathogenic strains are, at least in part, to be blamed for the low translation rate from in vitro to in vivo studies [111]. Even when adequate information is available, various bacterial strains have not yet been cultivated in these models.
The absence of appropriate formulations for disease-related growth media imposes further challenges on establishing representative in vitro model systems. Biofilms in vivo often grow in nutrient and gas deficient environments, and this deficiency may differentiate gene expression [112]. In addition, different nutrient compositions can lead to altered biofilm formation and virulence, as were shown for P. aeruginosa [113,114]. In this sense, the use of artificial growth media that are rich in nutrients may lead to clinically irrelevant phenotypes. As a consequence, efforts to better represent in vivo conditions range from artificial medium customized for a particular disease [115,] [116] to the use of patient-derived biological fluid [117]. For example, the use of a synthetic CF sputum medium resulted in a similar in vitro gene expression profile to that observed in expectorated CF sputum [118]. Finally, host- or host tissue-related factors are also missing in the aforementioned in vitro biofilm models, including a lack of 3D host tissue structure and the exclusion of host cells, particularly immune cells. Both of these parameters can alter nutrient and gas distribution and abundancy, therefore implicating their inclusion on future models [119,120].
Although both MTP-based systems and flow-based systems have clear limitations, they have been utilized for in vitro biofilm research for decades and have contributed tremendously to our understanding of biofilm biology. The ability to adjust individual variables while maintaining other experimental parameters produces a well-defined environment in which the effects of a single element on biofilm development can be systemically studied. However, it must be recognized that this reductionist approach has its limitations, as in vivo environments are exceptionally complex. When choosing in vitro models, it is important to realize that no single biofilm model system is better than any other, with each model having its limiting factors. However, a specific model may be more appropriated based on the questions being investigated and its clinical relevance of each model. For example, MTP-based systems are well-suited for high-throughput screening when a large number of variables are being investigated or a library of compounds is being tested. When investigating the processes involved in initial bacterial attachment, flow cell systems incorporate fluid flow and provide the opportunity for real-time observations using microscopy methods. Drip flow systems are preferred for studying biofilm heterogeneity, and CDC biofilm reactors have proved to be effective in assessing biofilm formation on biologically relevant materials and evaluating the effects of surface modification on biofilm formation.
2.2. Differences between biofilms produced in simple in vitro models and in vivo models
The hallmark of in vivo biofilms includes aggregated bacteria enclosed in a matrix of extracellular materials, which are extremely tolerant to host immune systems and antibiotic treatments. The formation, maturation, and dispersion of biofilms are the result of dynamic interactions between the complex bacteria communities and host environments, which are very hard to mimic. Despite the continuous efforts to optimize these simple in vitro systems, emerging evidence has shown that significant differences exist between in vitro and in vivo biofilms. One of the most striking differences lies in the size of the biofilms. A recent survey of biofilm sizes in vivo revealed that biofilms have approximate diameters ranging from a few μm to up to 1000 μm, which occurs in the presence of abiotic surfaces, such as implants [26]. However, even these surface-associated biofilms are at least two orders of magnitude smaller than biofilms observed in vitro, which range from 1 cm2 in MTP based systems to 10 cm2 in flow-based systems. In addition to size differences, the shape of the biofilms is also different. In the case of P. aeruginosa, an opportunistic pathogen commonly involved in CF, the classic “mushroom” structure is found in vitro [121] but has yet to be observed in vivo [27]. This change in morphology in biofilms in vivo has not been fully understood, but it is likely due to a combination of both nutrient/oxygen depletion and the presence of host immune systems (e.g., PMNs). In particular, the host immune component is typically missing in almost all in vitro model systems. One classic example that highlights the important role of the host immune response in biofilm formation comes from a study investigating P. aeruginosa biofilm formation in the lungs of CF patients [21]. By using a specific P. aeruginosa PNA fluorescence in situ hybridization probe, biofilms were observed as aggregated structures surrounded by pronounced PMN inflammation in the respiratory zone. Further confirming this observation, bacteria and immune cell interactions were also demonstrated both in vitro [38,122] and in vivo in mouse models [123]. In addition to their direct bactericidal activities via phagocytosis, immune cells can also regulate biofilm formation and growth by oxygen and nutrient consumption [124]. As a consequence, the heterogeneous growth patterns for in vivo biofilms have been linked to local concentrations of PMNs [125].
Another important discrepancy between most in vitro and in vivo biofilms is that multispecies bacterial communities are often present in vivo while most in vitro model systems include only one strain of bacteria. In a study examining the bacteriology of diabetic foot wounds, the presence of two or more bacterial isolates were identified in over 80% of the wounds [126]. In another example, over 92 oral species of bacteria were identified in early dental biofilms from 11 healthy human subjects, with streptococci being the most abundant species [127]. Although not much is known about the interspecies interactions in polymicrobial biofilm formation, we have begun to appreciate the importance of these interactions in altering biofilm virulence and persistence. For a detailed understanding of the role of polymicrobial interactions during human infections and potential preventative and curative strategies against such diseases, we refer the reader to a comprehensive review by Peters and colleagues [128].
Other factors that may also contribute to the differences seen between in vitro and in vivo biofilms include the complex chemical landscape biofilm encountered in vivo [26], varying biofilm durations with chronic infections in vivo lasting much longer [27], and a lack of abiotic surfaces in most in vivo infections with the exception of device-associated infections [129]. Due to these glaring discrepancies, it is unclear whether such in vitro biofilms sufficiently represent in vivo biofilms and whether results obtained from in vitro systems are translatable from a drug development perspective. Some of these differences can be, at least in part, mitigated by taking advantage of more sophisticated, 3D organoid and/or organ-on-a-chip platforms, which will be described next.
3. In vitro 3D organoid and microfluidic models
3.1. 3D organoid models
The history of the term “organoid” can be traced back to 1907, when Henry Van Peters Wilson from the University of North Carolina at Chapel Hill proposed that a dissociated individual silica sponge cell could self-organize and regenerate a whole functional organism [130]. Since then, numerous 3D culturing methods have been developed and utilized to generate organoids of various origins [131]. Typically, organoids are in vitro 3D models that are cultured in a Matrigel, a gel-like scaffold material that mimics the extracellular matrix (ECM) environment and provides essential growth cues for 3D organoid culture [132]. Distinguished from immortalized cell line cultures, organoids are able to maintain the cellular heterogeneity and exhibit organ-like functionality similar to that of the target host tissue under long-term culture conditions [133].
To date, 3D organotypic cultures, including 3D cultures derived from either pluripotent stem cells (PSCs) or adult stem cells (ASCs), have been widely applied in both tissue engineering and drug delivery. The incorporation of stem cells in organoids was first developed by the Japanese researcher Sasai when he demonstrated that 3D cerebral cortex tissue could be generated from embryonic stem cells (ESC) using an efficient 3D aggregate culture (the SFEBq method). Later, Sato and his colleagues generated intestinal organoids from adult single Lgr5 stem cells when cultured 3D in Matrigel. They subsequently developed a new method, the R-spondin method, which employed growth factors to induce key endogenous niche signals for the eventual development of human intestinal and other organoids from organs harboring Lgr5+ stem cells [134].
Generally, ESCs and induced pluripotent stem cells (iPSC) that can self-organize in 3D culture have been mainly employed to develop organoids due to their self-renewal and differentiation capabilities [135,136]. On the other hand, ASC or primary cell-derived organoids have the capability of developing more mature phenotypes compared with iPSC and therefore have also been applied in many models including gastrointestinal, lung and liver organoids [135]. For instance, gastrointestinal infection is one of the most investigated disease models and several organoid models have been described in recent years. Using crypt-derived mouse intestinal organoids, Zhang et al. investigated the pathophysiological interactions between Salmonella and epithelial cells by visualizing post-infection morphologic changes of the organoids through immunofluorescence. The disruption of ZO-1 in Salmonella infected organoids was observed. In addition, by comparing the levels of expression of the tight junction proteins ZO-1, Occludin, Claudin-2, Claudin-7, and the NF-kB pathway in infected organoids to non-infected control groups, they discovered that salmonella infection induced the disruption of epithelial tight junctions and activated the NF‐κB inflammation pathway in infected organoids [137]. Most importantly, this Salmonella–host interactions in vitro recapitulated many of the characteristics observed in vivo in a Salmonella-colitis animal model, including bacterial invasion, tight junction disruptions, and host inflammatory responses [[138], [139], [140]].
Organoid models generated from either gastric primary cells or gastric epithelial stem cells were also used to study the host response to Helicobacter pylori infection [141,142] as it is one of the main risks for gastric adenocarcinoma and causes a number of gastric diseases. In one example, human gastric primary cells were used to investigate Helicobacter pylori infections and the mechanisms of tumorigenesis. Primary cells were isolated from healthy human gastric glands and grown in Matrigel containing defined growth factors, developmental regulators, and apoptosis inhibitors. The differentiation of spheroids to gastric organoids was observed when Wnt3A and R-spondin1 were withdrawn from the medium, which was confirmed by the downregulation of stem cell markers (eg., CD44 and LGR5) and the upregulation of gastric differentiation markers (eg., TFF1 and GKN1, 2). The actively replicating 3D cultures were then transferred to a 2D environment to allow for the easier manipulation of the experimental conditions. Interestingly, important characteristics of the fully functional gastric epithelium, such as well polarized cells, presence of tight junction markers and differentiation gene expression patterns were maintained even in 2D cultures, as confirmed by both microarrays and immunofluorescence [143]. Once exposed to gastric pathogens, H. pylori infected primary cells exhibited the hallmarks of bacterial infection and the up-regulation of the NF-κB signaling pathway indicating the activation of host responses. Importantly, the use of primary and untransformed cells instead of commonly used gastric adenocarcinoma-derived cell lines in this 2D/3D organoid to demonstrate similar host-bacteria interactions observed in vivo may indicate that this may serve as a better model to study H. pylori infections including how it may lead to gastric disease or even gastric adenocarcinoma. Another gastrointestinal organoid system to study H. pylori infection involves the use of gastric epithelial stem cells [142]. In this organoid system, the gastric epithelial stem cells were differentiated into four lineages and exhibited a repetitive structure of the gland and pit domains. Wnt is suggested to be very important in the regulation stem cell differentiation. When Wnt is silenced, these gastric epithelial stem cells only differentiated toward the pit but not gland lineage. By monitoring the expression of cytokine mRNA, it was found that cells from the gland region had a stronger inflammatory response to H. pylori infections and a more robust NF-kB activation compared to pit lineages. Although still at an early stage, this organoid model clearly demonstrated the potential of establishing patient specific disease models for studying H. pylori infections and other gastric pathologies.
Not until recently have organoids of complicated tissues, such as the brain, been generated using human pluripotent stem cells grown under agitation in Matrigel substrates. Following similar protocols, several disease models have been generated to study cancer as well as many infectious diseases of both bacterial and viral origin, allowing for the examination of host responses and cell–microorganism interactions [144]. For instance, organoids have been used to study viral infections, such as ZIKA virus infections (ZIKV). Brain organoids were developed using iPSCs during the ZIKV outbreak in 2016. Derived from embryoid bodies, neuroectodermal tissues were embedded in Matrigel scaffolds and transferred to a spinning bioreactor for nutrient absorption and tissue growth. The reported culture protocol resulted in a rapid yet comprehensive development of brain tissue, in which various discrete but interdependent brain regions were observed [145,146]. Subsequent experiments have investigated the impact of a ZIKV infection on established brain organoids using immunocytochemistry and electron microscopy, and concluded that ZIKV targets human brain cortical progenitor cells, leading to reduced brain cell viability, elevated cell apoptosis and autophagy, and ultimately interferences with neurogenesis and neurodevelopment [147,148].
Taking advantages of the improved tissue organization and integration, 3D organoid models have gained wide attention and have surmounted many limitations present in conventional 2D models, including insufficient replication of organ structures and the microenvironment [149]. Nevertheless, while 3D organoids are capable of replicating 3D organ structure and mimicking its physiological functions in vitro, the integration and reconstitution of features, such as tissue–tissue interfaces, chemical gradients, and bio-mechanical cues provided by the surrounding microenvironment, remain a significant challenge. To address these issues, microfabrication and microfluidic techniques have emerged and have been leveraged to better recapitulate the microenvironment of living organs by incorporating cell-cell interactions, and chemical and bio-mechanical cues [150,151]. Consequently, these microengineered biomimetic models may be a more accurate representation of whole human organs, providing valuable information that can more efficiently guide the design and execution of subsequent in vivo studies [55].
3.2. Microfluidic models and organ-on-a-chip systems for infection and inflammation studies
3.2.1. What is an organ-on-a-chip?
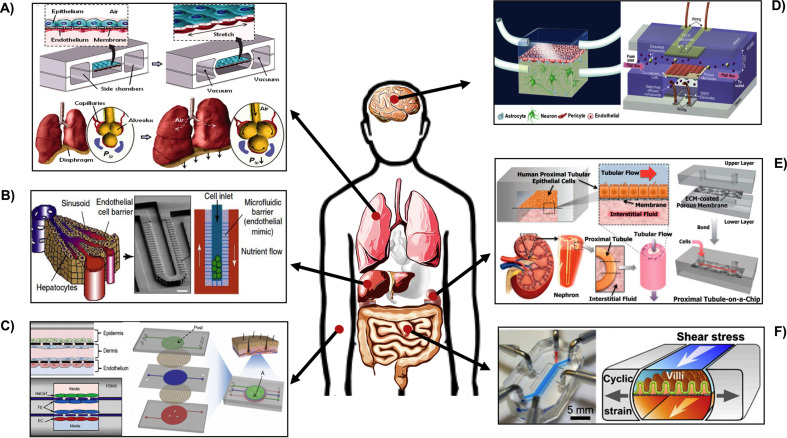
In short, organ-on-a-chip systems are functional microchips that house living cells and mimic the structure and function of human organs. Organ-on-a-chip systems are often fabricated out of PDMS and biodegradable poly(dl-lactide-co-glycolide) (PLGA) [152] using techniques such as replica modeling, soft lithography and microcontact printing [153,154]. Recently, these microfabrication techniques have greatly benefited from the development of integrated circuit technology and wafer fabrication facilities in electrical engineering, and many of the challenging facing organ-on-a-chip systems (such as miniaturization and reproduction of complex architectures similar to human tissues) have been addressed. In order to mimic the in vivo microenvironment to the highest extent, using biomaterials to fabricate organ-on-a-chip systems provide opportunities to allow higher precision and accuracy [155]. For instance, Sudo et al. implemented a microfluidic platform that incorporated a type-I collagen gel scaffold between two microfluidic channels under static or flow conditions. By co-culturing hepatocytes and vascular cells on each sidewall of the collagen scaffold, vascularization of liver tissues in 3D culture microenvironments was observed. Later pioneering studies further advanced these microfabrication technologies by incorporating both biological and mechanical cues. For example, the first human lung-on-a-chip developed in 2010 was a biomimetic, microfluidic system that reconstituted the critical functions of the human alveolar-capillary interface, especially the mechanical strains present in vivo [156]. The additional hollow vacuum chambers specifically induced lung-like stretching of epithelial and endothelial layers [157] (Fig. 2 A). Since then, many organ-on-a-chip models, including liver-on-chip [158], skin-on-chip [159], intestine-on-chip [160], blood-brain-barrier (BBB)-on-chip [161,162] and kidney-on-chip devices [163] have emerged (Fig. 2A–F). Following these “bottom-up” or “reverse engineering” approaches, tissue-tissue interfaces, biochemical and/or neuroelectrical cues and characteristic mechanical forces can be artificially introduced to provide organ-specific physical microenvironments [164].
Fig. 2.
Microfluidic organ-on-a-chip systems mimicking various organ. (A) Lung-on-a-chip device with two vacuum chambers built-in to mimic the mechanical movement of lungs [165]. (B) Liver-on-a-chip device [158]. (C) Skin-on-a-chip device with 3 separate channels and 4 vertically stacked cell layers [159]. (D) BBB-on-a-chip device with various brain tissue cell co-cultures [161,162]. (E) Kidney-on-a-chip device [163]. (F) Gut-on-a-chip device with vacuum chambers built-in to mimic intestinal movements [160].
3.2.2. Lung-on-a-chip
As organ-on-a-chip systems provide a promising platform to model physiological and pathological functions of tissues and organs in vitro, it has been adapted in areas such as anti-tumor drug delivery [[166], [167], [168]], organ function mimicry [169], and membrane-based permeability and toxicology investigations [170], exhibiting great potential for investigating cellular mechanisms of organ physiology. In addition to these applications, organ-on-a-chip systems have also been used to study bacterial and viral infections in vitro. Using the aforementioned lung-on-a-chip model, the organ-on-a-chip model was used to study the innate cellular responses to pulmonary infection of E. coli. For this, green fluorescent protein (GFP)-modified E. coli were introduced to TNF-α activated human alveolar epithelial cells for five hours on the upper side of a PDMS membrane. After five hours of incubation, the presence of fluorescent-labeled neutrophils, indicative of activated human pulmonary microvascular endothelial cells, was assessed. Results showed that most of the bacteria were cleared by the neutrophils, indicating that this biomimetic microchip can effectively replicate, as well as record, the general immune response to microbial infections in human lung alveoli on a cellular level [165].
In 2015, another lung-on-a-chip technology investigated cell recruitment and migration during infection and immune responses [171]. Instead of introducing a pro-inflammatory mediator, tumor necrosis factor–α (TNF-α) was directly added to system to replicate the live, cell-produced immune responses that closely mimic in vivo conditions [172,173]. In this regard, MF2.2D9 T-cell hybridomas, IC-21 macrophages, immortalized B6 dendritic cells, and mycobacterium avium expressing CFP were first separately loaded into the microdevice. Long-term cell behavior was subsequently imaged in real time. To investigate cell behaviors under an inflammatory chemokine gradient, LPS and immunogenic peptide-loaded macrophages were mixed with I-Ab-peptide restricted MF2.2D9 cells and introduced to the infection compartment, whereas MF2.2D9 cells were loaded on the migratory compartment.
Although the movement of cells was not significant, real-time images demonstrated the migration and recruitment kinetics of MF2.2D9 cells towards the infection site 2–3 h after the initial loading of the cells. In addition, primary dendritic cells were also investigated with regard to their migration towards a loaded chemoattractant CCL19 or cell-induced gradients of cytokines and chemokines. Directional migration of mature dendritic cells was observed when CCL19 was loaded in the activator compartment. Similarly, a long lasting directional movement of immature dendritic cells towards activator compartments containing a co-culture of pro-inflammatory or non-activated mature dendritic cells and T cells was observed, demonstrating the chemotactic properties of these devices. Although bacteria were not directly involved in this study, this device has the potential to be a promising platform for studying host immune responses to infections in the lung.
3.2.3. Gut-on-a-chip
Two years after the generation of a lung-on-a-chip, Ingber et al. also demonstrated another microfluidic device, the “gut-on-a-chip” [174]. Similar to the design of the previous lung-on-a-chip device, this biomimetic gut-on-a-chip consisted of two microfluidic channels, separated by a layer of human intestinal epithelial (Caco-2) cells grown on a porous PDMS membrane coated with ECM proteins. The system similarly contained two vacuum controllers built on each side of the channel to mimic the complex physiological peristaltic motion of the living intestine by exerting cyclic strain (10%; 0.15 Hz). More importantly, in addition to Caco-2 cells, a common intestinal microbe, Lactobacillus rhamnosus GG (LGG) was co-cultured with the epithelium for over one week for the first time ever [175]. By monitoring transepithelial, amonipeptidase and LGG b-galactosidase activity and cell morphology, the integrity and viability of intestinal epithelial cell monolayers were confirmed. Interestingly, compared to Caco-2 cells grown on either a 2D static Transwell chamber (no flow) or in the microfluidic chip without cyclic strain, cells exposed to cyclic strain increased in height and polarization after 3 days, and tended to spontaneously form undulations and folds. Moreover, according to Trans-Endothelial Electrical Resistance (TEER) results, co-culturing the probiotic strains of bacteria did not affect intestinal epithelial integrity [176] while intestinal barrier function can be enhanced [174,177].
Although this original gut-on-a-chip was not employed to study bacterial infections, it set a foundation for co-culturing cells with bacteria strains to analyze the biological and mechanical conditions of inflammatory cells under microbiome infections. More recent studies from the same group leveraged this co-culture, microchip model of the human intestine to investigate how probiotics and antibiotics suppress villus injury as induced by pathogenic bacteria. Furthermore, they studied how immune cells, specific inflammatory cytokines, and peristalsis-like motion impact intestinal inflammation and the integrity of the epithelial barrier function in inflammatory bowel disease [160]. In this study, several different factors that can affect normal intestinal functionality, such as commensal E. coli microbes, lipopolysaccharide endotoxin (LPS), and peripheral blood mononuclear cells (PBMCs), were taken into consideration and investigated. For instance, the study revealed that entero-invasive E. coli (EIEC) would rapidly overgrow the apical surface of villi within 24 h and induce a loss of normal intestinal villus morphology and intestinal barrier function. In contrast, a non-pathogenic laboratory strain of E. coli and lipopolysaccharide endotoxin (LPS) did not alter the TEER value of the intestinal barrier system. In addition, although introducing PBMCs alone did not induce damage on the intestine model, a combination of non-pathogenic E. coli and LPS revealed a significant loss of intestinal barrier functions.
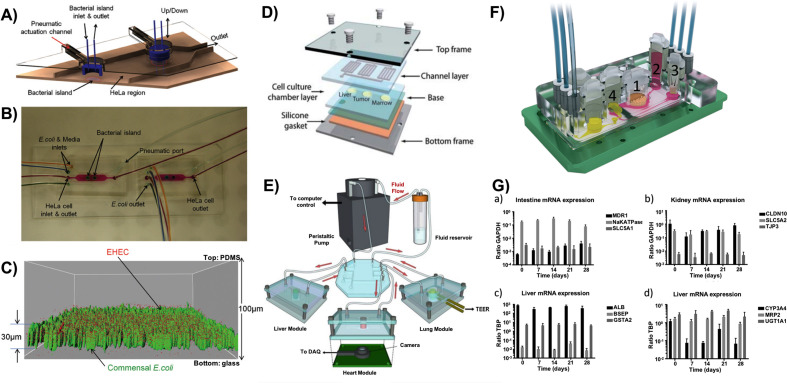
Other factors such as inflammatory cytokines (eg., IL-8, IL-1β, IL-6, and TNF-α), anti-inflammatory probiotic (VSL#3) and cyclic strains were also evaluated. Results revealed that both cyclic stress, which mimics physiological peristalsis-like mechanical motions, and the anti-inflammatory probiotics promote intestinal function as indicated by an increase of TEER value and a decrease of colonized bacteria; however, inflammatory cytokines impacted the intestine model even in the presence of immune cells [164]. Critically, this microfluidic complex provides a platform to analyze the interaction between multiple key factors, including normal tissue cells, immune cells, pathogenic and non-pathogenic bacteria, LPS, and cytokines in a separate or combined fashion. Consequently, this in vitro 3D model can serve as a promising platform to study and gain insights into human pathology and physiology. Another fundamental study investigated the effects of enterohemorrhagic E. coli (EHEC) colonization on the GI tract commensal microenvironment [178], where a co-culture microfluidic model was developed using HeLa cells and commensal E. coli biofilms (Fig. 3 A–C). By introducing EHEC to the established commensal environment, it demonstrated that the pathogenic colonization is strongly impacted by the signaling molecules present in the commensal microenvironment. In the experiment, wild-type commensal E. coli and E. coli BW25113 ΔtnaA (a strain that cannot produce indole, a signaling molecule that inhibits EHEC attachment) were co-cultured with HeLa cells, respectively, which was followed by exposure to EHEC infections. By comparing local exposures to pre-indole-treated EHEC bacteria, it was determined that local exposure of EHEC bacteria to commensal biofilms was more effective, possibly because of other bacterial signals during the infection, and/or the higher concentration of indole secreted by local commensal bacteria. These data provide a possibility to screen different GI microenvironment signals on pathogenic infectivity, as well as select potential probiotic strains to attenuate GI bacterial infections.
Fig. 3.
(A) Three-dimensional scheme of the co-culture device of epithelial cells and bacteria. (B) Micrograph of the co-culture device with color dyes showing the different regions (epithelial cell zone and bacterial islands). (C) Localization of EHEC (red) in E. coli BW25113 biofilms (green) [178]. (D) A schematic of multi-organs-on-a-chip cultured with liver, tumor and marrow [217]. (E) A schematic of multi-organ-on-a-chip cultured with liver, heart and lung tissues [218]. (F) A 3D view of the microfluidic four-organ-chip device cultured with intestine (1), liver (2), skin (3), and kidney (4) equivalents. (G) Gene expression in co-cultures of the four-organ-chip over 28 days [220]. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.4. Skin-on-a-chip
Being the largest organ in the human body, skin plays a critical role in wound healing and assessing drug bioavailability and absorption. Traditional monolayer skin equivalents used to study these processes often fail to mimic normal human skin barrier functions and properties due to excluded cell types and improper force application, thereby limiting physiological relevance [179]. As an improvement to traditional monolayer skin equivalents, 3D human skin equivalents better recapitulate natural skin tissue composition and function with a combination of endothelial cells, adipose tissue and immune cells added to the full-thickness skin models [181,182]. For example, Bellas et al. introduced a 3D full-thickness skin-equivalent in vitro model using silk and collagen as scaffolds. By combining 3D vascular adipose tissue cultured on collagen gels with the engineered epidermal-dermal tissue cultured on silk scaffolds, the proposed tri-layer tissue constructs expressed physiological morphologies of the epidermis and dermis and hence established a physiologically relevant skin-equivalent in vitro model [181]. However, these 3D systems are complicated to produce and might have high variability, resulting in inconsistent results. Skin-on-a-chip systems were developed to address these issues and have been established as one of the essential organ-on-a-chip systems [180]. Ideally, reconstructed human skin equivalents are multilayered with an engineered air-liquid interface that exposes the topical stratum corneum layer to air while immersing the dermal layer to the vasculature or medium [183]. Among several skin-on-a-chip systems reported in recent years [182,184,185], Ramadan et al. introduced a miniaturized skin-on-a-chip model with a co-culture of immortalized human keratinocytes (HaCaT) and a human leukemic monocyte lymphoma cell line (U937) [184]. This study aimed at investigating the effects of chemical and physical stimulation, such as bacterial LPS, on the function and integration of the skin barrier. Compared to the static models, the dynamic media perfusion combined with the air-liquid interface significantly improved tight junction formation and extended cell viability to 17 days. By comparing the expression of IL-6 and IL-1β after introducing LPS to HaCaT/U937 co-culture and mono-cultures, results also showed that keratinocytes formed a robust barrier and protected cells against LPS invasion. Another skin-on-a-chip model developed by Wufuer et al. which simulated inflammation, edema and drug absorption in vitro [159]. This proposed 3D model, which consisted of epidermal keratinocytes, fibroblasts and endothelial cell layer, was introduced to various doses of TNF-α perfused through the fibroblast channel to develop an on-a-chip skin inflammation model. By analyzing the expression of pro-inflammatory factors IL-1β, IL-6 and IL-8, results confirmed the pathological mechanism of TNF-α-induced inflammation through the NF-kB signaling pathway. Thus, this demonstrates a potential for applying this skin-on-a-chip equivalent for constructing in vitro skin disease models or for testing the toxicity of pharmaceutical agents.
Although skin-on-a-chip models have been extensively used for toxicology, pharmacology, and regenerative applications, few studies have investigated how microbes and biofilms are implicated in acute and chronic infections or how they delay the wound-healing process by inducing inflammation. Since acute wounds or chronic skin ulcers disrupt physical and chemical barriers of the skin, they provide an ideal growing environment for microbes [186]. Therefore, in addition to skin inflammation and cytotoxicity studies, these devices can also serve as a novel system to better understand the mechanisms of wound infections and wound healing, as well as to test the biocompatibility and efficacy of the antibiotics used to treat wound infections. In this regard, a few microfluidic wound models have been established for testing mechanobiological structures of the wound environment, as well as the behaviors of the bacteria/biofilm under antibiotic treatments [187,188]. In one study, an in vitro microfluidic wound model was developed to examine the effect of DispersinB and Gentamycin on Methicillin-resistant Staphylococcus pseudintermedius (MRSP) bacterial biofilms. A Y-shaped, collagen coated microfluidic channel served as an animal wound in vitro. Bacteria were labeled with fluorescent dyes to allow direct observation. Image analysis of the samples collected from the outlet showed a higher fluorescent intensity of MRSP biofilms removed by DispersinB-Gentamycin [188]. In addition, a similar device was reported to study the growth and detachment of S. epidermidis biofilms under Dispersin B and Rifampicin treatment. Bacteria that are released upon treatment were collected and quantified by a colony counting assay. Results proved the combined delivery of DispersinB and rifampicin was effective in removing biofilms formed by S. epidermidis, as no bacterial dispersal was detected by the end of the treatment [187].
Despite the fact that skin-on-a-chip systems have contributed significantly in skin regeneration and drug permeability test, the application of these systems in skin infection modeling is still limited and therefore requires further development. Moreover, as most of the current 2D and 3D skin wound infection models are static in vitro models [107,189], the dynamic interactions between bacteria and host cells during the wound-healing process remains unclear. With the advancement of novel biomaterials and microfluidic systems [[190], [191], [192], [193]], new approaches, such as 3D bioprinting technology, can potentially be applied to fabricate skin-on-a-chip systems with an ECM embedded and spatial heterogeneity incorporated. This may further facilitate the development of more representative microfluidic skin disease models in the future [194].
3.2.5. BBB-on-a-chip
Delivering pharmaceutical agents into the brain has been one of the most intensively investigated topics in recent years. The BBB, formed mainly by endothelial cells, maintains the integrity and homeostasis of the central nervous system (CNS), yet inhibits efficient drug delivery since free diffusion of substances from the circulating blood into the brain parenchyma is highly restricted [195]. Still, the BBB has been identified as the main target for brain drug delivery, as drug delivery into the CNS requires transport across the BBB. Many in vitro BBB models, including 2D, 3D, and BBB-on-a-chip models, have been developed to characterize drug permeability [196,197], intercellular signaling, which mediates neuroinflammation [198], and mechanisms of brain tumor development and brain infections [199,200]. For example, one study conducted by Eugenin et al. examined the role of human immunodeficiency virus (HIV)-infected astrocytes in BBB disruption using a 2D Transwell model [201]. Another study described by Cutting et al. examined selective autophagy activities in host defenses after BBB penetration of Group B Streptococcus (GBS), one of the leading meningeal pathogens, both in a 2D model and in vivo [202]. However, although many studies have been carried out using different in vitro BBB models, few studies have used this advantageous technology for brain infection studies. One of the underlying reasons for their limited use in this area may be due to the toxicity that results from co-culturing brain endothelial cells with infectious pathogens. With this, Brown et al. introduced a BBB-on-a-chip microfluidic device consisting of separate vascular and brain channels, separated by a porous PDMS membrane [202]. This not only enables cell-to-cell interaction between brain tissue cells, but also allows for the independent perfusion of both compartments. A follow-up study reported by the same group then co-cultured primary human brain-derived microvascular endothelial cells (hBMVEC) with human induced pluripotent stem cell (hiPSC)-derived human cortical neurons, pericytes and astrocytes to mimic the neurovascular unit [162,203]. After stimulating the BBB model with LPS and cytokine solutions containing TNF-α, IL-1β, and MCP1&2, the authors examined the TEER value, tight junction integration, and metabolites generated by each preparation. The data suggested that the BBB integrity was initially disrupted by LPS, as indicated by reduced tight junction formation and increased membrane permeability, but recovered (though not fully) to the pre-exposure level in a time- and LPS dose-dependent manner. In addition, metabolites obtained from each channel predicted metabolic network activity using biologically driven computational analysis. By comparing pathway activity between the brain and vasculature, results suggested that each uses different proteomic and metabolic pathways to induce inflammation. Moreover, under the circumstance where the same pathway was involved, the vasculature remained in a pro-inflammatory state whereas the other parts of BBB started to rebound. With cell viability maintained in the microchannels, these microfluidic devices allow for the in vitro investigation of tissue and organ function in response to various stimuli. Hence, they may serve as a suitable model for use in studies that incorporate normal tissue cells with environmental toxins and pathogens, providing a better understanding of mechanism(s) of action [150].
3.2.6. More microfluidic systems and their applications
Besides organ-on-a-chip systems, other microfluidic systems are being developed in order to investigate bacteria behaviors within their natural habitats: biofilms [[204], [205], [206], [207]]. One study investigated biofilm morphology with a microfluidic system that mimics a natural habitat, such as a sequence of corners caused by biofilm streamers and a constant flow of P. aeruginosa [208]. Using 3D porous materials made from transparent Nafion, the microfluidic system served as artificial soil. When flowing through these soil-like porous materials, P. aeruginosa biofilms tended to form 3D streamers and, as a result, caused rapid clogging, which disrupted the constant flow. The study also investigated the effects of gene expression profiles on the formation of biofilm streamers. For instance, ΔpelA (deficient in EPS matrix production) did not produce a significant biofilm, but ΔflgK (a non-motile flagellar mutant) produced biofilm streamers similar to the wild type. In addition to infections of bacteria origin, viral infection such as h hepatitis B virus (HBV) is another major health concern today. Towards this end, a 3D microfluidic primary human hepatocyte (PHH) culture was developed as a physiological relevant preclinical platform for studying HBV infections [209]. Notably, this well-established microfluidic system was able to recapitulate the hepatic sinusoid microarchitectures, including functional bile canaliculi and complete cell polarization, and extended the culture period to at least 3 weeks. More importantly, the culture system closely mimicked the HBV infection processes in vivo when infected with patient-derived HBV, including HBV replication, suppression of type I and III IFN and ISG expression of innate immune response, plus the maintenance of HBV covalently closed circular DNA (cccDNA). This developed 3D liver-on-a-chip microfluidic system provides a way to further expand this platform to other organ-on-a-chip models, and to study not only HBV or virus infection but other pathogens and microbial infections as well.
Other than previously described microfluidic devices that focus on tissue infection and inflammation, another important application of organ-on-a-chip systems is to recapitulate cancer growth and monitor therapeutic responses [210,211]. In general, microfluidic models of blood vessel systems, such as tumor blood vessels, can be employed to assess nanocarrier function or screen drug candidates to seek out new opportunities in cancer treatments [212]. To date, several types of 3D in vitro tumor-on-a-chip models have been established in an effort to elucidate tumor-microvascular interactions while mimicking the tumor microenvironment. One study employed previously described organ-on-a-chip technology to recapitulate and investigate human non-small-cell lung cancer (NSCLC) growth and invasion patterns as well as the tumor cell responses to therapeutic cues associated with breathing motions [213]. In addition to single organ-on-a-chip, multi-organs-on-a-chip, also known as body-on-a-chip, have been developed to understand the physiological coupling between different organs in vitro, study drug metabolism, and generate pharmacokinetic (PK) and pharmacodynamics (PD) models [[214], [215], [216], [217], [218]] (Fig. 3 D&E). One study integrated a micro cell culture analog (CCA) system with a fluorescent oxygen sensing system [219] that can mathematically analyze the adsorption, distribution, metabolism, elimination and toxicity (ADMET) of chemicals in vitro [215]. More recently, the capabilities of these multi-organs-on-a-chip have been extended to analyze physiological coupling between different organs and systemic ADMET profiling for in vitro drug candidate testing [220]. For example, intestine, skin, liver and kidney cells were co-cultured on chips with an interconnected fluidic environment that enabled a reproducible tissue culture for 28 days (Fig. 3F&G). To best mimic in vivo conditions, pharmaceutical agents were first administered to intestinal tissues through an isolated medium reservoir, and the subsequent distribution of drugs was then carried out by circulating medium into liver tissues to mimic first-pass metabolism. Finally, secondary metabolism and final excretion of the drug was successfully executed by the kidney equivalent. Altogether, the linked four-organ-on-a-chip system provided a thorough evaluation of physiological homeostasis, barrier integrities, molecular transportation, and pharmacokinetic and pharmacodynamic parameters such as toxicity, tolerable doses and time course of drug candidates. Consequently, these proposed studies provide potential platforms for the future investigation of anti-inflammation and anti-infection drug responses.
3.2.7. Challenges and limitations
Nevertheless, although organ-on-a-chip systems show a promising future, they exhibit several shared limitations which require further attention. One of the most frequent questions asked about this novel technique is, do these in vitro models accurately mimic animal or human in vivo systems? First of all, from a technical point of view, improving microfabrication methods for future advancements in microchips will improve the efficiency and validity of organ-on-a-chip systems. Further, studies have to be conducted to show an organ-on-a-chip matches an in vivo response. Although PDMS, the most common organ-on-a-chip material, is a suitable material due its clear color, flexibility, and cost, it is lipophilic and can absorb both small organic chemicals during the fabrication process as well as hydrophobic drugs and compounds introduced to the device [221]. Substitutions for PDMS are still under investigation and candidates such as polystyrene (PS), which has a high modulus of elasticity, have been proposed for fabricating lung-on-a-chip systems that require significant mechanical deformation [165,222]. None-the-less, it is doubtful than any polymer can accurately mimic the natural ECM of the organ it is intending to mimic. Even if protein layers are formed on such polymers, the bioactivity and conformation of those proteins will be different than a natural tissue.
From a biological standpoint, culturing cells in organ-on-a-chip systems, especially for studying infectious diseases, remains challenging [220]. Since the in vitro cell culture period for current organ-on-a-chip devices is limited to 4 weeks, they are not suitable for chronic or long-term disease modeling [223]. Meanwhile, with the introduction of infectious bacteria to tissue cell culture, how to keep the host tissue cells and bacteria in distinct regions of the microfluidic device without contamination until the healthy cells reach confluency remains a challenge [178]. Secondly, although incorporating various cell types to individual chips has already been accomplished by several groups [220,224], it is still beneficial to generate a standard protocol and a universal medium for culturing different cell types together. In addition, due to low culture volumes and cell numbers in organ-on-a-chip microdevices, appropriate organ scaling also needs to be addressed to accurately replicate physiologically relevant responses in vivo and ensure efficient detection sensitivity [55,221]. Consequently, compromises between accurately replicating physiological complexity and controlling interactions with applicable readouts is unavoidable. These challenges and limitations have to be addressed through either further validation and comparison or by using other models involving real tissues to ensure appropriate and accurate measurement and analysis.
4. In vivo models
Despite their instrumental roles in expanding our knowledge on critical biofilm biology in vitro, these in vitro model systems have their limitations—notably their failure to fully recapitulate the native host environment. This has led to the development of a new wave of more sophisticated in vivo models designed to better represent physiopathological conditions in humans. These range from non-mammalian models that allow for high-throughput screening to sophisticated mammalian models. To overcome the high cost and ethical concerns associated with mammalian model systems, non-mammalian models such as Drosophila melanogaster (fruit fly) and Danio rerio (zebrafish) are increasingly used in studying bacterial colonization and biofilm development [225,226] in the presence of host immune systems. These models are advantageous compared to mammalian models as they are low cost, easily maintained, and have high-throughput capabilities [61,62]. In recognition of the importance of interspecies communications during biofilm development, non-mammalian models are now even being adapted to study polymicrobial infections [227]. For example, oropharyngeal species that were beneficial to the flies in single species infection were identified to enhance P. aeruginosa virulence in a co-infection model, underscoring the importance of microbe-microbe interactions [228]. In addition, as the genomes for most of these systems have already been fully sequenced, genetic manipulations, such as knock-in and knock-out models, can be readily created to study the genetic basis of biofilm development and virulence. Despite these advantages, non-mammalian hosts have limited similarities to humans and limited lifespans (and therefore experiment duration), both of which can negatively impact their clinical translatability.
In this context, mammals are superior as they offer the closest environment to that of human hosts. Tremendous efforts have been made to develop mammalian models that are truly reflective of biofilm infections in animals, ranging from rodents to larger species such as sheep and pigs [229,230]. As a consequence, a large number of in vivo infection models are now available for targeting a wide range of both tissue-specific infections and device-associated infections. Evidence shows that these in vivo animal models produce very similar biofilms as those found in human infections: bacteria aggregates segregated by host materials. For a comprehensive list of available in vivo models of bacterial infections, we refer readers to a recent review [66].
Historically, procedures used to inspect biofilm development in these in vivo model systems required the retrieval of the infected tissues or devices for downstream analysis. This makes it extremely difficult to study the early stages of biofilm formation and its kinetics during development. Recently, advancements in highly sensitive imaging techniques combined with the ability to engineer bioluminescent bacteria strains has begun to afford continuous monitoring of biofilm infections in vivo [231]. In general, in vivo models are advantageous as they provide opportunities to investigate important questions regarding biofilm pathogenesis while taking into account the important host-microbe interactions, which are very difficult to model in in vitro models. Therefore, they allow for the best transferability compared to simple in vitro models or organs-on-chips.
Nonetheless, the use of mammalian in vivo models is restricted in some cases due to ethical considerations [63]. Projects involving the use of mammalian models have to first be evaluated based on the “three R rule”: mammalian animal models should only be used when in vitro models or non-mammalian models are not capable of addressing a specific scientific question; design experiments to minimize the number of animals required to obtain necessary information; and to alleviate potential pain or suffering for these animals whenever they have to be involved. This partly explains why in vitro models and in vivo non-mammalian models are still heavily used in the scientific community to answer important questions on biofilm biology and to identify potential therapeutic strategies. In addition, species differences, in particular their immune responses, still exist between in vivo mammalian models and humans, especially in murine models [64].
Fortunately, porcine models provide better translational potential than murine models due to their anatomic, physiological and immunological similarities to humans [229]. For example, similar dermal properties and wound healing processes such as re-epithelialization, scarring, and tissue granulation makes porcine models preferable to study chronic wound infections. Delayed wound healing was successfully linked to increasing biofilm characteristics of the wound infection in a porcine model [232]. However, porcine models are more expensive and less accessible than murine models [232]. Compared to most in vitro systems, in vivo models may also suffer from large variability between experiments and animals, as experimental parameters are often less controlled. Finally, mammalian models are often limited to an acute or sub-chronic experimental duration when modeling biofilm infections, due to host responses and ethical concerns. For example, the alginate bead rat models that mimic chronic lung infections have a limited lifetime of 1–3 weeks [233]. Although this issue could be partly mitigated by repeated bacteria exposure, host resistance to re-infection prevents this option [65]. While these animal models integrate the important interplay between the host immune systems and the pathogenic bacteria, the lack of chronicity, in addition to species differences, may explain the high failure rates in translation from animal models to the clinic.
5. Ex vivo models
Clearly, the applications of both in vitro and in vivo model systems have their caveats. In vitro model systems often fail to consider the extremely complex microenvironment, a large part of which remains unexplored, experienced by bacteria in human hosts. In vivo model systems have been invaluable tools to validate and complement in vitro findings. Yet, they are more expensive, low-throughput and their translatability is still debatable due to species differences. These limitations have prompted us to develop ex vivo systems, which sought to decrease the knowledge gap between in vitro and in vivo models.
In ex vivo models, tissues or organs are extracted from animals or humans and are cultured in an artificial environment using in vitro methods for further experimentation. One important advantage of ex vivo systems over traditional in vitro systems or even organ-on-a-chip systems is that it preserves the surface topography and 3D architecture of the native tissues. A growing body of evidence clearly supports that nano- and microscale surface topography has a huge influence on both bacterial attachment [234] and bacterial signaling [235] during biofilm formation. For instance, using a microfluidic device to control spatial structure and chemical communication, it was found that stable coexistence of interacting bacteria requires a defined microscale structure [236]. Recent advances in material sciences also revealed that a reduction of bacterial adhesion can be achieved via the control of surface topography [5], further confirming the role of physiochemical regulation of biofilm formation. Growing ex vivo samples in vitro also allows for more controlled and reproducible experimental conditions, and permits real-time monitoring of the biofilm progression using techniques like optical coherence tomography [58]. This can be particular useful in studying biofilm kinetics from time points as early as bacterial invasion and for assessing the effectiveness of antimicrobials against biofilms at different stages [237]. Lastly, the use of ex vivo tissues allows experiments to be performed in a more physiologically relevant environment that would otherwise be restricted using in vivo models due to ethical issues. For these reasons, many ex vivo biofilm model systems are developed using tissues from both animals and human donors, including ex vivo lung, skin, intestinal segment, dental and mucosal models. As a disadvantage, maintaining ex vivo models for a prolonged period is still a challenge. Depend on the size and geometry of the ex vivo tissues, an adequate supply of nutrients and oxygen throughout the tissue may also be an issue. The following discussion will focus on the two most commonly investigated models, the ex vivo lung model and ex vivo skin model, while highlighting the advantages and limitations of ex vivo models.
5.1. Ex vivo lung model
Chronic lung infections such as those associated with CF and tuberculosis (TB) are highly antibiotic resistant and are often lethal. The development of effective preventative or curative strategies against these dangerous diseases relies on a deep understanding of disease pathogenesis and progression. Many research efforts often focus on the key pathogens, for example P. aeruginosa in CF lung infection and Mycobacterium tuberculosis in TB, using in vitro and in vivo models. While both model systems have their merits in expanding our understanding of bacteria growth, virulence and persistence, each model has certain limitations as discussed before. To overcome these challenges, cheap, high-throughput models of chronic lung infections that recapitulate the native physicochemical environment are critically needed to bridge the gap between in vitro and in vivo model systems.
Towards this end, ex vivo lung models have been developed using tissues derived from rodents [238], pigs [239] and human donors [240]. Of these sources, pigs are arguably the best choice even though tissues from human donors are the most clinically relevant. First, porcine lungs are anatomically very similar to a humans, and this spatial structure is retained in ex vivo porcine models. Recent evidence suggested that this spatial environment could have a significant impact on bacterial interactions, growth and virulence. Second, pig lungs are cheap and are readily available from butchers. Therefore, the use of pig lungs poses much less ethical concerns compared to the use of mice or human lungs. Finally, a single porcine lung can produce dozens of lung samples that can be kept in culture for up to several weeks [239]. Combined with being inexpensive and readily available, these ex vivo models can potentially be used in a high-throughput fashion.
Two common preparation protocols are widely used to prepare ex vivo lung slices. The first method (Fig. 4 A) begins with surface decontamination, followed by dissection of tissues into cubes of approximately equal sizes [59,239]. In pigs, surface decontamination could be carried out by briefly searing the ventral surface of the pleura (<1s) with a hot pallet knife [239]. Lung tissues prepared using this method were then infected with P. aeruginosa and cultured in an artificial sputum medium to mimic the chemical environment of the CF lungs. Using this ex vivo pig lung model, P. aeruginosa growth, quorum sensing (QS), virulence factor production, and tissue damage were successfully quantified in a spatially structured environment that closely mimicked a chronically infected CF lung [239]. In addition, the ability to conveniently assess biofilm evolution at various times post-inoculation using CLSM opened up doors to evaluate the time-dependent therapeutic window against otherwise difficult to treat diseases. Similar techniques are also applicable to lung tissues from human donors. In a recent study, a human ex vivo lung tissue culture model was used to characterize the initial phase of mycobacterial infections and it was discovered that the infection of different cell types in early mycobacterial infections is bacteria species dependent [59]. Unfortunately, the ex vivo lung model prepared following this procedure also has limitations, the most important of which is the high variance in the data obtained, which most likely resulted from tissue heterogeneity and inconsistent cutting.
Fig. 4.
Ex vivo lung and skin models. A) Schematic of the final protocol for preparation, infection, and culture of ex vivo pig lung [239]; B) Workflow of precision cut lung slices: agarose embedding and cutting and 200× SEM image precision cut murine lung slices of 200 μm in thickness [243]; C) Schematic diagram of the assembled model to study anaerobic bacteria [244]; and D) BO-Drum skin culture model [57].
The development of the second preparation protocol, the precision cut lung slices (PCLS, Fig. 4B) method [241], improves the consistency of cutting and offers the possibility of studying thin tissue cultures. PCLS has been successfully applied to tissues from various sources (mice, pigs and human donors) and successfully used to model diseases like CF lung infections and TB [238,239]. In this setup, lungs are first subjected to infusion with a low percentage (0.75%–1.75%), low-melting point, agarose solution through airways and pulmonary arteries and are then hardened by cooling to facilitate cutting and slicing. Depending on the experiment needs, the lungs can be cut into cubes or slices of predetermined thickness using a vibratome.
One advantage of PCLS is that up to 30 slices can be prepared from one mouse lung, and many more can be obtained from lungs of a larger species such as rats, pigs or humans, resulting in a significant reduction in the number of animals needed [242]. Similar to other ex vivo models, PLCS retains much of the cellular diversity and spatial structure found in native lung. In particular, ciliary function is preserved in PLCS and can be confirmed microscopically via ciliary motility [60]. This provides the opportunity to investigate the interactions of cilia with infectious agents and allows us to begin to understand their important role in disease development and progression. PCLS is particularly useful in modeling TB due to the significant differences in mycobacteria-induced pathology, relative resistance observed in murine models, and the high cost associated with non-human primate models. In vitro models of TB are also limited and hard to compare due to the use of different cell types, bacteria strains, infection doses and culturing media in disease modeling [59]. PLCS, on the other hand, preserves the original cell population, structural integrity, and metabolic and transport functions of native lungs and can be invaluable in gaining new insight into disease mechanisms. Mycobacterial infection of PLCS induced TNF-α production, which is consistent with previous in vitro and in vivo studies, further demonstrating their potential in bridging gaps between in vitro and in vivo models [59].
5.2. Ex vivo skin model
Skin wounds and compromised wound healing are of major concern for public health, affecting more than 6.5 million patients in the United States alone. Their treatments represent a significant socioeconomic burden, nationally costing an excess of $25 billion in 2009, and this number is rapidly growing due to increasing health care costs, an aging population, and the prevalence of obesity and diabetes [245]. High bacterial burden, especially in the form of biofilms, is thought to be one of the underlying factors resulting in the non-healing of chronic wounds [246]. Studies investigating wound healing in humans are limited due to ethical considerations. This leads to a dependence on both in vitro and in vivo model systems, especially when disease pathogenesis is investigated. In this context, robust and easy-to-use experimental models are paramount to both gain a deeper understanding of infection pathophysiology and to develop effective therapeutic strategies.
Wound healing is often mimicked in vitro by creating defects on cell monolayers [247]. These 2D models, however, do not recapitulate the complex, multicellular processes during wound infection and wound healing [248]. 3D systems have been developed by culturing cells in 3D matrices, such as hydrogels, to improve model complexity; however, phenotypical changes of contractile fibroblasts were observed in these 3D matrices due to altered mechanical tension and the presence of a non-physiological level of proteins [249,250]. Similar to other in vitro models, host immune responses and other systemic interactions are also missing. This is important to note, considering chronic wound healing is a long-term, coherent process among cells, growth factors, cytokines and ECM proteins.
Alternatively, animal models of chronic skin wound healing have been developed to generate important information of host-bacteria interactions and evaluate treatment strategies in a clinically relevant environment. However, animal models are more expensive and are not compatible with high-throughput screening of a large number of therapeutic strategies. In addition, anatomical and physiological differences between most animals (with the exception of pigs) and human skin, combined with artificially-induced nonhuman pathology, invariably result in different healing kinetics and unique complications [57]. Pig skin is anatomically similar to human skin, and wound healing in pigs has been found to be similar to that of humans [251]. Unfortunately, the use of pigs is associated with higher costs and requires a specialized facility for animal keeping. Application of the treatment is also potentially a challenge in animal models, as preventative measures have to be taken to prevent animals from licking the infected site [252].
In light of these limitations, ex vivo models using pig skin or skin from human donors as substrates for bacterial attachment and as the primary source of nutrition have emerged as a cheap and high-throughput alternative to closely mimic in vivo physiochemical environments. For example, pig skins were obtained directly from slaughterhouses and were sterilized with chlorine gas without affecting the histological properties of the epidermis and dermis [253]. Sterile skin explants were then inoculated with clinical isolates P. aeruginosa wild type strain PAO1 and S. aureus ATCC 35556 (SA35556) to produce the most clinically relevant biofilms. Matured biofilm formed on these explants had increased tolerance to antibiotics. Skin penetration into the dermal matrix was similar to those observed in a human wound bed, suggesting their potential to model chronic wound infections in humans and serve as a screening tool to discover new antimicrobials. For example, the porcine skin explant model has been used to evaluate the ability of surfactant-based wound dressings to reduce biofilms, which provided the first evidence that poloxamer gels play a significant role to sensitize viable bacterial biofilms [254]. Similarly, the antimicrobial efficacy of antimicrobial dressings can be assessed against PAO1 biofilms at different levels of maturity (0–3days) using this ex vivo model [255]. Results obtained in this model were similar to that observed in an in vivo pig burn wound model, further confirming a good correlation between the ex vivo and in vivo models. Other advantages of using pig skin to model chronic wounds are that it is cheap, readily available, can be used fresh or frozen, and is similar to human skin anatomy and physiology. However, most of these models only involve biofilms formed by P. aeruginosa, the applicability of this model for clinical isolates to establish mature biofilms remains to be seen.
Ex vivo skin models have also been developed to study anaerobic bacterial infections, as anaerobic bacteria form a significant proportion of the microbial population in chronic wound infections [256]. For instance, a skin explant on a surgical gauze was placed in between a sterile agarose plug and an agarose pedestal (Fig. 4C), which was then allowed to equilibrate in 5% CO2 for 4 h to create a microenvironment with restricted oxygen supply [244]. After inoculation with a Fastidious Anaerobic Agar plug confluent with Dichelobacter nodosus (D. nodosus), the plate was incubated anaerobically. Using this model, the anaerobic bacteria D. nodosus could be cultured and was subsequently found to alter the expression of key inflammatory markers within the skin.
To standardize ex vivo skin models and allow skin explants to be cultured at the air-liquid interface, a BO-Drum® system (Fig. 4D) was developed as a robust, easy to use and reusable ex vivo full-skin culture system [57]. Cultured skin explants at an air-liquid interface allows for the natural maturation of keratinocytes and helps to preserve the skin barrier function [257]. Additionally, the separation of dermal and epidermal layers restricts bacteria and/or treatment to specified areas without worrying about cross contamination. Other advantages of this system include defined tissue tension and viability of tissue for up to 4 weeks.
5.3. Limitations of ex vivo models
Although ex vivo models provide a cheap and high-throughput alternative to in vivo models, they share some common limitations. Similar to in vitro models, one of the major disadvantages of the ex vivo model is the lack of natural immune systems. Migration of cells from blood into the lungs or skin during immune responses cannot be assessed. Culturing conditions in ex vivo models can also deviate from the natural environment found in animal models, although synthetic media has been developed to mimic the native environment. Finally, the lifespan of ex vivo models is often limited compared with the timespan of chronic infections.
6. Summary and future outlook
With the emergency of antibiotic resistance bacteria, the need for new antimicrobials is more critical than ever. As a consequence, model systems that are representative of native disease conditions are required not only to elucidate mechanisms of disease pathogenesis, but also to determine the safety and efficacy new antimicrobials. Although animal models are still considered as the “gold standard” for reflecting and predicting human responses, emerging evidence points to the significant anatomical, physiological and pathological differences among different species, leading to poor clinical translatability. In addition, in vivo models often lack the ability to provide controllable experimental conditions for a mechanistic understanding of disease etiology in a high-throughput manner. More recently, the use of animals for biomedical research has also been under serious scrutiny due to ethical concerns. Consequently, simple in vitro models (such as MTP-based and flow-based systems) are still heavily relied upon due to their low-cost, easy set-up and amenability to high throughput designs. They contribute to most of our mechanistic understanding of the bacterial infection etiology and virulence. However, many of these in vitro methods contain nutrients, fluid flow, surfaces and microorganisms that are not representative of in vivo environments, which put into question their clinical relevance. Efforts to negate these differences include the use of synthetic media that mimic a nutrient environment in vivo, whereas the employment of surface-independent methods produced biofilms of similar size, shape and antibiotic tolerance to those observed in vitro [264]. Nonetheless, most of these simple in vitro models contains only one bacteria species and therefore lack microbe-microbe or host-microbe interactions that are important in disease pathogenesis, progression and virulence.
Conversely, although conventional 2D models of cells cultured in Transwell plates provide the opportunity to incorporate host components and easy access to manipulating culturing parameters, they are unable to replicate 3D organ structure, integrate physiological functions and present in vivo environmental conditions such as blood vessel fluid flow, shear stress and cyclic stress/stretch [164,265]. As a result, improvements have been made in an effort to not only provide a simplified platform for culturing tissue-like and even organ-like structures, but also to successfully replicate the functionality of human organ systems and their microenvironments [53,266,267]. Many groups have confirmed that bioengineered systems, such as in vitro 3D organoids and organ-on-a-chip system, and ex vivo tissue models, enable mimicry of complex organ pathophysiology and allow an in-depth understanding of the mechanism of actions (see Table 2 ). These systems are therefore more suitable for the development of human-relevant disease models and for the prediction of drug efficacy and toxicity in patients. In particular, ex vivo models of infections attempt to combine the best of both in vitro and in vivo systems. In addition to retaining the 3D structure of native substrates and original cell types, ex vivo studies can be carried out with a greater control over experimental conditions, allowing mechanistic investigations to be studied with more clarity. It is worth noting that the lifespan of ex vivo models is still limited, which may restrict their applications in studying chronic infections. The rapid advancements in sophisticated microfluidic systems that better mimic nutrient and oxygen supply or even physical forces may be a remedy to prolong the tissue cultivation period [55].
Table 2.
Examples of established organ infection and inflammation models.
| Device Types | Description of the stated infection models | Cell/Tissue types | Incorporated bacteria/virus types | Refs | |
|---|---|---|---|---|---|
| Lung | Organ-on-a-chip | Using a lung-on-a-chip model to mimic the innate cellular response to pulmonary infection of E. coli | Human alveolar epithelial cells | Green fluorescent protein (GFP) modified E. coli | [165] |
| Ex vivo model | Ex vivo pig lung model was established to quantify P. aeruginosa growth, virulence and signaling in an environment that mimics infected cystic fibrosis lung | Porcine lung explants | P. aeruginosa | [239] | |
| Ex vivo model | Human ex vivo lung tissue culture model was established to characterize the initial phase of mycobacterial infections | Human lung tissues | Mycobacterial species (M. tuberculosis, M. avium, and M. abscessus) | [59] | |
| Ex vivo model | Precision cut lungs slices were used to model M. tuerculosis pathogenesis and bridge studies in vitro and in vivo | Precision cut lung slices from mice | M. tuerculosis strain H37Rv and M. bovis BCG | [238] | |
| Ex vivo model | Ex vivo viral infection of human lung tissues to study viral replication, tissue tropism and tissue activation | Human lung tissues | Influenza virus; adenovirus 7, and coronaviruses | [[258], [259], [260]] | |
| Gut (Intestine) | 3D organoid | Using 3D organoid and immunofluorescence techniques to visualize post-infection morphologic changes of small intestine | Crypt-derived mouse small intestinal cells | Salmonella | [137] |
| organ-on-a-chip | Reconstituting human intestinal inflammation and injury on-chip | Human colorectal carcinoma-derived (Caco-2) intestinal epithelial cells co-cultured with human capillary endothelial cells or human lymphatic microvascular endothelial cells | Nonpathogenic green fluorescent protein-labeled E. coli (GFP-EC) and pathological enteroinvasive E. coli (EIEC) | [160] | |
| Ex vivo model | Ex vivo human intestinal model to study Entamoeba histolytica pathogenesis | Human colon explant | E. histolytica WT strain HM1:IMSS | [261] | |
| Ex vivo model | Ex vivo mice intestinal tract was used to study the adhesion, colonization and pathology of Blastocystis spp. | Mice intestinal tract | Blastocystis spp. ST7-H and ST7-b | [262] | |
| Skin | Microfluidic Model | Examine the behavior of the bacteria/biofilm under antibiotic treatment | N/A | MRSP and S. epidermidis biofilms | [187,188] |
| Ex vivo model | Ex vivo porcine skin explant model was used to establish mature bacterial biofilm and assess effects of antimicrobial agents on these biofilms | Porcine skin | P. aeruginosa wild type strain PAO1, P. aeruginosa mutant PAO-JP1, and S. aureus ATCC 35556 | [253] | |
| Ex vivo model | 3D skin explant model to study anaerobic bacterial infection | Ovine interdigital skin | D. nodosus | [244] | |
| Brain | 2D model | Using a 2D model to study the autophagy activities in host defenses when bacteria enters the BBB | Human brain microvascular endothelial cells (hBMECs) | Group B Streptococcus (GBS) | [202] |
| 3D Organoid | Employing forbraine organoids for modeling ZIKV exposure | Human-induced pluripotent stem cells (iPSCs) | ZIKV | [147,148] | |
| Ex vivo model | Using organotypic brain slice culture to model viral encephalitis | Mice organotypic brain slice culture | Reovirus serotype 3 strain Abney (T3A) | [263] |
For future perspectives, it is important to keep in mind that a majority of chronic infections harbor polymicrobial communities, thus, incorporation of multiple bacterial species in infection models will likely produce data that are more clinically relevant. One of the scarcities of current models involving polymicrobial infections is likely due to the difficulties in interpreting data results from complex host-microbe or microbe-microbe interactions. To improve our understanding of specific interactions between cell-cell and cell-bacteria, and the effect of pharmaceutical agents on these ex vivo and in vitro models, sophisticated and well-developed simulation/analyzation methods are required. Significant progress can potentially be made by integrating computational modeling tools (such as metabolic multi-cells modeling) which allow for the prediction of bacterial behaviors in the context of complex host dynamics [268,269]. Specifically, for in vitro systems such as organ-on-a-chip, developing computational programs that can continuously monitor the cell and bacteria behaviors will give us the opportunity to study the mechanisms behind natural reactions, drug treatments, and pathological responses comprehensively [270]. In addition, the development in nanotechnology and nanofabrication can also be of tremendous help in advancing our understanding of multispecies interactions as they allow unprecedented control of the bacterial microenvironment at the nanoscale, permitting examination of cellular interactions at a single cell level [271]. A better understanding of the spatial and temporal interactions between microbes, hosts and their environment will not only help advance disease modeling in vitro, ex vivo and in vivo, but may also open up new ways of therapeutic interventions. In an era of personalized medicine, the establishment of patient-specific disease models that can mimic multi-site or whole-body pathology and physiology, faithfully recapitulate the complex organ-level interactions, rapidly evaluate systemic responses to drug candidates and provide high-throughput analysis on their safety and efficacy may be of interest to clinical, pharmaceutical and biotech industries. For instance, recent biotechnology breakthrough of iPSC technology can be included in developing patient-specific neural, liver, cardiac, and brain tissue models with viral or bacterial infections. The patient-specific iPSC-derived in vitro disease models will provide a versatile and non-invasive platform, which allow for the investigation of patient-pathogen interactions and the discovery of personalized medicine [272]. Conversely, by exposing patient-derived viruses (such as HBV) or bacteria to an established in vitro model, it provides us an opportunity to analyze the patient-specific immune responses towards the infection and thus enables the understanding and discovering of the immune evasion pathways and biomarkers [209]. Additionally, for current models representing chronic diseases, how to maintain both structural integrity and biological viability after long-term culture and exposure to pharmaceutics remains unsolved. From a therapeutic development point of view, most of the existing in vitro and ex vivo models are still prototypes that still need substantial validation and standardization from international regulatory bodies to define results obtained from these assays.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.biomaterials.2018.10.030.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Section 3: Current Antibiotic Resistance Threats in the United States, by Microorganism, (n.d.). https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf#page=11 (accessed July 1, 2018).
- 2.Mi G., Shi D., Herchek W., Webster T.J. Self-assembled arginine-rich peptides as effective antimicrobial agents. J. Biomed. Mater. Res. Part A. 2017;105:1046–1054. doi: 10.1002/jbm.a.35979. [DOI] [PubMed] [Google Scholar]
- 3.Darouiche R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004;350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 4.Moran E., Byren I., Atkins B.L. The diagnosis and management of prosthetic joint infections. J. Antimicrob. Chemother. 2010;65:iii45–iii54. doi: 10.1093/jac/dkq305. [DOI] [PubMed] [Google Scholar]
- 5.Mi G., Shi D., Wang M., Webster T.J. Reducing bacterial infections and biofilm formation using nanoparticles and nanostructured antibacterial surfaces. Adv. Healthc. Mater. 2018:1800103. doi: 10.1002/adhm.201800103. [DOI] [PubMed] [Google Scholar]
- 6.Zimlichman E., Henderson D., Tamir O., Franz C., Song P., Yamin C.K., Keohane C., Denham C.R., Bates D.W. Health care-associated infections: AMeta-analysis of costs and financial impact on the US health care system. JAMA Intern. Med. 2013;173:2039–2046. doi: 10.1001/jamainternmed.2013.9763. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS. 2013;121:1–58. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 8.Römling U., Kjelleberg S., Normark S., Nyman L., Uhlin B.E., Åkerlund B. Microbial biofilm formation: a need to act. J. Intern. Med. 2014;276:98–110. doi: 10.1111/joim.12242. [DOI] [PubMed] [Google Scholar]
- 9.Breyer M.D., Look A.T., Cifra A. From bench to patient: model systems in drug discovery. Dis. Model. Mech. 2015;8:1171–1174. doi: 10.1242/dmm.023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langhans S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018;9:6. doi: 10.3389/fphar.2018.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ball S.E., Scatina J.A., Sisenwine S.F., Fisher G.L. The application of in vitro models of drug metabolism and toxicity in drug discovery and drug development. Drug Chem. Toxicol. 1995;18:1–28. doi: 10.3109/01480549509017855. [DOI] [PubMed] [Google Scholar]
- 12.Vela J.M., Maldonado R., Hamon M. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2014. In Vivo Models for Drug Discovery. [DOI] [Google Scholar]
- 13.López Hernández Y., Yero D., Pinos-Rodríguez J.M., Gibert I. Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Front. Microbiol. 2015;6:38. doi: 10.3389/fmicb.2015.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zak T. O ’reilly. MINIREVIEW animal models in the evaluation of antimicrobial agents. Antimicrob. Agents Chemother. 1991;35:1527–1531. doi: 10.1128/aac.35.8.1527. https://pdfs.semanticscholar.org/543f/fd46a397ea7c09202085a1b4aaf2b0fa2a3a.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zschaler J., Schlorke D., Arnhold J. Differences in innate immune response between man and mouse. Crit. Rev. Immunol. 2014;34:433–454. doi: 10.1615/CritRevImmunol.2014011600. [DOI] [PubMed] [Google Scholar]
- 16.Fischer K. Animal testing and marketing bans of the EU cosmetics legislation. Eur. J. Risk Regul. 2015;6:613–621. doi: 10.1017/S1867299X00005158. [DOI] [Google Scholar]
- 17.Ho W.J., Pham E.A., Kim J.W., Ng C.W., Kim J.H., Kamei D.T., Wu B.M. Incorporation of multicellular spheroids into 3-D polymeric scaffolds provides an improved tumor model for screening anticancer drugs. Cancer Sci. 2010;101:2637–2643. doi: 10.1111/j.1349-7006.2010.01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drewitz M., Helbling M., Fried N., Bieri M., Moritz W., Lichtenberg J., Kelm J.M. Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol. J. 2011;6:1488–1496. doi: 10.1002/biot.201100290. [DOI] [PubMed] [Google Scholar]
- 19.Blevins S.M., Bronze M.S. Robert Koch and the “golden age” of bacteriology. Int. J. Infect. Dis. 2010;14:e744–e751. doi: 10.1016/j.ijid.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Murray C.J.L., Lopez A.D. Alternative projections of mortality and disability by cause 1990-2020: global burden of disease study. Lancet. 1997;349:1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 21.Hoiby N., Flensborg E.W., Beck B., Friis B., V Jacobsen S., Jacobsen L. Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of Pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. Scand. J. Respir. Dis. 1977;58:65–79. [PubMed] [Google Scholar]
- 22.Lam J., Chan R., Lam K., Costerton J.W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H., Moser C., Wang H.Z., Høiby N., Song Z.J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015;7:1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macià M.D., Rojo-Molinero E., Oliver A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 2014;20:981–990. doi: 10.1111/1469-0691.12651. [DOI] [PubMed] [Google Scholar]
- 25.Mah T.F.C., O'Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 26.Bjarnsholt T., Alhede M., Alhede M., Eickhardt-Sørensen S.R., Moser C., Kühl M., Jensen P.Ø., Høiby N. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Roberts A.E.L., Kragh K.N., Bjarnsholt T., Diggle S.P. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J. Mol. Biol. 2015;427:3646–3661. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Potera C. Forging a link between biofilms and disease. Science (80-. ) 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- 29.Römling U., Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012;272:541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- 30.Høiby N., Bjarnsholt T., Moser C., Bassi G.L., Coenye T., Donelli G., Hall-Stoodley L., Holá V., Imbert C., Kirketerp-Møller K., Lebeaux D., Oliver A., Ullmann A.J., Williams C. ESCMID study group for biofilms (ESGB), consulting external expert Werner Zimmerli, ESCMID* guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015;21:S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 31.Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., Ray S.M., Thompson D.L., Wilson L.E., Fridkin S.K. Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flemming H.C., Neu T.R., Wozniak D.J. The EPS matrix: the “house of biofilm cells”. J. Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart P.S. Diffusion in biofilms. J. Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderl J.N., Franklin M.J., Stewart P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000;44:1818–1824. doi: 10.1128/AAC.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pages J.-M., Lavigne J.-P., Leflon-Guibout V., Marcon E., Bert F., Noussair L., Nicolas-Chanoine M.-H. Efflux pump, the masked side of ß-lactam resistance in Klebsiella pneumoniae clinical isolates. PloS One. 2009;4:e4817. doi: 10.1371/journal.pone.0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjarnsholt T., Jensen P.Ø., Burmølle M., Hentzer M., Haagensen J.A.J., Hougen H.P., Calum H., Madsen K.G., Moser C., Molin S., Høiby N., Givskov M. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 37.Alhede M., Bjarnsholt T., Jensen P., Phipps R.K., Moser C., Christophersen L., Christensen L.D., van Gennip M., Parsek M., Høiby N., Rasmussen T.B., Givskov M. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155:3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 38.Jesaitis A.J., Franklin M.J., Berglund D., Sasaki M., Lord C.I., Bleazard J.B., Duffy J.E., Beyenal H., Lewandowski Z. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 2003;171:4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 39.Armbruster C.E., Hong W., Pang B., Weimer K.E.D., Juneau R.A., Turner J., Edward Swords W. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in Polymicrobial Otitis media occurs via interspecies quorum signaling. MBio. 2010;1 doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer J., Kazmerzak K., Hansen M.C., Kolenbrander P.E. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 2001;69:5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balaban N.Q., Merrin J., Chait R., Kowalik L., Leibler S. Bacterial persistence as a phenotypic switch. Science (80-. ) 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 42.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 43.Fisher R.A., Gollan B., Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 44.Danese P.N. Antibiofilm approaches: prevention of catheter colonization. Chem. Biol. 2002;9:873–880. doi: 10.1016/S1074-5521(02)00192-8. [DOI] [PubMed] [Google Scholar]
- 45.Król J.E., Nguyen H.D., Rogers L.M., Beyenal H., Krone S.M., Top E.M. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl. Environ. Microbiol. 2011;77:5079–5088. doi: 10.1128/AEM.00090-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zak O., Sande M.A. Academic Press; 1999. Handbook of Animal Models of Infection : Experimental Models in Antimicrobial Chemotherapy. [Google Scholar]
- 47.Pletzer D., Mansour S.C., Wuerth K., Rahanjam N., Hancock R.E.W. New mouse model for chronic infections by gram-negative bacteria enabling the study of anti-infective efficacy and host-microbe interactions. MBio. 2017;8 doi: 10.1128/mBio.00140-17. e00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kukavica-Ibrulj I., Levesque R.C. Animal models of chronic lung infection with Pseudomonas aeruginosa : useful tools for cystic fibrosis studies. Lab. Anim. 2008;42:389–412. doi: 10.1258/la.2007.06014e. [DOI] [PubMed] [Google Scholar]
- 49.Cole S.J., Records A.R., Orr M.W., Linden S.B., Lee V.T. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect. Immun. 2014;82:2048–2058. doi: 10.1128/IAI.01652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalton T., Dowd S.E., Wolcott R.D., Sun Y., Watters C., Griswold J.A., Rumbaugh K.P. An In Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PloS One. 2011;6 doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korgaonkar A., Trivedi U., Rumbaugh K.P., Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coenye T., Nelis H.J. In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Methods. 2010;83:89–105. doi: 10.1016/j.mimet.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 53.Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 54.Yin X., Mead B.E., Safaee H., Langer R., Karp J.M., Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esch E.W., Bahinski A., Huh D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015;14:248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reif R. The body-on-a-chip concept: possibilities and limitations. EXCLI J. 2014;13:1283–1285. http://www.ncbi.nlm.nih.gov/pubmed/26417343 [PMC free article] [PubMed] [Google Scholar]
- 57.Steinstraesser L., Sorkin M., Niederbichler A.D., Becerikli M., Stupka J., Daigeler A., Kesting M.R., Stricker I., Jacobsen F., Schulte M. A novel human skin chamber model to study wound infection ex vivo. Arch. Dermatol. Res. 2010;302:357–365. doi: 10.1007/s00403-009-1009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lenton P., Rudney J., Chen R., Fok A., Aparicio C., Jones R.S. Imaging in vivo secondary caries and ex vivo dental biofilms using cross-polarization optical coherence tomography. Dent. Mater. 2012;28:792–800. doi: 10.1016/j.dental.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganbat D., Seehase S., Richter E., Vollmer E., Reiling N., Fellenberg K., Gaede K.I., Kugler C., Goldmann T. Mycobacteria infect different cell types in the human lung and cause species dependent cellular changes in infected cells. BMC Pulm. Med. 2016;16 doi: 10.1186/s12890-016-0185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kennedy J.L., Koziol-White C.J., Jeffus S., Rettiganti M.R., Fisher P., Kurten M., Eze A., House S., Sikes J.D., Askew E., Putt C., Panettieri R.A., Jones S.M., Kurten R.C. Effects of rhinovirus 39 infection on airway hyperresponsiveness to carbachol in human airways precision cut lung slices. J. Allergy Clin. Immunol. 2018;141:1887–1890.e1. doi: 10.1016/j.jaci.2017.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Squiban B., Kurz C.L. C. elegans: an all in one model for antimicrobial drug discovery. Curr. Drug Targets. 2011;12:967–977. doi: 10.2174/138945011795677854. [DOI] [PubMed] [Google Scholar]
- 62.Letamendia A., Quevedo C., Ibarbia I., Virto J.M., Holgado O., Diez M., Izpisua Belmonte J.C., Callol-Massot C. Development and validation of an automated high-throughput system for zebrafish in vivo screenings. PloS One. 2012;7 doi: 10.1371/journal.pone.0036690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Festing S., Wilkinson R. The ethics of animal research. Talking Point on the use of animals in scientific research. EMBO Rep. 2007;8:526–530. doi: 10.1038/sj.embor.7400993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seok J., Warren H.S., Cuenca A.G., Mindrinos M.N., V Baker H., Xu W., Richards D.R., McDonald-Smith G.P., Gao H., Hennessy L., Finnerty C.C., López C.M., Honari S., Moore E.E., Minei J.P., Cuschieri J., Bankey P.E., Johnson J.L., Sperry J., Nathens A.B., Billiar T.R., West M.A., Jeschke M.G., Klein M.B., Gamelli R.L., Gibran N.S., Brownstein B.H., Miller-Graziano C., Calvano S.E., Mason P.H., Cobb J.P., Rahme L.G., Lowry S.F., V Maier R., Moldawer L.L., Herndon D.N., Davis R.W., Xiao W., Tompkins R.G. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moser C., Jensen P.O., Kobayashi O., Hougen H.P., Song Z., Rygaard J., Kharazmi A., Høiby N. Improved outcome of chronic Pseudomonas aeruginosa lung infection is associated with induction of a Th1-dominated cytokine response. Clin. Exp. Immunol. 2002;127:206–213. doi: 10.1046/j.1365-2249.2002.01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lebeaux D., Chauhan A., Rendueles O., Beloin C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens. 2013;2:288–356. doi: 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fletcher M. The effects of culture concentration and age, time, and temperature on bacterial attachment to polstyrene. Can. J. Microbiol. 1977;23:1–6. doi: 10.1139/m77-001. [DOI] [Google Scholar]
- 68.Tunney M.M., Ramage G., Field T.R., Moriarty T.F., Storey D.G. Rapid colorimetric assay for antimicrobial susceptibility testing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004;48:1879–1881. doi: 10.1128/AAC.48.5.1879-1881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Das T., Sehar S., Koop L., Wong Y.K., Ahmed S., Siddiqui K.S., Manefield M. Influence of Calcium in Extracellular DNA Mediated Bacterial Aggregation and Biofilm Formation. PloS One. 2014;9 doi: 10.1371/journal.pone.0091935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Carvalho F.G., Puppin-Rontani R.M., de Fúcio S.B.P., Negrini T. de C., Carlo H.L., Garcia-Godoy F. Analysis by confocal laser scanning microscopy of the MDPB bactericidal effect on S. mutans biofilm CLSM analysis of MDPB bactericidal effect on biofilm. J. Appl. Oral Sci. 2012;20:568–575. doi: 10.1590/S1678-77572012000500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geilich B.M., Gelfat I., Sridhar S., van de Ven A.L., Webster T.J. Superparamagnetic iron oxide-encapsulating polymersome nanocarriers for biofilm eradication. Biomaterials. 2017;119:78–85. doi: 10.1016/J.BIOMATERIALS.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 72.Mampel J., Spirig T., Weber S.S., Haagensen J.A.J., Molin S., Hilbi H. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 2006;72:2885–2895. doi: 10.1128/AEM.72.4.2885-2895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ceri H., Olson M.E., Stremick C., Read R.R., Morck D., Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olson M.E., Ceri H., Morck D.W., Buret A.G., Read R.R. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 2002;66:86–92. http://www.ncbi.nlm.nih.gov/pubmed/11989739 [PMC free article] [PubMed] [Google Scholar]
- 75.Allison K.R., Brynildsen M.P., Collins J.J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Azeredo J., Azevedo N.F., Briandet R., Cerca N., Coenye T., Costa A.R., Desvaux M., Di Bonaventura G., Hébraud M., Jaglic Z., Kačániová M., Knøchel S., Lourenço A., Mergulhão F., Meyer R.L., Nychas G., Simões M., Tresse O., Sternberg C. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017;43:313–351. doi: 10.1080/1040841X.2016.1208146. [DOI] [PubMed] [Google Scholar]
- 77.Harrison J.J., Ceri H., Yerly J., Stremick C.A., Hu Y., Martinuzzi R., Turner R.J. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary biofilm device. Biol. Proced. Online. 2006;8:194–213. doi: 10.1251/bpo127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chavant P., Gaillard-Martinie B., Talon R., Hébraud M., Bernardi T. A new device for rapid evaluation of biofilm formation potential by bacteria. J. Microbiol. Methods. 2007;68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 79.Tasse J., Croisier D., Badel-Berchoux S., Chavanet P., Bernardi T., Provot C., Laurent F. Preliminary results of a new antibiotic susceptibility test against biofilm installation in device-associated infections: the Antibiofilmogram ®. Pathog. Dis. 2016;74:ftw057. doi: 10.1093/femspd/ftw057. [DOI] [PubMed] [Google Scholar]
- 80.Sulaeman S., Le Bihan G., Rossero A., Federighi M., Dé E., Tresse O. Comparison between the biofilm initiation of Campylobacter jejuni and Campylobacter coli strains to an inert surface using BioFilm Ring Test ®. J. Appl. Microbiol. 2010;108:1303–1312. doi: 10.1111/j.1365-2672.2009.04534.x. [DOI] [PubMed] [Google Scholar]
- 81.Olivares E., Badel-Berchoux S., Provot C., Jaulhac B., Prévost G., Bernardi T., Jehl F. The BioFilm ring test: a rapid method for routine analysis of Pseudomonas aeruginosa biofilm formation kinetics. J. Clin. Microbiol. 2016;54:657–661. doi: 10.1128/JCM.02938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Badel S., Laroche C., Gardarin C., Bernardi T., Michaud P. New method showing the influence of matrix components in leuconostoc mesenteroides biofilm formation. Appl. Biochem. Biotechnol. 2008;151:364–370. doi: 10.1007/s12010-008-8199-y. [DOI] [PubMed] [Google Scholar]
- 83.Renier S., Chagnot C., Deschamps J., Caccia N., Szlavik J., Joyce S.A., Popowska M., Hill C., Knøchel S., Briandet R., Hébraud M., Desvaux M. Inactivation of the SecA2 protein export pathway in Listeria monocytogenes promotes cell aggregation, impacts biofilm architecture and induces biofilm formation in environmental condition. Environ. Microbiol. 2014;16:1176–1192. doi: 10.1111/1462-2920.12257. [DOI] [PubMed] [Google Scholar]
- 84.Harrison J.J., Ceri H., Yerly J., Stremick C.A., Hu Y., Martinuzzi R., Turner R.J. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary biofilm device. Biol. Proced. Online. 2006;8:194–213. doi: 10.1251/bpo127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pamp S.J., Sternberg C., Tolker-Nielsen T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytom. Part A. 2009;75A:90–103. doi: 10.1002/cyto.a.20685. [DOI] [PubMed] [Google Scholar]
- 86.McCoy W.F., Bryers J.D., Robbins J., Costerton J.W. Observations of fouling biofilm formation. Can. J. Microbiol. 1981;27:910–917. doi: 10.1139/m81-143. [DOI] [PubMed] [Google Scholar]
- 87.Harrison J.J., Ceri H., Yerly J., Stremick C.A., Hu Y., Martinuzzi R., Turner R.J. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary biofilm device. Biol. Proced. Online. 2006;8:194–213. doi: 10.1251/bpo127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Prijck K., De Smet N., Rymarczyk-Machal M., Van Driessche G., Devreese B., Coenye T., Schacht E., Nelis H.J. Candida albicans biofilm formation on peptide functionalized polydimethylsiloxane. Biofouling. 2010;26:269–275. doi: 10.1080/08927010903501908. [DOI] [PubMed] [Google Scholar]
- 89.Nava-Ortiz C.A.B., Burillo G., Concheiro A., Bucio E., Matthijs N., Nelis H., Coenye T., Alvarez-Lorenzo C. Cyclodextrin-functionalized biomaterials loaded with miconazole prevent Candida albicans biofilm formation in vitro. Acta Biomater. 2010;6:1398–1404. doi: 10.1016/j.actbio.2009.10.039. [DOI] [PubMed] [Google Scholar]
- 90.Raad I., Hanna H., Dvorak T., Chaiban G., Hachem R. Optimal antimicrobial catheter lock solution, using different combinations of minocycline, EDTA, and 25-percent ethanol, rapidly eradicates organisms embedded in biofilm. Antimicrob. Agents Chemother. 2007;51:78–83. doi: 10.1128/AAC.00154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Curtin J., Cormican M., Fleming G., Keelehan J., Colleran E. Linezolid compared with eperezolid, vancomycin, and gentamicin in an in vitro model of antimicrobial lock therapy for Staphylococcus epidermidis central venous catheter-related biofilm infections. Antimicrob. Agents Chemother. 2003;47:3145–3148. doi: 10.1128/AAC.47.10.3145-3148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolfaardt G.M., Lawrence J.R., Robarts R.D., Caldwell S.J., Caldwell D.E. Multicellular organization in a degradative biofilm community. Appl. Environ. Microbiol. 1994;60:434–446. doi: 10.1128/aem.60.2.434-446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bakker D.P., Van der Plaats A., Verkerke G.J., Busscher H.J., Van der Mei H.C. Comparison of velocity profiles for different flow chamber designs used in studies of microbial adhesion to surfaces. Appl. Environ. Microbiol. 2003;69:6280–6287. doi: 10.1128/AEM.69.10.6280-6287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tolker-Nielsen T., Sternberg C. Growing and analyzing biofilms in flow chambers. Curr. Protoc. Microbiol. 2011 doi: 10.1002/9780471729259.mc01b02s21. 1B.2.1–1B.2.17, (Chapter 1) [DOI] [PubMed] [Google Scholar]
- 95.Klausen M., Heydorn A., Ragas P., Lambertsen L., Aaes-Jørgensen A., Molin S., Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 96.Klausen M., Aaes-Jørgensen A., Molin S., Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 97.Kommerein N., Doll K., Stumpp N.S., Stiesch M. Development and characterization of an oral multispecies biofilm implant flow chamber model. PloS One. 2018;13:e0196967. doi: 10.1371/journal.pone.0196967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crusz S.A., Popat R., Rybtke M.T., Cámara M., Givskov M., Tolker-Nielsen T., Diggle S.P., Williams P. Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling. 2012;28:835–842. doi: 10.1080/08927014.2012.716044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Donlan R.M., Piede J.A., Heyes C.D., Sanii L., Murga R., Edmonds P., El-Sayed I., El-Sayed M.A. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl. Environ. Microbiol. 2004;70:4980–4988. doi: 10.1128/AEM.70.8.4980-4988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paredes J., Becerro S., Arizti F., Aguinaga A., Del Pozo J.L., Arana S. Real time monitoring of the impedance characteristics of Staphylococcal bacterial biofilm cultures with a modified CDC reactor system. Biosens. Bioelectron. 2012;38:226–232. doi: 10.1016/j.bios.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 101.Paredes J., Becerro S., Arana S. Label-free interdigitated microelectrode based biosensors for bacterial biofilm growth monitoring using Petri dishes. J. Microbiol. Methods. 2014;100:77–83. doi: 10.1016/j.mimet.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 102.Paredes J., Becerro S., Arizti F., Aguinaga A., Del Pozo J.L., Arana S. Interdigitated microelectrode biosensor for bacterial biofilm growth monitoring by impedance spectroscopy technique in 96-well microtiter plates. Sensor. Actuator. B Chem. 2013;178:663–670. doi: 10.1016/J.SNB.2013.01.027. [DOI] [Google Scholar]
- 103.Tolker-Nielsen T., Sternberg C. Growing and analyzing biofilms in flow chambers. Curr. Protoc. Microbiol. 2011;74:4463–4471. doi: 10.1002/9780471729259.mc01b02s21. [DOI] [PubMed] [Google Scholar]
- 104.Rasmussen K., Lewandowski Z. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng. 1998;59:302–309. doi: 10.1002/(SICI)1097-0290(19980805)59:3<302::AID-BIT6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 105.Sharma P., Rozenbaum R.T., Woudstra W., de Jong E.D., van der Mei H.C., Busscher H.J., Sharma P. A constant depth film fermenter to grow microbial biofilms. Protoc. Exch. 2017 doi: 10.1038/protex.2017.024. [DOI] [Google Scholar]
- 106.Harrison J.J., Turner R.J., Ceri H. High-throughput metal susceptibility testing of microbial biofilms. BMC Microbiol. 2005;5:53. doi: 10.1186/1471-2180-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McBain A.J. Chapter 4 in vitro biofilm models. An overview. Adv. Appl. Microbiol. 2009;69:99–132. doi: 10.1016/S0065-2164(09)69004-3. [DOI] [PubMed] [Google Scholar]
- 108.Williams D.L., Bloebaum R.D. Observing the biofilm matrix of staphylococcus epidermidis ATCC 35984 grown using the CDC biofilm reactor. Microsc. Microanal. 2010;16:143–152. doi: 10.1017/S143192760999136X. [DOI] [PubMed] [Google Scholar]
- 109.Goeres D.M., Hamilton M.A., Beck N.A., Buckingham-Meyer K., Hilyard J.D., Loetterle L.R., Lorenz L.A., Walker D.K., Stewart P.S. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat. Protoc. 2009;4:783–788. doi: 10.1038/nprot.2009.59. [DOI] [PubMed] [Google Scholar]
- 110.Sharma P., Rozenbaum R.T., Woudstra W., de Jong E.D., van der Mei H.C., Busscher H.J., Sharma P. A constant depth film fermenter to grow microbial biofilms. Protoc. Exch. 2017 doi: 10.1038/protex.2017.024. [DOI] [Google Scholar]
- 111.Buhmann M.T., Stiefel P., Maniura-Weber K., Ren Q. In Vitro biofilm models for device-related infections. Trends Biotechnol. 2016;34:945–948. doi: 10.1016/j.tibtech.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 112.Raad I., Hanna H., Dvorak T., Chaiban G., Hachem R. Optimal antimicrobial catheter lock solution, using different combinations of minocycline, EDTA, and 25-percent ethanol, rapidly eradicates organisms embedded in biofilm. Antimicrob. Agents Chemother. 2007;51:78–83. doi: 10.1128/AAC.00154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Palmer K.L., Aye L.M., Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ochsner U.A., Vasil M.L. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Palmer K.L., Mashburn L.M., Singh P.K., Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harrison J.J., Ceri H., Yerly J., Stremick C.A., Hu Y., Martinuzzi R., Turner R.J. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary biofilm device. Biol. Proced. Online. 2006;8:194–213. doi: 10.1251/bpo127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science (80-. ) 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 118.Turner K.H., Wessel A.K., Palmer G.C., Murray J.L., Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hirschfeld J. Dynamic interactions of neutrophils and biofilms. J. Oral Microbiol. 2014;6:1–10. doi: 10.3402/jom.v6.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nix D.E., Goodwin S.D., Peloquin C.A., Rotella D.L., Schentag J.J. Antibiotic tissue penetration and its relevance: impact of tissue penetration on infection response. Antimicrob. Agents Chemother. 1991;35:1953–1959. doi: 10.1128/AAC.35.10.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 122.Günther F., Wabnitz G.H., Stroh P., Prior B., Obst U., Samstag Y., Wagner C., Hänsch G.M. Host defence against Staphylococcus aureus biofilms infection: phagocytosis of biofilms by polymorphonuclear neutrophils (PMN) Mol. Immunol. 2009;46:1805–1813. doi: 10.1016/j.molimm.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 123.Van Gennip M., Christensen L.D., Alhede M., Phipps R., Jensen P.Ø., Christophersen L., Pamp S.J., Moser C., Mikkelsen P.J., Koh A.Y., Tolker-Nielsen T., Pier G.B., HØiby N., Givskov M., Bjarnsholt T. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 2009;117:537–546. doi: 10.1111/j.1600-0463.2009.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kolpen M., Hansen C.R., Bjarnsholt T., Moser C., Christensen L.D., Van Gennip M., Ciofu O., Mandsberg L., Kharazmi A., Döring G., Givskov M., Høiby N., Jensen P. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax. 2010;65:57–62. doi: 10.1136/thx.2009.114512. [DOI] [PubMed] [Google Scholar]
- 125.Kragh K.N., Alhede M., Jensen P., Moser C., Scheike T., Jacobsen C.S., Poulsen S.S., Eickhardt-Sørensen S.R., Trøstrup H., Christoffersen L., Hougen H.P., Rickelt L.F., Kühl M., Høiby N., Bjarnsholt T. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect. Immun. 2014;82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Citron D.M., Goldstein E.J.C., Merriam C.V., Lipsky B.A., Abramson M.A. Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J. Clin. Microbiol. 2007;45:2819–2828. doi: 10.1128/JCM.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Heller D., Helmerhorst E.J., Gower A.C., Siqueira W.L., Paster B.J., Oppenheim F.G. Microbial diversity in the early in vivo-formed dental biofilm. Appl. Environ. Microbiol. 2016;82:1881–1888. doi: 10.1128/AEM.03984-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peters B.M., Jabra-Rizk M.A., O'May G.A., William Costerton J., Shirtliff M.E. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kirketerp-Møller K., Jensen P., Fazli M., Madsen K.G., Pedersen J., Moser C., Tolker-Nielsen T., Høiby N., Givskov M., Bjarnsholt T. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 2008;46:2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.V Wilson H. A new method by which sponges may be artificially reared. Science (80-. ) 1907;25:912–915. doi: 10.1126/science.25.649.912. [DOI] [PubMed] [Google Scholar]
- 131.Lancaster M.A., Knoblich J.A. Organogenesisin a dish: modeling development and disease using organoid technologies. Science (80-. ) 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 132.Yin X., Mead B.E., Safaee H., Langer R., Karp J.M., Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Iakobachvili N., Peters P.J. Humans in a dish: the potential of organoids in modeling immunity and infectious diseases. Front. Microbiol. 2017 doi: 10.3389/fmicb.2017.02402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sato T., Vries R.G., Snippert H.J., Van De Wetering M., Barker N., Stange D.E., Van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 135.Takebe T., Zhang B., Radisic M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell. 2017;21:297–300. doi: 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 136.Clevers H. Modeling development and disease with organoids. Cell. 2016 doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 137.Zhang Y.G., Wu S., Xia Y., Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host–bacterial interactions. Physiol. Rep. 2014;2 doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tam M.A., Rydström A., Sundquist M., Wick M.J. Early cellular responses to Salmonella infection: dendritic cells, monocytes, and more. Immunol. Rev. 2008 doi: 10.1111/j.1600-065X.2008.00679.x. [DOI] [PubMed] [Google Scholar]
- 139.Lu R., Wu S., Liu X., Xia Y., Zhang Y.G., Sun J. Chronic effects of a salmonella type III secretion effector protein AvrA in vivo. PloS One. 2010 doi: 10.1371/journal.pone.0010505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martinez Rodriguez N.R., Eloi M.D., Huynh A., Dominguez T., Lam A.H.C., Carcamo-Molina D., Naser Z., Desharnais R., Salzman N.H., Porter E. Expansion of paneth cell population in response to enteric Salmonella enterica Serovar Typhimurium infection. Infect. Immun. 2012 doi: 10.1128/IAI.05638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schlaermann P., Toelle B., Berger H., Schmidt S.C., Glanemann M., Ordemann J., Bartfeld S., Mollenkopf H.J., Meyer T.F. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016 doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bartfeld S., Bayram T., Van De Wetering M., Huch M., Begthel H., Kujala P., Vries R., Peters P.J., Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015 doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Boxberger H.-J., Sessler M.J., Grausam M.C., Becker H.-D., Meyer T.F. Isolation and culturing of highly polarized primary epithelial cells from normal human stomach (antrum) as spheroid-like vesicles. Methods Cell Sci. 1997;19:169–178. doi: 10.1023/A:1009751913391. [DOI] [Google Scholar]
- 144.Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 145.Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M., Knoblich J.A., Lachmann R., Pääbo S., Huttner W.B., Treutlein B. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. 2015:201520760. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Garcez P.P., Loiola E.C., Da Costa R.M., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., Rehen S.K. Zika virus: Zika virus impairs growth in human neurospheres and brain organoids. Science (80-. ) 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 148.Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L.M., Guimarães K.P., Benazzato C., Almeida N., Pignatari G.C., Romero S., Polonio C.M., Cunha I., Freitas C.L., Brandaõ W.N., Rossato C., Andrade D.G., Faria D. de P., Garcez A.T., Buchpigel C.A., Braconi C.T., Mendes E., Sall A.A., Zanotto P.M.D.A., Peron J.P.S., Muotri A.R., Beltrao-Braga P.C.B.B. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pampaloni F., Reynaud E.G., Stelzer E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 150.Huh D., Hamilton G.A., Ingber D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huh D., Kim H.J., Fraser J.P., Shea D.E., Khan M., Bahinski A., Hamilton G.A., Ingber D.E. Microfabrication of human organs-on-chips. Nat. Protoc. 2013;8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- 152.King K.R., Wang C.C.J., Kaazempur-Mofrad M.R., Vacanti J.P., Borenstein J.T. Biodegradable microfluids. Adv. Mater. 2004;16:2007–2012. doi: 10.1002/adma.200306522. [DOI] [Google Scholar]
- 153.Therriault D., White S.R., Lewis J.A. Chaotic mixing in three-dimensional microvascular networks fabricated by direct-write assembly. Nat. Mater. 2003;2:265–271. doi: 10.1038/nmat863. [DOI] [PubMed] [Google Scholar]
- 154.Bernard A., Renault J.P., Michel B., Bosshard H.R., Delamarche E. Microcontact printing of proteins. Adv. Mater. 2000;12:1067–1070. doi: 10.1002/1521-4095(200007)12:14<1067::AID-ADMA1067>3.0.CO;2-M. [DOI] [Google Scholar]
- 155.Joshi P.N. Lab-on-a-Chip Fabr. Appl. InTech; 2016. Cells and organs on chip — a revolutionary platform for biomedicine; pp. 77–123. [DOI] [Google Scholar]
- 156.Tilles A.W., Baskaran H., Roy P., Yarmush M.L., Toner M. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a microchannel flat-plate bioreactor. Biotechnol. Bioeng. 2001;73:379–389. doi: 10.1002/bit.1071. [DOI] [PubMed] [Google Scholar]
- 157.Takayama S., Ostuni E., Qian X., McDonald J.C., Jiang X., LeDuc P., Wu M.H., Ingber D.E., Whitesides G.M. Topographical micropatterning of poly(dimethylsiloxane) using laminar flows of liquids in capillaries. Adv. Mater. 2001;13:570–574. doi: 10.1002/1521-4095(200104)13:8<570::AID-ADMA570>3.0.CO;2-B. [DOI] [Google Scholar]
- 158.Lee P.J., Hung P.J., Lee L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 159.Wufuer M., Lee G.H., Hur W., Jeon B., Kim B.J., Choi T.H., Lee S.H. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016;6:37471. doi: 10.1038/srep37471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim H.J., Li H., Collins J.J., Ingber D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Booth R., Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB) Lab Chip. 2012;12:1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 162.Brown J.A., Codreanu S.G., Shi M., Sherrod S.D., Markov D.A., Neely M.D., Britt C.M., Hoilett O.S., Reiserer R.S., Samson P.C., McCawley L.J., Webb D.J., Bowman A.B., McLean J.A., Wikswo J.P. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. J. Neuroinflammation. 2016;13:306. doi: 10.1186/s12974-016-0760-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Paoli R., Samitier J. Mimicking the kidney: a key role in organ-on-chip development. Micromachines. 2016;7:126. doi: 10.3390/mi7070126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ingber D.E. Reverse engineering human pathophysiology with organs-on-chips. Cell. 2016;164:1105–1109. doi: 10.1016/j.cell.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 165.Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Yuan Hsin H., Ingber D.E. Reconstituting organ-level lung functions on a chip. Science (80-. ) 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bersini S., Jeon J.S., Dubini G., Arrigoni C., Chung S., Charest J.L., Moretti M., Kamm R.D. A microfluidic 3D invitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35:2454–2461. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lee D.W., Choi Y.S., Seo Y.J., Lee M.Y., Jeon S.Y., Ku B., Kim S., Yi S.H., Nam D.H. High-throughput screening (HTS) of anticancer drug efficacy on a micropillar/microwell chip platform. Anal. Chem. 2014;86:535–542. doi: 10.1021/ac402546b. [DOI] [PubMed] [Google Scholar]
- 168.Zhang B., Cheng Y., Wang H., Ye B., Shang L., Zhao Y., Gu Z. Multifunctional inverse opal particles for drug delivery and monitoring. Nanoscale. 2015;7:10590–10594. doi: 10.1039/c5nr02324f. [DOI] [PubMed] [Google Scholar]
- 169.Bowdler A. Humana Press; 2002. The Complete Spleen: Structure, Function and Clinical Disorders. [Google Scholar]
- 170.Hatherell K., Couraud P.O., Romero I.A., Weksler B., Pilkington G.J. Development of a three-dimensional, all-human in vitro model of the blood-brain barrier using mono-, co-, and tri-cultivation Transwell models. J. Neurosci. Methods. 2011;199:223–229. doi: 10.1016/j.jneumeth.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 171.Gopalakrishnan N., Hannam R., Casoni G.P., Barriet D., Ribe J.M., Haug M. Halaas, Infection and immunity on a chip: a compartmentalised microfluidic platform to monitor immune cell behaviour in real time. Lab Chip. 2015;15:1481–1487. doi: 10.1039/c4lc01438c. [DOI] [PubMed] [Google Scholar]
- 172.Tong Z.Q., Balzer E.M., Dallas M.R., Hung W.C., Stebe K.J., Konstantopoulos K. Chemotaxis of cell populations through confined spaces at Single-Cell resolution. PloS One. 2012;7 doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ambravaneswaran V., Wong I.Y., Aranyosi A.J., Toner M., Irimia D. Directional decisions during neutrophil chemotaxis inside bifurcating channels. Integr. Biol. 2010;2:639–647. doi: 10.1039/c0ib00011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kim H.J., Huh D., Hamilton G., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 175.Shanahan F. The host-microbe interface within the gut. Bailliere’s Best Pract. Res. Clin. Gastroenterol. 2002;16:915–931. doi: 10.1053/bega.2002.0342. [DOI] [PubMed] [Google Scholar]
- 176.Fang H.W., Bin Fang S., Chiau J.S.C., Yeung C.Y., Chan W.T., Bin Jiang C., Cheng M.L., Lee H.C. Inhibitory effects of Lactobacillus casei subsp. rhamnosus on Salmonella lipopolysaccharide-induced inflammation and epithelial barrier dysfunction in a co-culture model using Caco-2/peripheral blood mononuclear cells. J. Med. Microbiol. 2010;59:573–579. doi: 10.1099/jmm.0.009662-0. [DOI] [PubMed] [Google Scholar]
- 177.Dai C., Zhao D.H., Jiang M. VSL#3 probiotics regulate the intestinal epithelial barrier in vivo and in vitro via the p38 and ERK signaling pathways. Int. J. Mol. Med. 2012;29:202–208. doi: 10.3892/ijmm.2011.839. [DOI] [PubMed] [Google Scholar]
- 178.Kim J., Hegde M., Jayaraman A. Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions. Lab Chip. 2010 doi: 10.1039/b911367c. [DOI] [PubMed] [Google Scholar]
- 179.Leroy M., Labbé J.F., Ouellet M., Jean J., Lefèvre T., Laroche G., Auger M., Pouliot R. A comparative study between human skin substitutes and normal human skin using Raman microspectroscopy. Acta Biomater. 2014;10:2703–2711. doi: 10.1016/j.actbio.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 180.Ronaldson-Bouchard K., Vunjak-Novakovic G. Organs-on-a-Chip: a fast track for engineered human tissues in drug development. Cell Stem Cell. 2018;22:310–324. doi: 10.1016/j.stem.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bellas E., Seiberg M., Garlick J., Kaplan D.L. In vitro 3D full-thickness skin-equivalent tissue model using silk and collagen biomaterials. Macromol. Biosci. 2012;12:1627–1636. doi: 10.1002/mabi.201200262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Monfort A., Soriano-Navarro M., García-Verdugo J.M., Izeta A. Production of human tissue-engineered skin trilayer on a plasma-based hypodermis. J. Tissue Eng. Regen. Med. 2013;7:479–490. doi: 10.1002/term.548. [DOI] [PubMed] [Google Scholar]
- 183.Abaci H.E., Gledhill K., Guo Z., Christiano A.M., Shuler M.L. Pumpless microfluidic platform for drug testing on human skin equivalents. Lab Chip. 2015;15:882–888. doi: 10.1039/c4lc00999a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ramadan Q., Ting F.C.W. In vitro micro-physiological immune-competent model of the human skin. Lab Chip. 2016;16:1899–1908. doi: 10.1039/c6lc00229c. [DOI] [PubMed] [Google Scholar]
- 185.Mori N., Morimoto Y., Takeuchi S. Skin integrated with perfusable vascular channels on a chip. Biomaterials. 2017;116:48–56. doi: 10.1016/j.biomaterials.2016.11.031. [DOI] [PubMed] [Google Scholar]
- 186.Hampton S. Bacteria and wound healing. J. Community Nurs. 2007;21:32–40. doi: 10.1097/00001432-200404000-00004. [DOI] [Google Scholar]
- 187.Wright E., Neethirajan S., Weng X. Microfluidic wound model for studying the behaviors of Pseudomonas aeruginosa in polymicrobial biofilms. Biotechnol. Bioeng. 2015 doi: 10.1002/bit.25651. [DOI] [PubMed] [Google Scholar]
- 188.Terry J., Neethirajan S. A novel microfluidic wound model for testing antimicrobial agents against Staphylococcus pseudintermedius biofilms. J. Nanobiotechnol. 2014 doi: 10.1186/1477-3155-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.van der Meer A.D., Vermeul K., Poot A.A., Feijen J., Vermes I. A microfluidic wound-healing assay for quantifying endothelial cell migration. AJP Hear. Circ. Physiol. 2010;298:H719–H725. doi: 10.1152/ajpheart.00933.2009. [DOI] [PubMed] [Google Scholar]
- 190.Sriram G., Alberti M., Dancik Y., Wu B., Wu R., Feng Z., Ramasamy S., Bigliardi P.L., Bigliardi-Qi M., Wang Z. Full-thickness human skin-on-chip with enhanced epidermal morphogenesis and barrier function. Mater. Today. 2018;21:326–340. doi: 10.1016/j.mattod.2017.11.002. [DOI] [Google Scholar]
- 191.van Midwoud P.M., Janse A., Merema M.T., Groothuis G.M.M., Verpoorte E. Comparison of biocompatibility and adsorption properties of different plastics for advanced microfluidic cell and tissue culture models. Anal. Chem. 2012;84:3938–3944. doi: 10.1021/ac300771z. [DOI] [PubMed] [Google Scholar]
- 192.Halldorsson S., Lucumi E., Gómez-Sjöberg R., Fleming R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 193.Łopacińska J.M., Emnéus J., Dufva M. Poly(Dimethylsiloxane) (PDMS) Affects Gene Expression in PC12 Cells Differentiating into Neuronal-Like Cells. PloS One. 2013;8 doi: 10.1371/journal.pone.0053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mohammadi M.H., Heidary Araghi B., Beydaghi V., Geraili A., Moradi F., Jafari P., Janmaleki M., Valente K.P., Akbari M., Sanati-Nezhad A. Skin diseases modeling using combined tissue engineering and microfluidic technologies. Adv. Healthc. Mater. 2016 doi: 10.1002/adhm.201600439. [DOI] [PubMed] [Google Scholar]
- 195.Sun C., Ding Y., Zhou L., Shi D., Sun L., Webster T.J., Shen Y. Noninvasive nanoparticle strategies for brain tumor targeting. Nanomed. Nanotechnol. Biol. Med. 2017;13:2605–2621. doi: 10.1016/j.nano.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 196.Shi D., Sun L., Mi G., Sheikh L., Bhattacharya S., Nayar S., Webster T.J. Controlling ferrofluid permeability across the blood-brain barrier model. Nanotechnology. 2014;25 doi: 10.1088/0957-4484/25/7/075101. [DOI] [PubMed] [Google Scholar]
- 197.Kaisar M.A., Sajja R.K., Prasad S., Abhyankar V.V., Liles T., Cucullo L. New experimental models of the blood-brain barrier for CNS drug discovery. Expert Opin. Drug Discov. 2017;12:89–103. doi: 10.1080/17460441.2017.1253676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cho H., Seo J.H., Wong K.H.K., Terasaki Y., Park J., Bong K., Arai K., Lo E.H., Irimia D. Three-dimensional blood-brain barrier model for in vitro studies of neurovascular pathology. Sci. Rep. 2015;5:15222. doi: 10.1038/srep15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ruck T., Bittner S., Meuth S.G. Blood-brain barrier modeling: challenges and perspectives. Neural Regen. Res. 2015;10:889–891. doi: 10.4103/1673-5374.158342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Terrell-Hall T.B., Ammer A.G., Griffith J.I.G., Lockman P.R. Permeability across a novel microfluidic blood-tumor barrier model. Fluids Barriers CNS. 2017;14:3. doi: 10.1186/s12987-017-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Eugenin E.A., Clements J.E., Zink M.C., Berman J.W. Human immunodeficiency virus infection of human astrocytes disrupts blood-brain barrier integrity by a gap junction-dependent mechanism. J. Neurosci. 2011;31:9456–9465. doi: 10.1523/JNEUROSCI.1460-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cutting A.S., Del Rosario Y., Mu R., Rodriguez A., Till A., Subramani S., Gottlieb R.A., Doran K.S. The role of autophagy during group B streptococcus infection of blood-brain barrier endothelium. J. Biol. Chem. 2014;289:35711–35723. doi: 10.1074/jbc.M114.588657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Achyuta A.K.H., Conway A.J., Crouse R.B., Bannister E.C., Lee R.N., Katnik C.P., Behensky A.A., Cuevas J., Sundaram S.S. A modular approach to create a neurovascular unit-on-a-chip. Lab Chip. 2013;13:542–553. doi: 10.1039/c2lc41033h. [DOI] [PubMed] [Google Scholar]
- 204.Drescher K., Shen Y., Bassler B.L., Stone H.A. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl. Acad. Sci. 2013 doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kolter R., Greenberg E.P. Microbial sciences: the superficial life of microbes. Nature. 2006 doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 206.Hall-Stoodley L., Stoodley P., Kathju S., Høiby N., Moser C., William Costerton J., Moter A., Bjarnsholt T. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 2012 doi: 10.1111/j.1574-695X.2012.00968.x. [DOI] [PubMed] [Google Scholar]
- 207.Donlan R.M., Costerton J.W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002 doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rusconi R., Lecuyer S., Guglielmini L., Stone H.A. Laminar flow around corners triggers the formation of biofilm streamers. J. R. Soc. Interface. 2010 doi: 10.1098/rsif.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ortega-Prieto A.M., Skelton J.K., Wai S.N., Large E., Lussignol M., Vizcay-Barrena G., Hughes D., Fleck R.A., Thursz M., Catanese M.T., Dorner M. 2018. 3D Microfluidic Liver Cultures as a Physiological Preclinical Tool for Hepatitis B Virus Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tsai H.F., Trubelja A., Shen A.Q., Bao G. Tumour-on-a-chip: microfluidic models of tumour morphology, growth and microenvironment. J. R. Soc. Interface. 2017;14 doi: 10.1098/rsif.2017.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Albanese A., Lam A.K., Sykes E.A., V Rocheleau J., Chan W.C.W. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat. Commun. 2013;4 doi: 10.1038/ncomms3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Caballero D., Blackburn S.M., De Pablo M., Samitier J., Albertazzi L. Tumour-vessel-on-a-chip models for drug delivery. Lab Chip. 2017;17:3760–3771. doi: 10.1039/c7lc00574a. [DOI] [PubMed] [Google Scholar]
- 213.Hassell B.A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C.S., Ingber D.E. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–516. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 214.Esch M.B., Mahler G.J., Stokol T., Shuler M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab a Chip - Miniaturisation Chem. Biol. 2014;14:3081–3092. doi: 10.1039/c4lc00371c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sin A., Chin K.C., Jamil M.F., Kostov Y., Rao G., Shuler M.L. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol. Prog. 2004;20:338–345. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- 216.Wagner I., Materne E.M., Brincker S., Süßbier U., Frädrich C., Busek M., Sonntag F., Sakharov D.A., Trushkin E.V., Tonevitsky A.G., Lauster R., Marx U. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip. 2013;13:3538–3547. doi: 10.1039/c3lc50234a. [DOI] [PubMed] [Google Scholar]
- 217.Sung J.H., Kam C., Shuler M.L. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 218.Skardal A., Murphy S.V., Devarasetty M., Mead I., Kang H.W., Seol Y.J., Zhang Y.S., Shin S.R., Zhao L., Aleman J., Hall A.R., Shupe T.D., Kleensang A., Dokmeci M.R., Jin Lee S., Jackson J.D., Yoo J.J., Hartung T., Khademhosseini A., Soker S., Bishop C.E., Atala A. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017;7:8837. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kostov Y., Harms P., Randers-Eichhorn L., Rao G. Low-cost microbioreactor for high-throughput bioprocessing. Biotechnol. Bioeng. 2001;72:346–352. doi: 10.1002/1097-0290(20010205)72:3<346::AID-BIT12>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 220.Maschmeyer I., Lorenz A.K., Schimek K., Hasenberg T., Ramme A.P., Hübner J., Lindner M., Drewell C., Bauer S., Thomas A., Sambo N.S., Sonntag F., Lauster R., Marx U. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 221.Low L.A., Tagle D.A. Organs-on-chips: progress, challenges, and future directions. Exp. Biol. Med. 2017;242:1573–1578. doi: 10.1177/1535370217700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Young E.W.K. Cells, tissues, and organs on chips: challenges and opportunities for the cancer tumor microenvironment. Integr. Biol. 2013;5:1096. doi: 10.1039/c3ib40076j. [DOI] [PubMed] [Google Scholar]
- 223.Kim S., Takayama S. Organ-on-a-chip and the kidney. Kidney Res. Clin. Pract. 2015;34:165–169. doi: 10.1016/j.krcp.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Materne E.-M., Maschmeyer I., Lorenz A.K., Horland R., Schimek K.M.S., Busek M., Sonntag F., Lauster R., Marx U. The Multi-organ Chip - a Microfluidic Platform for Long-term Multi-tissue Coculture. J. Vis. Exp. 2015 doi: 10.3791/52526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Edwards S., Kjellerup B.V. Exploring the applications of invertebrate host-pathogen models for in vivo biofilm infections. FEMS Immunol. Med. Microbiol. 2012;65:205–214. doi: 10.1111/j.1574-695X.2012.00975.x. [DOI] [PubMed] [Google Scholar]
- 226.Gabrilska R.A., Rumbaugh K.P. Biofilm models of polymicrobial infection. Future Microbiol. 2015;10:1997–2015. doi: 10.2217/fmb.15.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Murray J.L., Connell J.L., Stacy A., Turner K.H., Whiteley M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 2014;52:188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sibley C.D., Duan K., Fischer C., Parkins M.D., Storey D.G., Rabin H.R., Surette M.G. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008 doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jensen L.K., Johansen A.S.B., Jensen H.E. Porcine models of biofilm infections with focus on pathomorphology. Front. Microbiol. 2017;8:1961. doi: 10.3389/fmicb.2017.01961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Boase S., Jervis-Bardy J., Cleland E., Pant H., Tan L., Wormald P.J. Bacterial-induced epithelial damage promotes fungal biofilm formation in a sheep model of sinusitis. Int. Forum Allergy Rhinol. 2013;3:341–348. doi: 10.1002/alr.21138. [DOI] [PubMed] [Google Scholar]
- 231.Lorenz U., Schäfer T., Ohlsen K., Tiurbe G.C., Bühler C., Germer C.T., Kellersmann R. In vivo detection of staphylococcus aureus in biofilm on vascular prostheses using non-invasive biophotonic imaging. Eur. J. Vasc. Endovasc. Surg. 2011;41:68–75. doi: 10.1016/j.ejvs.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 232.Roche E.D., Renick P.J., Tetens S.P., Ramsay S.J., Daniels E.Q., Carson D.L. Increasing the presence of biofilm and healing delay in a porcine model of MRSA-infected wounds. Wound Repair Regen. 2012;20:537–543. doi: 10.1111/j.1524-475X.2012.00808.x. [DOI] [PubMed] [Google Scholar]
- 233.Pedersen S.S., Shand G.H., Hansen B.L., Hansen G.N. Induction of experimental chronic Pseudomonas aeruginosa lung infection with P. aeruginosa entrapped in alginate microspheres. APMIS. 1990;98:203–211. doi: 10.1111/j.1699-0463.1990.tb01023.x. [DOI] [PubMed] [Google Scholar]
- 234.Hsu L.C., Fang J., Borca-Tasciuc D.A., Worobo R.W., Moraru C.I. Effect of micro- and nanoscale topography on the adhesion of bacterial cells to solid surfaces. Appl. Environ. Microbiol. 2013;79:2703–2712. doi: 10.1128/AEM.03436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bhattacharjee A., Khan M., Kleiman M., Hochbaum A.I. Effects of growth surface topography on bacterial signaling in coculture biofilms. ACS Appl. Mater. Interfaces. 2017;9:18531–18539. doi: 10.1021/acsami.7b04223. [DOI] [PubMed] [Google Scholar]
- 236.Kim H.J., Boedicker J.Q., Choi J.W., Ismagilov R.F. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl. Acad. Sci. 2008 doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wolcott R.D., Rumbaugh K.P., James G., Schultz G., Phillips P., Yang Q., Watters C., Stewart P.S., Dowd S.E. Biofilm maturity studies indicate sharp debridement opens a time- dependent therapeutic window. J. Wound Care. 2010;19:320–328. doi: 10.12968/jowc.2010.19.8.77709. [DOI] [PubMed] [Google Scholar]
- 238.Carranza-Rosales P., Carranza-Torres I.E., Guzmán-Delgado N.E., Lozano-Garza G., Villarreal-Treviño L., Molina-Torres C., Vargas-Villarreal J., Vera-Cabrera L., Castro-Garza J. Modeling tuberculosis pathogenesis through ex vivo lung tissue infection. Tuberculosis. 2017;107:126–132. doi: 10.1016/j.tube.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Harrison F., Muruli A., Higgins S., Diggle S.P. Development of an ex vivo porcine lung model for studying growth Virulence, and signaling of pseudomonas aeruginosa. Infect. Immun. 2014;82:3312–3323. doi: 10.1128/IAI.01554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jäger J., Marwitz S., Tiefenau J., Rasch J., Shevchuk O., Kugler C., Goldmann T., Steinert M. Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect. Immun. 2014;82:275–285. doi: 10.1128/IAI.00703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ebsen M., Mogilevski G., Anhenn O., Maiworm V., Theegarten D., Schwarze J., Morgenroth K. Infection of murine precision cut lung slices (PCLS) with respiratory syncytial virus (RSV) and chlamydophila pneumoniae using the Krumdieck technique. Pathol. Res. Pract. 2002;198:747–753. doi: 10.1078/0344-0338-00331. [DOI] [PubMed] [Google Scholar]
- 242.Henjakovic M., Martin C., Hoymann H.G., Sewald K., Ressmeyer A.R., Dassow C., Pohlmann G., Krug N., Uhlig S., Braun A. Ex vivo lung function measurements in precision-cut lung slices (PCLS) from chemical allergen-sensitized mice represent a suitable alternative to in vivo studies. Toxicol. Sci. 2008;106:444–453. doi: 10.1093/toxsci/kfn178. [DOI] [PubMed] [Google Scholar]
- 243.Bennett R.D., Ysasi A.B., Belle J.M., Wagner W.L., Konerding M.A., Blainey P.C., Pyne S., Mentzer S.J. Laser microdissection of the alveolar duct enables single-cell genomic analysis. Front. Oncol. 2014;4:260. doi: 10.3389/fonc.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Maboni G., Davenport R., Sessford K., Baiker K., Jensen T.K., Blanchard A.M., Wattegedera S., Entrican G., Tötemeyer S. A novel 3D skin explant model to study anaerobic bacterial infection. Front. Cell. Infect. Microbiol. 2017;7:404. doi: 10.3389/fcimb.2017.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Nuutila K., Katayama S., Vuola J., Kankuri E. Human wound-healing research: issues and perspectives for studies using wide-scale Analytic platforms. Adv. Wound Care. 2014;3:264–271. doi: 10.1089/wound.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.James G.A., Swogger E., Wolcott R., deLancey Pulcini E., Secor P., Sestrich J., Costerton J.W., Stewart P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 247.Calderon M., Lawrence W.T., Banes A.J. Increased proliferation in keloid fibroblasts wounded in vitro. J. Surg. Res. 1996;61:343–347. doi: 10.1006/jsre.1996.0127. [DOI] [PubMed] [Google Scholar]
- 248.Edmondson R., Broglie J.J., Adcock A.F., Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Abbott R.D., Kaplan D.L. Strategies for improving the physiological relevance of human engineered tissues. Trends Biotechnol. 2015;33:401–407. doi: 10.1016/j.tibtech.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Schindler M., Nur-E-Kamal A., Ahmed I., Kamal J., Liu H.Y., Amor N., Ponery A.S., Crockett D.P., Grafe T.H., Chung H.Y., Weik T., Jones E., Meiners S. Living in three dimensions: 3D nanostructured environments for cell culture and regenerative medicine. Cell Biochem. Biophys. 2006;45:215–227. doi: 10.1385/CBB:45:2:215. [DOI] [PubMed] [Google Scholar]
- 251.Sullivan T.P., Eaglstein W.H., Davis S.C., Mertz P. The pig as a model for human wound healing. Wound Repair Regen. 2001;9:66–76. doi: 10.1046/j.1524-475X.2001.00066.x. [DOI] [PubMed] [Google Scholar]
- 252.Rubinchik E., Pasetka C. Ex vivo skin infection model. Methods Mol. Biol. 2010;618:359–369. doi: 10.1007/978-1-60761-594-1_22. [DOI] [PubMed] [Google Scholar]
- 253.Yang Q., Phillips P.L., Sampson E.M., Progulske-Fox A., Jin S., Antonelli P., Schultz G.S. Development of a novel ex vivo porcine skin explant model for the assessment of mature bacterial biofilms. Wound Repair Regen. 2013;21:704–714. doi: 10.1111/wrr.12074. [DOI] [PubMed] [Google Scholar]
- 254.Yang Q., Larose C., Della Porta A.C., Schultz G.S., Gibson D.J. A surfactant-based wound dressing can reduce bacterial biofilms in a porcine skin explant model. Int. Wound J. 2017;14:408–413. doi: 10.1111/iwj.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Phillips P.L., Yang Q., Davis S., Sampson E.M., Azeke J.I., Hamad A., Schultz G.S. Antimicrobial dressing efficacy against mature Pseudomonas aeruginosa biofilm on porcine skin explants. Int. Wound J. 2015;12:469–483. doi: 10.1111/iwj.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Bowler P.G., Duerden B.I., Armstrong D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Prunieras M., Regnier M., Woodley D. Methods for cultivation of keratinocytes with an air-liquid interface. J. Invest. Dermatol. 1983;81:S28–S33. doi: 10.1111/1523-1747.ep12540324. [DOI] [PubMed] [Google Scholar]
- 258.Hocke A.C., Becher A., Knepper J., Peter A., Holland G., Tönnies M., Bauer T.T., Schneider P., Neudecker J., Muth D., Wendtner C.M., Rückert J.C., Drosten C., Gruber A.D., Laue M., Suttorp N., Hippenstiel S., Wolff T. Emerging human middle east respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am. J. Respir. Crit. Care Med. 2013;188:882–886. doi: 10.1164/rccm.201305-0954LE. [DOI] [PubMed] [Google Scholar]
- 259.Wu W., Booth J.L., Duggan E.S., Patel K.B., Coggeshall K.M., Metcalf J.P. Human lung innate immune cytokine response to adenovirus type 7. J. Gen. Virol. 2010;91:1155–1163. doi: 10.1099/vir.0.017905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Weinheimer V.K., Becher A., Tönnies M., Holland G., Knepper J., Bauer T.T., Schneider P., Neudecker J., Rückert J.C., Szymanski K., Temmesfeld-Wollbrueck B., Gruber A.D., Bannert N., Suttorp N., Hippenstiel S., Wolff T., Hocke A.C. Influenza A viruses target type II pneumocytes in the human lung. J. Infect. Dis. 2012;206:1685–1694. doi: 10.1093/infdis/jis455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bansal D., Ave P., Kerneis S., Frileux P., Boché O., Baglin A.C., Dubost G., Leguern A.S., Prevost M.C., Bracha R., Mirelman D., Guillén N., Labruyère E. An ex-vivo human intestinal model to study Entamoeba histolytica pathogenesis. PLoS Negl. Trop. Dis. 2009;3:e551. doi: 10.1371/journal.pntd.0000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ajjampur S.S.R., Png C.W., Chia W.N., Zhang Y., Tan K.S.W. Ex vivo and in vivo mice models to study blastocystis spp. adhesion, colonization and pathology: closer to proving koch's postulates. PloS One. 2016;11:e0160458. doi: 10.1371/journal.pone.0160458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Dionne K.R., Leser J.S., Lorenzen K.A., Beckham J.D., Tyler K.L. A brain slice culture model of viral encephalitis reveals an innate CNS cytokine response profile and the therapeutic potential of caspase inhibition. Exp. Neurol. 2011;228:222–231. doi: 10.1016/j.expneurol.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Alhede M., Kragh K.N., Qvortrup K., Allesen-Holm M., van Gennip M., Christensen L.D., Jensen P.Ø., Nielsen A.K., Parsek M., Wozniak D., Molin S., Tolker-Nielsen T., Høiby N., Givskov M., Bjarnsholt T. Phenotypes of non-attached pseudomonas aeruginosa aggregates resemble surface attached biofilm. PloS One. 2011;6 doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Fang Y., Eglen R.M. Three-dimensional cell cultures in drug discovery and development. SLAS Discov. 2017;22:456–472. doi: 10.1177/1087057117696795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sato T., Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science (80-. ) 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 267.Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kim H.U., Sohn S.B., Lee S.Y. Metabolic network modeling and simulation for drug targeting and discovery. Biotechnol. J. 2012;7:330–342. doi: 10.1002/biot.201100159. [DOI] [PubMed] [Google Scholar]
- 269.Ilie O., van Loosdrecht M.C.M., Picioreanu C. Mathematical modelling of tooth demineralisation and pH profiles in dental plaque. J. Theor. Biol. 2012;309:159–175. doi: 10.1016/j.jtbi.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 270.Avci H., Do ˘ Gan F., Uzel G.¨, Erol S., Akpek A. 2017. Recent Advances in Organ-on-a-chip Technologies and Future Challenges: a Review. [DOI] [Google Scholar]
- 271.Hol F.J.H., Dekker C. Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science. 2014;80–:346. doi: 10.1126/science.1251821. [DOI] [PubMed] [Google Scholar]
- 272.Trevisan M., Sinigaglia A., Desole G., Berto A., Pacenti M., Palù G., Barzon L. Modeling viral infectious diseases and development of antiviral therapies using human induced pluripotent stem cell-derived systems. Viruses. 2015;7:3835–3856. doi: 10.3390/v7072800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.