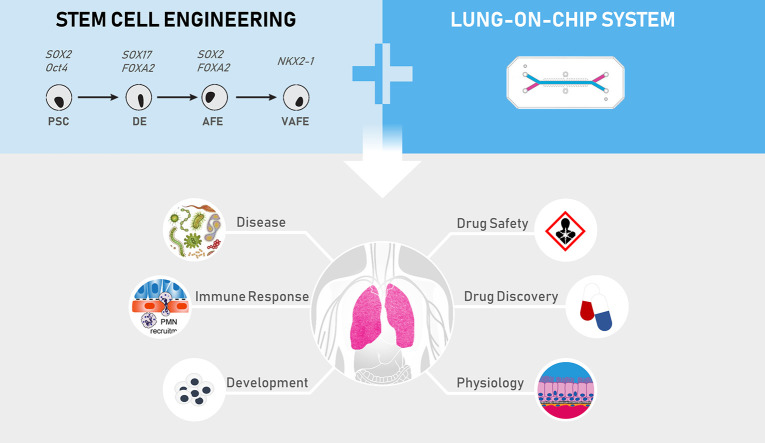
Abstract
Pathologies of the respiratory system such as lung infections, chronic inflammatory lung diseases, and lung cancer are among the leading causes of morbidity and mortality, killing one in six people worldwide. Development of more effective treatments is hindered by the lack of preclinical models of the human lung that can capture the disease complexity, highly heterogeneous disease phenotypes, and pharmacokinetics and pharmacodynamics observed in patients. The merger of two novel technologies, Organs-on-Chips and human stem cell engineering, has the potential to deliver such urgently needed models. Organs-on-Chips, which are microengineered bioinspired tissue systems, recapitulate the mechanochemical environment and physiological functions of human organs while concurrent advances in generating and differentiating human stem cells promise a renewable supply of patient-specific cells for personalized and precision medicine. Here, we discuss the challenges of modeling human lung pathophysiology in vitro, evaluate past and current models including Organs-on-Chips, review the current status of lung tissue modeling using human pluripotent stem cells, explore in depth how stem-cell based Lung-on-Chips may advance disease modeling and drug testing, and summarize practical consideration for the design of Lung-on-Chips for academic and industry applications.
Keywords: Organs-on-Chips, Lung-on-Chips, Lung Chip, Stem cells, Precision medicine, Tissue engineering, Respiratory drug testing, Respiratory disease modeling, Drug discovery and development
Graphical abstract
1. Introduction
The lungs constitute the largest tissue interface between the human body and its environment. Exposed to an average of seven liters of inhaled air per minute, lungs are in direct contact with countless noxious particles, viral or bacterial pathogens, hazardous chemicals or toxic gases. As formulated by Green et al., “Each day a surface as large as a tennis court is exposed to a volume of air and contaminants that would fill a swimming pool” [1]. Not surprisingly the lungs are susceptible to numerous deadly acute and chronic conditions that constitute an immense global health burden. Indeed, respiratory diseases are directly responsible for one in six deaths worldwide [2], and chronic respiratory conditions affect more than one billion people worldwide [3]. In Europe alone, the total cost of respiratory disease amounts to more than €380 billion annually [4]. Among all respiratory diseases, five account for the majority of morbidity and mortality worldwide: chronic obstructive pulmonary disease (COPD), asthma, acute respiratory infections, tuberculosis, and lung cancer [5]. Recent reports suggest that onset or aggravation of respiratory disease caused by in- and outdoor air pollution, is responsible for 6.5 million deaths annually, a number likely to increase even further in the future [6]. It is therefore imperative to develop new therapeutic strategies for lung diseases, but this requires new tools to better study respiratory diseases and understand the underlying mechanisms.
Studying lung disease pathogenesis and drug efficacy, as well as inhalation toxicology, necessitate physiologically relevant models of human lung tissue. While animals, especially rodents, have provided seminal insight into lung physiology and pathophysiology, they are limited in recapitulating the development, structure, disease symptoms, and responses of the human respiratory system, providing a strong rationale for developing and investigating human in vitro lung models for disease modeling, drug discovery, and drug testing [7,8]. For instance, the timing of lung developmental events differs markedly between mice and humans. The prenatal saccular stage, in which alveolar sacs with distinguishable alveolar cell types form and surfactant secretion begins, takes place relatively earlier, and postnatal differentiation of immature saccules continues for a relatively longer time in humans compared to mice. This different pace of lung development results in a greater degree of branching and complexity of human distal lung structures including respiratory bronchioles, alveolar ducts and associated alveoli [9]. Cellular composition also differs between mouse and human lung. For example, in the mouse airways, mucus-producing goblet cells are rare and secretory club cells (formerly known as Clara cells) are abundant, whereas the opposite applies to human airways [10]. Further, many gene mutations induce different, if any, respiratory symptoms in mice compared to humans [11]. While rodents remain the main animal model for pre-clinical studies, other non-rodent species such as guinea pigs, dogs, sheep, pigs and non-human primates which more closely mimic human lung physiology, are also used in preclinical studies. However, ethical and financial issues as well as non-availability of species-specific reagents often preclude their use for routine experimentation. Taken together, these limitations demonstrate that animals are imperfect models for a range of human lung diseases and their drug treatment, necessitating the need for human-specific preclinical models of the lung.
Standard 2D culture of cancer or immortalized cell lines still represents the most common alternative to animal models for the study of tissue pathophysiology and response to pharmacological agents. The great advantage of such cell lines, the ease of use for high-throughput experiments, is clouded by their limited physiological relevance and clinical predictivity. In the past 10 years, advances in tissue engineering and soft lithography techniques have converged to give rise to Organs-on-Chips, miniaturized microengineered cell culture systems that recreate key functional and micro-environmental features of human organs in vitro [12]. Importantly, the idea is not to rebuild an entire human lung with its intricate architecture as so far this remains technically not feasible and would also greatly complicate the experimental manipulation, analysis, and interpretation of the engineered system. Rather, the promise and great benefit of Organs-on-Chips lies in their ability to recreate well-defined functional units of the lung, such as the alveolar epithelium-blood capillary interface, or the mucociliary barrier of the airways. Each specific Lung-on-Chip model can then be used to isolate, amplify, and systematically combine specific cellular and acellular components of the tissue and dissect their interaction as well as individual roles in health and disease processes.
Concurrent with the advances in Organs-on-Chips technology, the field of developmental biology has made tremendous progress towards efficient culture and differentiation of stem cell-derived human lung tissue in the form of static 2D cultures or 3D organoids [7,13]. While stem cell technology enables precise modeling of virtually any human tissue, and long-term cultures of patient-derived cells, Organs-on-Chips provide the cell microenvironment, biomechanical forces, vascular perfusion and circulation of immune cells, tissue relevant cyto-architecture, and sampling capabilities that organoids lack. It is therefore possible that combining both technologies will help to study human lung development and pathophysiology, responses to inhaled toxicants, evaluate drugs pharmacodynamics and pharmacokinetics (PK/PD), and discover new diagnostics and therapeutics.
The purpose of this review is to provide a comprehensive survey of existing state-of-the-art Organs-on-Chip systems that model human lung tissue and envision how this innovative technology can converge with the field of lung stem cells to establish highly relevant models of lung development, respiratory diseases and drug PK/PD.
2. Modeling the human lung
The difficulty of in vitro tissue modeling lies in the challenge to identify and then faithfully recapitulate the essential structural and functional elements of human tissue that govern healthy and pathological organ responses. Especially in the case of complex organs such as the human lungs, the challenge is increased by the incomplete understanding of the organ's morphology and physiology as exemplified by the unclear role that several of the 40 different resident cells play in lung homeostasis [14]. While the lungs exert an essential but seemingly simple vital function by providing a constant supply of oxygen and removal of CO2 through gas exchange between the inhaled air and circulating blood, lung morphology is complex and consists of distinct units with specific physiological roles.
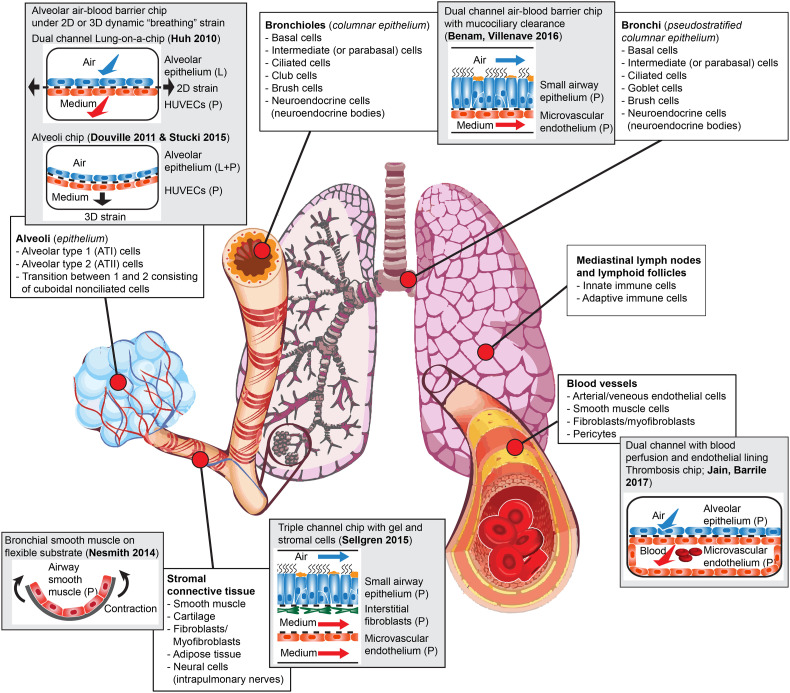
These units include regional sections that can be defined by their respective cellular composition and function, such as the trachea, bronchi, bronchioles or small airways, alveoli or air sacs, stromal connective tissue, blood vessels, and hematopoietic and lymphoid tissue, and functional units such as the epithelial mesenchymal trophic unit (EMTU) [15] or the alveolar-capillary interface (Fig. 1 ). Together, these units build the organ's architecture and enable the process of gas exchange. Specifically, inhaled air enters the respiratory tract through the nasal and oral passages and flows through the conducting airways until it reaches the distal airways and alveoli. The conducting airways are composed of the trachea that divides into bronchi and bronchioles. The bronchioles lead to the alveoli that constitute most of the lung surface area and enable gas exchange with the circulating blood. The conducting airways are lined with highly specialized cells including ciliated cells, mucus producing cells called goblet cells, and club cells, while underlying basal cells act as progenitors. The relative proportion of these cells depends on their location in the respiratory tract [14,16]. For instance, while club cells are almost nonexistent in the upper airways lining, they constitute a large part of the terminal and respiratory bronchioles cell population [16]. Similarly, goblet cells account for 10 to 15% of epithelial cells in large airways but are almost absent from terminal and respiratory bronchioles [16]. This characteristic cell distribution can be altered during chronic diseases. For example, increased goblet cells number in the airway lining has been observed in patients with obstructive respiratory diseases such as asthma and COPD [17,18].
Fig. 1.
Cell population diversity and location in human lungs and corresponding on-chip models. For each on-chip model, cell origin is indicated (L = human cell line; P = human-derived primary cells).
The distal, respiratory part of the lungs contains an average of 450 million pulmonary alveoli constituted by thin, flat, non-dividing alveolar type 1 (ATI) cells that enable gas exchange, and small, cuboidal alveolar type 2 (ATII) cells that secrete pulmonary surfactant and can divide and differentiate to replace damaged alveolar cells [19]. In addition, the human lungs are home to numerous resident and circulating immune cells including macrophages, innate lymphoid cells, mast cells, lymphocytes, eosinophils and neutrophils that contribute to the protection from pathogens and noxious particles continuously entering the lungs [20]. Further, the submucosa surrounding the conducting and respiratory airway epithelium includes airway smooth muscle cells, fibroblasts, myofibroblasts and neural cells that play major roles in health and disease [21,22]. Sub-mucosal cells also play critical roles in many respiratory diseases. For instance, secretion of pro-inflammatory mediators and hyper-responsiveness to external stimuli by airway smooth muscle cells contribute to tissue inflammation and lead to bronchoconstriction and airflow obstruction in asthma [23]. Similarly, uncontrolled activation of lung fibroblasts plays a central role in the development of pulmonary fibrosis [24]. The submucosa also harbors a network of blood and lymphatic vessels comprised of endothelial cells and pericytes that help recruit circulating immune cells during the inflammatory response. Hence, the human lung is a complex tissue comprised of numerous populations of cells that interact and depend on each other to grow and function properly [25].
The high diversity in cell types, numbers, function and tissue morphology among the different lung regions and disease states is an essential parameter to consider when designing an in vitro model of the human lungs and precludes a one-model-fits-all approach. Instead, the appropriate model will depend on the experimental questions one wishes to answer, with the goal of recapitulating the key morphological and functional features of the organ unit. Specifically, this might require the spatiotemporal interaction of different cell types involved in the function or pathophysiology of interest. An effective strategy is to initially create a simple, well characterized base model and increase complexity by adding cellular components one by one, in order to reveal their individual contributions.
2.1. Current in vitro models of the human lung
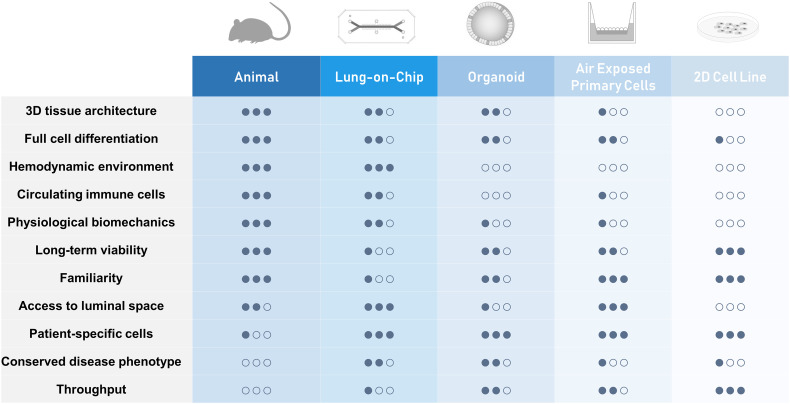
Given the technical and ethical difficulties of using real human organs to carry out research, scientists have largely relied on simpler and smaller surrogate models to gain insight into human physiology and pathophysiology. Modeling human diseases in general, and lung diseases in particular, largely depends on immortalized cell lines or primary cells combining one or more cell types, cultured in a static, 2D environment, and on animal models (Fig. 2 ). However, while cell monolayers are easy and cheap to use and amenable to high throughput studies, they cannot replicate the functions of many tissue-specific, differentiated cell types or faithfully predict in vivo tissue functions and drug effects observed in the native 3-dimensional human organ [26]. On the other hand, although animal models provide access to 3D native tissues and are indeed used for modeling human diseases and assessing efficacy of therapeutics in a number of tissues and organs, they are often poor predictors of clinical success due to species-species differences in mechanisms of action or toxicities of drugs [27]. For instance, failure was reported for 18 out of 23 Phase II/III clinical trials testing 17 distinct therapeutic anticancer vaccines [28] due to elevated levels of circulating immunosuppressive cytokines and various immunological checkpoints in humans that may not be present in rodents [29]. Lung specific studies equally suffer from these limitations and over-reliance on animals to model complex human respiratory diseases such as asthma have contributed to the lack of new efficacious treatment despite huge research efforts, as animal models have a long record of failing to predict clinical efficacy of novel therapies in human [30]. This is also true for several other respiratory diseases, including cystic fibrosis. The DeltaF508 mutation of the cystic fibrosis transmembrane conductance regulator (CFTR) is the most common genetic cause of human cystic fibrosis; however, mice with the DeltaF508 mutation do not have any respiratory abnormalities [31]. Also, investigations of human respiratory infections are hindered by the lack of human pathogen-specific receptor in animals. For instance, a majority of rhinovirus types uses human intercellular adhesion molecule 1 (ICAM-1) as an entry receptor but cannot bind the murine form, precluding infection of mouse airway cells [32], although transgenic mouse models expressing human ICAM-1 have recently been developed [33]. Animal models are often also not permissive for replication of human viruses [34,35]. The utility of animals to study human chronic diseases such as asthma and develop therapeutics is also controversial as most animal models do not naturally exhibit the asthma disease phenotypes observed in humans [36].
Fig. 2.
Comparison of experimental strategies for lung modeling.
The limited clinical predictive value of traditional cell culture and animal models have motivated the development of more complex human in vitro 2D or 3D models that incorporate multiple types of differentiated cells or involve cell patterning in order to be more representative of the morphology and function of human organs. Three dimensional cell culture usually relies on bioinspired scaffolds made of extracellular matrix (ECM) or synthetic polymers as well as specific growth factors or co-cultured mesenchymal cells which induce cells to polarize and differentiate into cellular structures resembling in vivo tissue morphology and function. When grown in ECM hydrogels, cells can self-assemble into 3D cellular clusters known as spheroids or organoids that contain multiple cells types resembling the organ tissue architecture and recapitulating some specific organ function [37]. Lung organoids derived from human primary lung tissues, embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) are also referred to as tracheospheres, bronchospheres, and pneumospheres (or alveolospheres), and have been successfully employed to study lung development and pathophysiology [19,[38], [39], [40]]. Nonetheless, they also have limitations [41]; organoids lack the native organ microenvironment and appropriate cyto-architecture that is often essential in disease development, tissue growth and repair. Another drawback of 3D organoid cultures is that analysis or stimulation of enclosed cells, particularly to measure transmembrane transporter activity, evaluate the transport and metabolism of drugs across the cell layer, or to quantify the release of inflammatory mediators or newly formed virions, is often precluded by the difficulty of accessing and sampling luminal contents. Also, exposing the organoids to uniform drug concentrations may be hampered by the slow diffusion and interference with the ECM hydrogel, or the technical difficulty of micro-injection into the luminal space. In addition, organoids usually lack tissue-tissue interfaces, such as the interface between vascular endothelium and surrounding stroma and parenchymal cells, which are essential for the morphology and function of virtually all organs. Finally, exposing epithelia to airborne substances to mimic environmental exposures or inhaled drug delivery is not readily feasible in organoid cultures. While it is possible to break up organoids and thereby expose their apical side to airborne substances, the mechanical forces used in the process, e.g. rapid pipetting and sudden changes in the media composition, may cause cellular stress and therefore alter responses. 3D models of the human airway epithelium based on air liquid interface cultures of primary epithelial cells in transwells inserts have also been shown to closely mimic the gene expression signature of the human airway epithelium in vivo [42,43] and to recapitulate several hallmarks of human airway epithelium in vivo, leading to seminal insights into numerous biological processes in lung development and pathophysiology [[44], [45], [46], [47], [48], [49]]. However, as for organoids, these models typically lack physiological fluid and solid mechanical cues, including shear flow and tensile and compressive stresses that cells experience in vivo. The absence of fluid flow also prevents perfusion of cultured cells with circulating blood and immune cells and limits modeling of inflammatory responses to infection or drugs. Though relatively novel and not yet as familiar to investigators, the emergent field of Organs-on-Chips promises to overcome the limitations of traditional in vitro model systems by offering microengineered features such as dynamic stresses and microscale flows. Thus, combining the best attributes of stem cell assays and Organs-on-Chips technologies might enable the design of a more powerful in vitro model of the human lung.
3. Organs-on-Chips technology
Recent advances in microfabrication and tissue engineering have made it possible to create Organs-on-Chips – i.e., continuously perfused microchannels lined by living human cells. This design can reconstitute key functional and microenvironmental features of whole organs including tissue-tissue interfaces, mechanical forces, fluid flow, and biochemical gradients [[50], [51], [52]]. Organs-on-Chips have been shown to reproduce complex integrated organ-level responses to pathogens and inflammatory cytokines, as well as nanoparticles and pharmaceuticals; they can also effectively recapitulate disease states and complex pathophysiological responses. Mechanically active Organs-on-Chips populated with human cells therefore promise to provide more predictive models and provide lower-cost alternatives to animal and clinical studies for disease modeling, drug testing and safety applications [26]. As discussed in the following, Organs-on-Chips aim to recreate the complex, dynamic state in which living cells function within the native human organ, including interactions with the substrate (extracellular matrix), tissue-tissue interface, mechanical forces, immune cells and blood components, and biochemical microenvironment, which are critical in lung health and disease.
3.1. Extracellular matrix
The ECM is the non-cellular component of a tissue that provides the structural scaffolding and biochemical and biomechanical support to the surrounding cells. In the lung and other organs, the ECM gives the tissue its physical and mechanical properties, contributes to tissue development, morphology and function, and influences cell shape and cell-cell interactions [53]. In the lungs, the ECM influences fundamental processes including cell signaling pathways [54], cell shape and function [55], cytoskeletal organization and differentiation [56], organogenesis [57], and wound healing [58]. The ECM composition is not only specific to each organ but is also unique to the different regions of a tissue. In the lungs, which is a relatively soft tissue with an elastic modulus ranging between 1 and 5 kPa [59], alveolar ECM is composed of a mix of collagen III, IV, V, laminin, fibronectin and elastin [60] whereas the ECM of the large airways includes collagens I, II (cartilage), V, laminin and fibronectin [61]. Further, extensive ECM remodeling has been implicated in many physiological and pathophysiological processes such as wound healing and pulmonary fibrosis [62]. Therefore, the ECM composition used in a model of the lungs must be carefully selected and will depend on the region and the disease state one seeks to model.
3.2. Cell-cell interactions
While lung ECM scaffolding modulates cellular function, shapes tissue architecture and precisely compartmentalizes lung cell populations, cell behavior is also determined by the constant interaction between neighboring cellular compartments. Interactions can be mediated through direct cell-to-cell contact, for example, between airway epithelial cells and fibroblasts during fibrosis [63] and between alveolar macrophages and alveolar epithelial cells in health and disease [64]. In addition, cellular interactions can also leverage soluble factors, such as in epithelial-endothelial cross talk during influenza virus-induced cytokine storms [65], endothelial influence on epithelial differentiation [66], and mesenchymal cell influence on epithelial development [15]. Such cell-to-cell cross talk can modulate tissue growth, differentiation, and cell activation mediating the recruitment of immune cells during inflammation. Such multi cell type interactions can also be recapitulated in Organs-on-Chips; for instance, co-culture of differentiated human primary airway epithelial cells and endothelium in close proximity results in cross-communication between both tissues following treatment with a pro-inflammatory stimulus [52]. Similarly, stimulation of the epithelium with a pro-inflammatory agent results in activation of the underlying endothelium, as indicated by overexpression of adhesion molecules such as ICAM-1, VCAM-1 and E-Selectin [52,67].
3.3. Mechanical forces
Despite extensive studies, the magnitude and nature of the mechanical forces experienced by the lung epithelial cells during development, breathing movements, and disease states are still not well understood and remain the object of intense research [68]. It is admitted that during normal breathing movements, lungs undergo dynamic deformation estimated to cause 4% stretch distension of the basement membrane [69,70], while deep inspiration may induce a 12% stretch distension [71]. During bronchoconstriction in e.g. asthma patients, compressive forces act on parts of the airway wall [72]. These mechanical forces strongly influence lung cells, including effects on growth and repair, surfactant release, injury, inflammation [[72], [73], [74], [75]], as well as tissue development from fetal to adult stage [73,76]. Additional mechanical forces that influence cell function and development include shear stresses induced by the bidirectional air flow in the lumen of the conducting airways and the blood flow in the capillaries. Endothelial cells are able to sense and adapt to variation in the blood-flow and the vascular wall serves as an interface between the blood and tissue and can respond to hemodynamic cues. The physiological shear stresses acting on the vascular wall have been show to modulate gene expression, cell morphology, and cell metabolism [[77], [78], [79]] of endothelial cells. Organs-on-Chips offer the possibility to apply and control physiological biomechanical forces, including breathing movements [67,80] and shear stresses resulting from air and blood flow [81,82] that cells experience in vivo.
3.4. Biochemical microenvironment
The biochemical surroundings of cells are composed of elements that are secreted or transported through the tissue. These include growth factors, hormones, dissolved gases, and small molecules such as salts and nutrients. In the lungs, biochemical mediators are central to processes ranging from tissue development and homeostasis [83], to inflammation and injury resolution [84]. Lung cells also play a significant role in the metabolism of xenobiotics and endogenous hormones such as serotonin, leading to degradation as well as activation of important biologic properties [85]. Xenobiotic-metabolizing cytochrome P450 enzymes expressed in bronchial and bronchiolar epithelium, club cells, type II alveolar cells, and alveolar macrophages in human lung activate environmental chemicals, modifying cell biochemical microenvironment and contributing to pathologies such as cancer and COPD [86]. Expression and activity of such enzymes is in part determined by epithelial differentiation, indicating the need for appropriate cellular differentiation in models [87]. Organs-on-Chips offer the possibility to accurately control the regional and temporal biochemical microenvironment of cells through controlled perfusion of growth medium and gases.
3.5. Immune cells and blood components
Circulating and resident immune cells are key effectors of inflammation and play a central role in the pathogenesis and resolution of respiratory diseases. Together with airway epithelial cells, lung resident and circulating immune cells, such as dendritic cells and macrophages initiate and orchestrate immune responses [88]. The ability to perfuse fluids through Organs-on-Chips not only allows dynamic supply of nutrients and gases, but also enables perfusion of immune cells under physiological conditions, such as shear stress experienced within microvessels [67,89]. This unique feature enables visualization and real time monitoring of the interaction between freshly isolated, circulating immune cells with lung endothelial and epithelial cells [52,67]. Whole human blood can also be perfused through Organs-on-Chips and, for example, thrombotic events have been modeled and utilized to assess drug delivery, toxicity, and efficacy [82,90].
4. A brief history of Organs-on-Chips
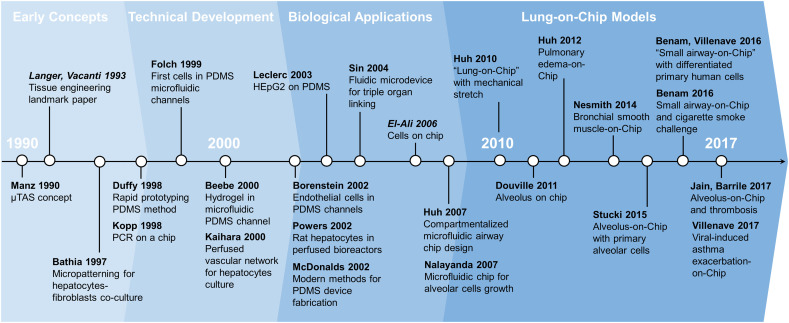
The field of Organs-on-Chips derives from “miniaturized total chemical analysis systems” or μTAS, microscale chemical platforms directly inspired from the development and miniaturization efforts of the electronic industry in the second half of the twentieth century [91,92] (Fig. 3 ). These μTAS, later regrouped under the name “lab-on-a-chip” systems, integrate fluidic microsystems into a single platform to perform several steps of a chemical assay [93,94]. Originally, these systems did not contain any living components. Adding living cells was facilitated when new methods to fabricate microscale fluidic channels were established as an alternative to fused silica capillaries [95]. Specifically, Organs-on-Chips emerged from the convergence of cellular micro-patterning methods designed to control cell shape and function with early miniaturized systems for electrophoresis [[96], [97], [98], [99]].
Fig. 3.
Timeline highlighting key studies of Organs-on-Chips technology development. Review articles are italicized.
Micro-engineered cellular systems were initially called “cells on chip” to illustrate the merger of cell biology with microfabrication methods adapted from the computer microchip industry [50,51,100]. The more recent denomination “Organs-on-Chips” implies the modeling of complex physiological organ-level function in microfabricated biochips. However, in place of silicon Organs-on-Chips are typically made from hydrogels [101] or a silicone elastomer called poly(dimethylsiloxane) (PDMS) which has played a central role in Organs-on-Chips sudden popularity [95,102]. PDMS offers numerous mechanical and chemical advantages over traditional micro-engineering materials such as glass and silicon [103]. First, PDMS is relatively inexpensive compared to silicon, allowing cheap prototyping. PDMS stiffness can also be easily modified by controlling the degree of cross-linking between the polymer chains, enabling the design of soft and stretchable surfaces similar to the mechanical environment of cells. Moreover, since PDMS forms a tight seal with glass and can be reversibly or irreversibly bound to plastic polymers, hybrid devices containing rigid parts can be constructed. Importantly, PDMS is non-toxic and easy to work with, and rapid prototyping methods involving soft lithography and replica molding permit the creation of inexpensive devices with complex flow channel designs [102,104]. Finally, PDMS is gas permeable, biocompatible and optically clear which makes it particularly well suited for growing living cells in enclosed fluidic microchannels and monitoring their behavior using various types of light microscopy.
Faced with the promises and challenges of tissue engineering, notably the need for cellular scaffolds and blood perfusion of engineered in vitro tissue, researchers in the mid-nineties and early 2000s started to adapt microfabrication approaches to culture human cells and engineer human tissues [[98], [99], [100],105,106]. Early attempts at cellular micro-patterning used PDMS microchannels sealed against a tissue-culture dish to support the alignment, perfusion, and growth of 3T3-J2 fibroblasts [97]. Later, microfluidic designs were modeled to incorporate blood capillary-like channels to support the culture of human endothelial cells [107,108], quickly followed by other cell types, including liver [[109], [110], [111], [112]], muscles [113], bones [114], brain [115], gut [116], and kidney [117,118]. In parallel, inter-connected culture chambers containing cells derived from different organs and later called “body-on-chip” systems were developed and applied for pharmacological studies [119,120].
Hence, Organs-on-Chips were originally developed to solve perfusion and scaffolding challenges encountered in the field of tissue engineering. However, their potential to recapitulate complex human organ-level functions became rapidly evident and led to an array of advanced models of human lung tissue, such as the “lung-on-a-chip”, “alveolus-on-a-chip” and “small airway-on-a-chip” [50,52,80,82,[121], [122], [123]]. Applications include the study of lung cells growth and injury [122,123], modeling of alveolar tissue-tissue interaction and inflammatory processes [50], responses of the alveolar epithelium to drugs, mechanical stresses [81,121], and pulmonary thrombotic events [82], as well as lung cancer [124]. Most recently, a functional dynamic model of human airways comprising well-differentiated airway epithelial cells at air-liquid interface has been developed. Importantly, the airway epithelium is supported by a porous membrane and interacts with a continuously perfused pulmonary microvascular endothelium underneath that experiences physiological shear stress allowing circulation of immune cells. This model has been leveraged to model human obstructive respiratory diseases and human viral infection, and test novel therapeutics [52,125].
5. Modeling human lung alveolus pathophysiology on-chip
Several attempts have been made to develop and characterize a functional alveolus unit on-Chip [67,80,82,[121], [122], [123], [124]]. Most of these attempts sought to recapitulate the alveolar-capillary interface by recreating the boundary between the lung's air sacs and its capillaries within a microengineered system. The majority of the systems reviewed here rely on a similar chip design: a microengineered chamber divided longitudinally into two parallel channels by a flexible, ECM-coated porous membrane that recreates the alveolar interstitium. Such designs support the growth of human alveolar cell lines [67,81,121] or primary [80,82,122,124] epithelial cells at an air-liquid interface, while human pulmonary microvascular endothelial cells (HMVECs) or umbilical vein endothelial cells (HUVECs) line the opposite side of the same membrane and are exposed to the laminar flow of culture medium. The use of elastic materials for membrane and chip housing enables linear or 3D cyclic stretch that recreates motions of a breathing lung and influences cell behavior [[73], [74], [75]].
These systems have been applied to replicate diseases of the lungs, investigate mechanical stress and cell damage, explore immune cells recruitment and extravasation, and test drug efficacy and toxicity. The first alveolus-on-a-chip, originally called “Lung-on-a-chip” has been used to recapitulate complex physiological mechanisms such as diapedesis of circulating human primary neutrophils following stimulation of the alveolar epithelium with TNF-α or infection with a strain of E. coli [67]. In this disease model of bacterial infection of the lung, the endothelium situated underneath the alveolar epithelium becomes activated, as indicated by a rapid increased expression of the adhesion molecule ICAM-1, and promotes adhesion and extravasation of perfused neutrophils. As Organs-on-Chips are made of PDMS, a transparent polymer, the whole physiological process can be observed by real time, high resolution microscopy [50]. Similarly, the “Lung-on-a-chip” recapitulated silica nanoparticle-induced toxicity and uncovered the critical role of breathing motions in production of reactive oxygen species (ROS) as well as the cellular uptake of nanoparticles and their transport across both cell layers into the vascular channel [50]. Another application of the original “Lung-on-a-chip” includes replication of human pulmonary edema through IL-2-induced lung toxicity [81]. IL-2 was shown to induce limited pulmonary vascular leakage into the air channel under static conditions whereas physiological breathing motions significantly increased vascular permeability, a response which could be attenuated pharmacologically by a TRPV4 inhibitor [81].
Another model of the lung alveolar epithelium on-chip has been applied to investigate the effects of mechanical strain and surface tension (propagation of the air–liquid interface) on cell death in A549 cells and murine type II alveolar epithelial cells [121]. Whereas the first Lung-on-Chip recapitulates a linear stretch, this alternative design contains a circular stretchable diaphragm whose downward deflection enables a 3-dimensional stretch of its tissue lining. More recently, efforts have been made to increase the chip's physiological relevance by using human primary alveolar epithelial cells and endothelial cells instead of cell lines [80,90,124]. Stucki et al. designed another version of an alveolus-on-a-chip that also supports a 3D cyclic deformation of human primary alveolar epithelial cells [80]. This study found that, compared to static conditions, cyclic stretch affects barrier permeability and increases the metabolic activity of primary alveolar epithelial cells and the release of inflammatory markers. A complex model of intravascular thrombosis utilizing human primary alveolar epithelial and human umbilical vascular endothelial cells has been also developed to assess antithrombic therapeutics [82]. In this model, all sides of the vascular channel are coated with ECM and seeded with endothelial cells to prevent clotting of the perfused human whole blood. The study shows that treatment of the alveolar lumen of the epithelium-endothelium co-culture with lipopolysaccharide (LPS) found in the outer membrane of Gram-negative bacteria, triggered thrombotic events. Conversely, LPS stimulation of endothelial cells alone did not lead to blood clot formation, thus highlighting the importance of recapitulating the tissue-tissue interface in a model of inflammation-mediated thrombosis. A model of lung cancer has also been developed in an alveolus-on-a-chip [124]. The model features an epithelium-endothelium interface with a small proportion of non-small cell lung cancer tumor cells in the alveolar space and was used to investigate the influence of the bio-mechanical microenvironment on tumor cell growth and migration. The study shows that cyclic stretch limits tumor cell invasion of the vascular compartment but also reduces the efficacy of the widely used class of cancer drugs, Tyrosine Kinase inhibitors, suggesting that local microenvironment cues may influence cancer cell growth and drug efficacy.
6. Modeling human airways pathophysiology on-chip
Based on similar designs and microfabrication techniques used for the compartmentalized microfluidic airway system [122] and later on for the alveolus-on-chip [67], several on-chip assays modeling various features of the human airways in health and disease have recently been developed [52,[126], [127], [128]]. For instance, the new “small airway-on-a-chip” recapitulates the human bronchial and bronchiolar epithelium by supporting the full differentiation of a columnar, pseudostratified, mucociliary, bronchiolar epithelium composed of human primary airway epithelial cells isolated from normal or diseased cells from patients. As in the alveolus model, the epithelium is underlined by a functional human pulmonary microvascular endothelium experiencing continuous fluid flow [52]. Human airway cells cultured on-chip at air liquid interface for three weeks reconstitute an in vivo-like epithelium composed of ciliated cells with physiological cilia beating frequency as well as goblet cells secreting mucus into the lumen resulting in robust mucociliary clearance [52]. Interestingly, the proportions of ciliated, goblet and basal cells inside the mature small-airway-on-a-chip are strikingly similar to those found in human lungs, suggesting that the reconstituted tissue closely recapitulates the morphology and functions of the human airway epithelium (Table 1 ). Perfusion of IL-13 through the vascular channel to recreate a microenvironment enriched with inflammatory Type 2 T helper (Th2) cells as observed in allergic asthma resulted in significant airway remodeling with goblet cells hyperplasia, increase of pro-inflammatory cytokines, and reduction of cilia beating frequency. This phenotype was suppressed following incubation with Tofacitinib, a Janus kinase (JAK) inhibitor that blocks cytokine signaling, which is a therapeutic prescribed for rheumatoid arthritis [52]. Using primary airway cells derived from COPD patients, the small-airway-on-a-chip was also leveraged to investigate exacerbations in COPD, and to measure human neutrophil recruitment to the activated endothelium following epithelial exposure to pro-inflammatory stimuli. In addition, because the small-airway-on-a-chip has a separate air channel, it is possible to circulate air-borne pollutants through the epithelial chamber and evaluate, for example, the response of epithelial cells to cigarette smoke [126]. Recent advances in airway modeling also include a new on-chip model of viral-induced asthma exacerbations [125]. Derived from the original small airway-on-a-chip, this new chip supports infection of fully differentiated human airway epithelial cells with a human rhinovirus (HRV) to high titers and enables immune cell diapedesis through a 3 μm pore membrane. Stimulation with IL-13 altered HRV-induced pro-inflammatory and interferon responses and increased neutrophil recruitment to the vascular walls. This response was pharmacologically inhibited by a CXCR2 antagonist.
Table 1.
Comparison of structure and function between a human airway epithelium in vivo and the human Airway-Chip. Values for human airways were reported in [16,47,129,130]; values from the Airway-Chip were reported in [52,125].
| Parameters | Human airway | Airway chip (SD) |
|---|---|---|
| Mucociliary velocity | 40–150 μm/s | 103.5 μm/s (±46.1) |
| Cilia beating frequency (Hz) | 9–20 Hz | 16.35 (±2.6) |
| Ciliated cells (%) | ~30 | 29.3 (±1.9) |
| Goblet cells (%) | ~10–15 | 18.4 (±1.2) |
| Basal cells (%) | ~6–30 | 10.4 (±3.8) |
Alternative on-chip systems of the human airway epithelium include a triple channel chip design where an additional compartment containing fibroblasts is inserted between the epithelial and vascular channels [128,131]. This configuration, however, so far does not support full differentiation of the human primary airway epithelial cells but remains promising for investigating cross communication between the three different cell populations and their respective influence on each other's growth and differentiation. In addition, including fibroblasts may be useful for studying diseases such as idiopathic pulmonary fibrosis. Using a markedly different design approach, Nesmith et al. built a human airway musculature-on-a-chip consisting of ECM cantilevers actuated by primary human bronchial smooth muscle cells [127]. When stimulated with IL-13, increased cantilever curvature mimicked acetylcholine-induced hypercontractility observed in asthmatics, a response that could be reduced by a Rho kinase (ROCK) inhibitor.
To date, every Lung-on-Chip systems relies on cancer or immortalized cell lines or primary cells to reconstitute lung tissue in vitro. While cell lines are cost efficient and enable high-throughput studies, they are progressively being replaced by primary cells isolated directly from human or animal tissue as the biological relevance of cancerous or otherwise immortalized cells is increasingly questioned [132]. Primary cells offer a number of advantages compared to cell lines including recapitulation of original tissue characteristics, the ability to differentiate to in vivo-like tissue, preservation of donor disease phenotypes, increased predictiveness of human responses to drugs, control of cell source, and increased donor diversity to reflect natural diversity of human population. Nonetheless, primary cells also have important drawbacks. First, primary cells usually have a limited lifespan, although methods to prolong culture have been developed [133,134]. Primary cells are also difficult and costly to genetically modify as transfection efficiency can be very low, hindering effective and economical gene editing approaches. Finally, primary cell diversity can also be an obstacle to long term in vitro studies as cells from the same patients are hard to obtain. Human stem cells on the other hand can overcome some of these limitations of primary cells. For instance, because adult stem cells can be maintained indefinitely and are easy to transform, functional gene studies are particularly straightforward and inexpensive. Stem cells from a single patient can also be used to recreate virtually any cell type. This possibility is exceedingly valuable when the primary tissue is difficult to isolate, such as alveolar epithelial cells. Stem cells are also advantageous when autologous co-cultures systems (i.e. cells obtained from the same patient) are needed, for instance to insure compatibility when culturing epithelial and T cells together. It is therefore important to consider replacing primary cells in Organs-on-Chips with stem cells, either by differentiating 2D cultures of stem cells on-chip or by directly seeding 3D organoids inside the chips. Furthermore, combining Organs-on-chips with stem cell technology would potentially enable features that are currently missing in lung organoids and other stem cell-based systems, such as the recapitulation of dynamic physicochemical stem cells niches, access to luminal and vascular compartments, dynamic immune cell circulation, and controlled application of physiological mechanical cues (Table 2 ).
Table 2.
Advantages and limitations of Organs-on-Chips and stem cell organoids.
| Organs-on-Chips |
Stem cell organoids |
||
|---|---|---|---|
| Advantages | Limitations | Advantages | Limitations |
|
|
|
|
7. Human embryonic and induced pluripotent stem cells for human lung modeling
7.1. Differentiating human pluripotent stem cells into lung tissue
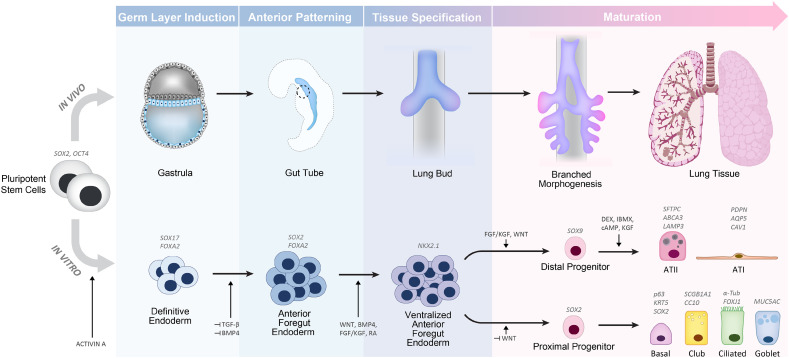
Lung cells derived from human pluripotent stem cells (PSCs), particularly human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), hold great potential to build advanced in vitro models of human lung tissue and to further our understanding of lung physiology and disease. While numerous attempts have been made to generate airway and distal lung epithelial cells from human PSCs, only recently has this undertaking gained significant traction. Initial studies first reported efficient embryonic induction into mesodermal and ectodermal lineages; however, maturation into the third endodermal germ layer remained limited [135]. While recreation of posterior endoderm cell derivatives that give rise to organs such as the liver, intestine, and pancreas became possible in the early 2000s [[136], [137], [138]], anterior foregut endoderm induction leading to lung tissues was not achievable until more recently. In 2011, a key study eventually elucidated a mechanism permitting stem cell differentiation into lung-specific endoderm precursor [139], thus leading to an effective strategy for ESC and iPSC generation of lung epithelial cells.
The basic protocol for generation of lung progenitor cells from ESCs and iPSCs in vitro is accomplished through directed differentiation, a process where in vivo tissue developmental stages are mimicked using controlled sequences of endogenous signaling factors [140] (Fig. 4 ). Pluripotent stem cells are first directed into definitive endoderm through activin-A simulation, followed by anterior foregut endoderm (AFE) induction through dual inhibition of the bone morphogenic protein (BMP) and transforming growth factor (TGF-β) signaling pathways [139]. Following AFE establishment, a ventral patterning step is needed mirroring the morphogenesis of the endoderm where the trachea and lung buds eventually emerge ventrally [141]. AFE ventralization is largely achieved through WNT, BMP, retinoic acid and fibroblast growth factor (FGF) signaling [66,139] yielding cells expressing transcription factor NKX2–1, the major marker for lung epithelial lineage (in addition to neural and thyroid tissue fate) [142]. This population of NKX2–1+cells is considered the primordial lung epithelial progenitor able to differentiate into proximal airway or distal lung bud lineages, primarily defined by SOX2 and SOX9 expression, respectively [143]. Terminal differentiation is then directed by further pathway signaling modification and determined by specific sets of markers. Methods used for specification to proximal airway or distal alveolar progenitors are discussed in the next paragraphs.
Fig. 4.
Overview of the major stages of lung development in humans corresponding to the directed differentiation pathways of pluripotent stem cells towards a lung epithelial fate. The various intermediate steps of development with key signaling factors and common markers are indicated.
7.2. Advances in alveolar generation
Over the past three years, notable progress has been made using NKX2-1+ lung progenitors to generate human alveolar epithelial cells, by leveraging strategies for maintaining human primary cells. The combination of glucocorticoids, growth factors, and cAMP effectors (dexamethasone, 8-br-cAMP, IBMX, and KGF/FGF7; collectively known as DCIK) [144,145] has been shown to induce alveolar maturation through activation of PKA and CDP-choline pathways which upregulate lamellar body surfactant production in ATII cells [146]. Recognized markers of the distal alveolar epithelium include Surfactant Protein C (SFTPC), Surfactant Protein B (SFTPB) HTII-280, ABCA3, and LAMP3/DC-LAMP for ATII cells, while Podoplanin, Caveolin (CAV1), and Aquaporin 5 (AQP5) primarily define ATI cells. Stimulation of two dimensional cultures of differentiated PSCs growing in 2D with DCIK, FGF10 and WNT activators leads to expression of ATI and ATII markers, mature phenotypic characteristics of lamellar bodies, and functional surfactant uptake capability [147]. Similarly, PSCs spheroid cultures also induce cell maturation of the alveoli epithelium. Interestingly, 3D co-culture with human fetal lung fibroblasts and stimulation with DCIK yielded ATII specific markers and lamellar bodies formation, although functional maturity was not confirmed [148]. Functional lamellar bodies through BODIPY-labeled SFTPB uptake was later shown by first generating PSC lung bud organoids, followed by Matrigel culture with a WNT activator, FGF10, KGF, BMP4 and RA in place of DCIK. ATII markers were found to be abundant in this protocol while ATI markers were minimally expressed [40]. Advanced maturation of ATII cells characterized by the capacity of lamellar bodies to process surfactant proteins and produce dipalmitoylphosphatidylcholine (DPPC) surfactant phospholipid was demonstrated by maturing NKX2-1+ cells in Matrigel with DCIK and temporally modulating WNT activity. Remarkably, mature forms of SFTPB and SFTPC could also be detected in the lamellar bodies, consistent with late-gestation primary fetal ATII cells [149].
Long-term expansion of alveolar epithelial cells, a challenge with distal lung cell cultures, was recently achieved using an organoid approach and a refined differentiation sequence after AFE induction [150]. This sequence differs from previous protocols by more closely mimicking the distal tip cell microenvironment through a preconditioning step of WNT activation, DAPT-mediated notch inhibition, and FGF10 plus KGF supplementation, resulting in SFTPC gene expression similar to levels found in the fetal lung. These cells were then matured using DCIK in organoids co-cultured with fibroblasts. Interestingly, co-cultures with fetal lung fibroblasts lines resulted in SFTPC expressing cells, though extremely variable (ranging from 2% to 51%), whereas incorporating a dermal fibroblast line showed no SFTPC induction. With passaging, the proportion of ATII cells in the culture increased to approximately 70%, and ATI-like cells were also present. Notably, induction of SFTPC expression was also achieved in fibroblast-free cultures using DCIK plus ROCK inhibition, WNT activation, and TGF-β inhibition, albeit at a lower efficiency of 23%.
7.3. Advances in proximal lung generation
Protocols for proximal airway differentiation also advanced in the past few years. Differentiation of PSCs into airway epithelial cells was achieved in spheroids cultures using FGF10, KGF, WNT agonist and Notch inhibition [151]. More recently, modifying WNT signaling in the directed differentiation strategy efficiently induced proximal fate, potentially explaining why previous studies encountered difficulty in maintaining airway epithelial markers in culture. Suppressing WNT signaling after ventralizing AFE promoted proximal fate and led to epithelial cells expressing the classic markers SCGB1A1/CC10 for club cells, MUC5AC in goblet cells, and p63 and Keratin 5 for basal cells [152].To promote cilia development of the airway, Notch inhibition in organoids or air-liquid interface in 2D cultures produced motile multiciliated cells characterized by acetylated alpha-tubulin (α-Tub) staining [[152], [153], [154]]. Another protocol used 2D air-liquid interface cultures with FGF18 stimulation to generate a mature and polarized epithelial layer, in which motile cilia and mucus could be observed at the apical surface [155]. It was shown that the scaffold and micro-environment of iPSC-derived lung progenitors is a major determinant for achieving a mature airway epithelial phenotype, as shown using human iPSC-derived lung organoids [156] provided with a bioartificial scaffold before transplantation to mouse epididymal fat pads [157]. Eight weeks following transplantation airway-like structures were observed only in organoids cultured and transplanted on the scaffold. For long-term expansion of iPSC-derived airway epithelial cells, it may be possible to incorporate findings from human primary cell cultures. Specifically, inhibition of TGF-β/BMP/SMAD signaling pathways was recently found to enable the long-term expansion of primary p63+ airway basal cells by inhibiting terminal differentiation and promoting self-renewal [134], pathways that may also be utilized to improve proximal PSC lung cultures.
7.4. Challenges
Tracing the respiratory developmental path has provided major insights into lung differentiation, yet the majority of our knowledge about lung development stems from rodent studies due to limited availability of human samples. Recently, new studies have been conducted examining human lung development using various gestational week human fetal lungs [158]. The genetic transcriptome for mouse lung development was found to be remarkably similar to humans; further, differences between both may help identify targets and develop better strategies to advance human lung modeling. For instance, lung bud tip signaling is associated with BMP4 and Sonic hedgehog (Shh) in the mouse, yet BMP2, BMP7, and Indian hedgehog (IHH) were found to be expressed in human tips. Additionally, human pseudoglandular tips were found to initially express both SOX2 and SOX9 rather than solely SOX9. Interestingly, these tip cells were used successfully as stem cells for forming organoids without mesenchymal cells under conventional FGF, WNT, BMP, and TGF-β signaling modifications [158].
Despite progress in recapitulating in vivo developmental pathways for lung differentiation, many signaling mechanisms still require further investigation. For instance, incorporating WNT signaling inhibition earlier in combination with TGF-β and BMP inhibition increased NKX2-1 expression, suggesting ventral patterning may begin before AFE [159]. Additionally, utilizing FGF2 and Sonic hedgehog (SHH) signaling for promoting AFE expressing NKX2-1 was successful, albeit with suboptimal efficiency [153]. Further, studies indicate that FGF signaling may be dispensable for lung specification, highlighting a need to explore the essential timing of FGF signaling [159,160].
While current differentiation protocols are effective for both ESCs and iPSC sources, high induction efficiency remains one of the major challenges. The directed differentiation strategy results in heterogeneous populations of NKX2-1+ cells with efficiencies widely ranging from 20% to 87%, an outcome that led to the questioning of NKX2-1 as a lung origination marker, but was recently confirmed [161]. Additional tools for purifying lung progenitor cells could ultimately provide the necessary supply for in vitro applications. Recently, this was accomplished with the discovery of highly specific cell surface markers CD47 [161] and Carboxypeptidase M (CPM). These markers allowed for live tracking and sorting resulting in an impressive ~90% purity of NKX2-1+ cells when differentiating into ventralized AFE [150].
Despite substantial progress in advancing the differentiation of lung stem cells, full maturation is still a key challenge as parameters defining fully differentiated epithelial cells are still evolving. For example, demonstrating SFTPC and SFTPB gene expression was initially sufficient to claim ATII cell differentiation. However, with the accelerated progress in lung PSC protocols, more strict metrics are now being considered to reach the ultimate goal of generating a supply of cells that recapitulates the same molecular, biochemical, phenotypical, and functional features found in the adult lung. Beers and Moodley, along with other leading experts and pioneers in the lung field, reviewed this concern and proposed a set of standards specific to alveolar cell generation in order to bridge lung biology with stem cell development [162]. In summary, for claiming terminal ATII cell maturation, cells need to demonstrate morphological similarity with in vivo tissue, a significantly mature surfactant system, characteristic lamellar body ultrastructure by transmission electron microscopy, functional maintenance and repair of barrier function, self-renewal and regeneration, and transcriptomic analysis comparable to isolated ATII cells. This initiative to assist alveolar stem cell development calls for highly sophisticated techniques and protocols that Lung-on-Chip platforms may assist with, and requires researchers to fully characterize cell populations. These notions are applicable for all lung epithelial generation and paramount for developing relevant and standardized in vitro applications.
7.5. Rationale for stem cell-derived Lung-on-Chip
Incorporation of stem cell-derived differentiated cultures into Lung-on-Chips models offers new opportunities to study lung development and to develop patient-specific disease models. It also enables investigators to include different cell types (e.g. airway epithelial and endothelial cells) derived from the same donor within one model. By closely recreating the spatiotemporal dynamics and heterogeneity found in the lung, Lung-on-Chip models may also be useful for appropriate differentiation and maturation of stem cell-derived lung cells. Indeed, chemical signals, structural cues, ECM, and additional cell types have all been shown to enhance maturation of lung epithelial PSCs: implantation into injured mice, scaffolding using decellularized lung matrix, or multi-culturing with lung mesenchyme and fibroblasts have led to the development of lung epithelia of all fates [40,148,156,157,159,161,163,164]. Furthermore, many lung PSC culture systems utilize a 3D system to create advanced lung organoids. Likewise, Lung-on-Chip models provide a dynamic microenvironment with increased cellular complexity, factors known to influence stem cell differentiation [165]. By delicately dissociating stem-cell derived organoids, it may be possible to seed matured lung cells onto the surface of the ECM-coated porous membrane forming a monolayer within the chip channels, thus supplying cells with a microenvironment characterized by physiological ECM, air-liquid interface, stretch, and fluid shear. This will also allow introducing added complexity from a vascular channel for additional cell types such as endothelial cells and fibroblasts, which can be derived from the same iPSC donors resulting in a patient-specific model. Further, fluid flow and apical/basal distinctions in the Lung-on-Chip can be utilized to finely tune the timing and distribution of signaling factors, and explore spatiotemporal responses to stimuli. The multitude of parameters Organs-on-Chips technology provides for isolating dynamic and spatiotemporally heterogeneous aspects of in vivo physiology holds great potential for fully maturing lung PSCs and advancing in vitro lung models.
8. Applications of stem cell-based human Lung-on-Chip systems in disease modeling and drug development
8.1. Respiratory disease modeling
Human microengineered systems, such as Organs-on-Chips, aim to provide additional insights into the pathogenesis of human tissues and enable the prediction of drug efficacy and safety in a physiologically relevant context. Once such benefits are clearly established, combining Organs-on-Chips with human stem cells in which all cell types come from the same donor would offer the opportunity to test compounds that correct patient specific phenotypes associated with defined genotypes or genes mutations, thus creating predictive platforms for the field of personalized medicine that offer complementary advantages to models based on animals and immortalized or primary cells [13]. While maturation potential and genetic stability of embryonic stem cells (ESCs) remain superior [166], tissue-derived induced pluripotent stem cells (iPSCs) present key advantages over ESCs, including renewable supply of patient-specific cells from individuals with acquired and genetic lung disease. The first 100 lung disease–specific iPSC lines were generated, including lines from individuals with cystic fibrosis and α1-antitrypsin deficiency, the two most common monogenic lung diseases [167]. Immediate research applications of these disease–specific cell lines include probing the relative contribution of somatically acquired versus genetic risk factors, genetic engineering to induce or repair putative disease-causing mutations, the comparison of tissues derived from patients and their healthy relatives, and the study of both disease pathology in different genetic backgrounds and their response to drugs [168]. Direct consequences for clinical research are imminent: because stem cells are derived from a specific patient, analysis of their response to various stimuli or drugs should predict individual patient responses. For example, if a compound improves a specific function in a model using iPSC derived from a patient, the same result may be achieved in the patient. Conversely, if adverse drug effects are identified in human iPSC-derived tissues with a specific genetic background, it might predict drug failure in the clinical trial for this subset of patients. In monoculture, iPSCs are most suitable for investigating monogenetic diseases that have complete penetrance and well-defined cellular phenotypes caused by the mutation (cell-autonomous disease), such as cystic fibrosis cells with mutations of the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which impair chloride ion transport in cells that express CFTR. However, by integrating multiple cell types, immune cells, and a dynamic mechanochemical environment, Organs-on-Chip technology provides the ability to also study non-cell-autonomous diseases such as asthma, in which environmental agents in concert with airway and immune cells generate a diversity of phenotypes. Lastly, Lung-on-Chip systems combined with iPSCs could facilitate research in rare pulmonary diseases for which there are no preclinical disease models [169]. In the following, we will discuss diseases that might benefit from stem cell-based Lung-on-Chip system. To provide a tangible example, specific research questions are outlined that could be studied in a Lung-on-Chip model of cystic fibrosis, but similar approaches are relevant to the other diseases discussed below.
8.1.1. Cystic fibrosis
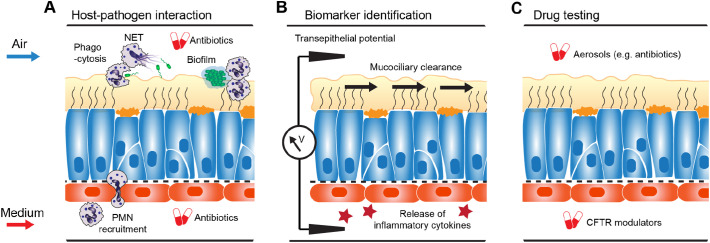
Mutations in the CFTR gene cause cystic fibrosis in humans, a debilitating disease characterized by persistent airway infections and permanent damage to the lung resulting from changes in chloride ion transport, mucus rheology, inflammation and bacterial adherence [170]. Over the past 20 years, there has been tremendous progress in alleviating the symptoms and even treating some of the underlying molecular causes of the disease, leading to a significance increase in life expectancy [171]. This advancement would have been impossible without the development of a host of preclinical assay in animal and traditional cell culture models [172]. However, there remains a lack of suitable model systems for certain mechanisms, including the predominant cause of pulmonary decline in CF patients, the infection with biofilm-forming phenotypes of Pseudomonas aeruginosa that emerge in the CF lung [173]. Mice with deletion of the CFTR gene do not develop the spontaneous lung disease or chronic bacterial infections seen in humans [174] and, unlike humans, CFTR mutant mice appear to have alternate chloride ion channels compensating for dysfunctional transport by CFTR [175]. Recent development of pig and ferret models of CF alleviate some of these limitations and have been shown to recapitulate key aspects of CF in humans, including persistent bacterial infection of the lung [176], however, these animal models do not replicate the highly specific association with P. aeruginosa seen in human patients, and are unlikely to support genotype-phenotype studies for most of the 2000 mutant CFTR alleles known to impair synthesis, trafficking, stability and/or ion gating function of CFTR [177,178]. Human Organs-on-Chips, whose vascular and interstitial flow channels support the controlled addition of circulating or resident immune cells as well as bacteria, are a promising new avenue for studying the interactions between microbes, epithelium, and immune system [179,180]. Among many applications, such a system could shed light on the development of the suspended 3D biofilm P. aeruginosa typically forms in affected patients, on the recruitment and response of polymorphonuclear leukocytes (PMN) to biofilms, the role of phagocytosis, neutrophil extracellular traps (NET) and neutrophil microvesicles on clearing, or failing to clear, P. aeruginosa, and the potential contribution of patient-specific (=CFTR-defective) PMNs to enabling chronic infection (Fig. 5A) [181].
Fig. 5.
Example application for Lung-on-Chip for disease modeling. Small airway epithelium derived from stem cells of cystic fibrosis patients can be used to study universal cystic fibrosis disease mechanisms as well as the role of specific CFTR mutations within the context of the dynamic micro-environment at the blood-mucus-air interface. For example, the chip can be used to study A, the interaction between PMNs, biofilm-producing bacteria such as P. aeruginosa, and antibiotics treatment; B, the sensitivity and patient-specificity of new and established markers of lung function, including transepithelial potential difference, mucociliary clearance, and biomarkers of inflammation; and C, the efficacy of delivery and action of drugs for the treatment of cystic fibrosis patients, such as CFTR modulators and antibodies.
As CF is characterized by heterogeneity in the therapeutic response rate, the interpretation of clinical trials can be problematic, suggesting that personalized treatment is required, based on appropriate endpoints and biomarkers [182]. Recently, a CFTR-dependent swelling-response of stem-cell derived gut organoids has been used as a bioassay to probe the disease phenotypes of different CFTR mutations and screen the mutation-specific efficacy of CFTR-restoring compounds [183]. Other, fluorescent signal-based assays enable phenotypic profiling by measuring CFTR-mediated conductance [184]. Stem-cell derived Lung-on-Chip models can augment these efforts by enabling the measurement of lung-specific functions, such as mucociliary transport, and clinically relevant endpoints, such as transepithelial potential difference and inflammation markers [185]. In addition to identifying new biomarkers most sensitive for classifying disease phenotypes and detecting drug responses, it could be probed, for example, how much functional CFTR needs to be present to normalize mucociliary clearance (Fig. 5B).
Another potential application of Lung-on-Chip models is to probe the pharmacokinetics and pharmacodynamics of treatments for cystic fibrosis patients, such as inhaled antibiotics [186], or CFTR modulators [187] (Fig. 5C). CFTR modulators demonstrated that direct repair of the malfunctioning protein can partly restore normal lung function [188]. However, this treatment requires continuous pharmacological intervention. Hence, great hopes are placed into restorative therapies which would bring longer-lasting relief. Recently, CRISPR-Cas9 genome editing technology was used to restore normal functional CFTR intestinal stem cell organoids, demonstrating the powerful potential of these systems for testing human gene therapy [189]. Moreover, in 2017 a RNA-editing strategy for the most common CF mutation [190] successfully completed the Phase I trial (NCT02532764). Extending the limited functional readouts provided by organoids, Lung-on-Chip models can be used for gauging the improvement of tissue-wide functions, such as mucociliary clearance and transepithelial conductance.
8.1.2. Congenital pediatric lung diseases
A range of pediatric lung diseases are due to mutations in genes encoding surfactant proteins (SP-C, SP-B) or factors required for surfactant trafficking (ABCA3), causing surfactant deficiency. Mouse models generated by gene deletion tend to recapitulate only a subset of the clinical spectrum observed in human populations, likely because of different lung physiology, and no therapy has been developed thus far [191]. Stem-cell based in vitro models of human lung development [40] provide a new system for studying etiology and pathogenesis of these and other pediatric congenital lung diseases, and might lead to novel therapeutic approaches.
8.1.3. Inflammatory obstructive lung diseases
Chronic obstructive lung disease (COPD) and asthma are the most prevalent of all chronic respiratory diseases worldwide, and they rank among the top 20 conditions causing disability globally [192]. Both conditions engender respiratory distress and chronic inflammation of the lung and are thought to result from environmental exposure in genetically susceptible individuals [193,194]. The symptoms can be further exacerbated by airborne particles, or viral and bacterial infection of the airways. Currently, there are many treatment options for managing severe asthma and COPD; however no treatments reduce disease progression [195,196]. To advance this research, a plethora of model systems has been developed over multiple decades, ranging from very simple models using human cells (e.g. bronchial epithelial cells and fibroblasts) in mono- or co-culture, whole tissue explants (biopsies, muscle strips, bronchial rings) to in vivo studies in animals or in humans [197]. While these platforms cover substantial grounds, the addition of dynamically perfused, patient-specific microtissues could help dissect the contributions of environmental factors, genetic predispositions, and acquired susceptibility. In fact, animal models do not naturally develop asthma [36], and even when they can be rendered sensitive to antigens, they fail to recapitulate all aspects of the human pathogenesis [[198], [199], [200]]. This is likely due to well-known differences in airways physiology, anatomy, and immunology between animals and humans [[201], [202], [203]]. For instance, secretory goblet cells are increased in size and number in COPD and asthmatic airways; this pathological hyperplasia is likely mediated by Notch signaling-directing differentiation of basal cells, the stem cells of the large airway, towards a secretory fate [204]. Mice, however, exhibit a much lower abundance of goblet cells than humans and therefore do not provide a good model for development and homeostasis of goblet cells in healthy and diseased human lungs. Traditional cell culture models using airway epithelial cells from asthmatics [205] or mimicking asthma by exposing normal epithelial cells to asthma-associated cytokines such as IL-13 [206], have helped reveal the role of inflammation and the innate immune system in asthma [207], however, the spatiotemporal dynamics of this process cannot easily be studied in either static culture nor in animal systems. Also, the origin of pathological inflammatory responses might be found in developmental events that are human- or even patient-specific, such as in utero fetal programming of gene expression involved in lung development [208], or signaling from the microbiome during a critical postnatal time window thought to promote immune tolerance [209]. Human primary cell 3D models of the airways have demonstrated a new avenue towards addressing these and other questions in vitro [52,210]. In the future, human stem-cell based Lung-on-Chip models that support the dynamic interaction with circulating immune cells, essential in asthma and COPD pathogenesis, could model inflammation and exacerbation, and lead to the evaluation of new therapies and treatment options.
8.1.4. Pulmonary fibrosis
In pulmonary fibrosis, normal parenchymal tissue is gradually replaced by fibrotic tissue as a result of scarring, resulting in functional impairment. Pulmonary fibrosis may develop secondary to other diseases, such as autoimmune disorders or hypersensitivity pneumonitis. Idiopathic pulmonary fibrosis (IPF) develops without a known cause, although the identification of disease-specific mutations in subsets of patients is clear evidence of a genetic predisposition [211]. IPF is a particular severe form of pulmonary fibrosis with a progressive decline in lung function, that causes the death of 80,000 US Americans and Europeans each year [212]. To date no therapy is available to reverse the disease process. In the recent years, two pharmaco-therapeutic agents, pirfenidone and nintedanib, have been developed that can halt disease progression, however use of both drugs is associated with adverse effects [213]. While the precise disease mechanisms are still unknown, it is believed that myofibroblasts and fibrosis progenitor cells are responsible for the fibrotic architectural remodeling in the lung [63,214]. Altered mucociliary properties could also contribute to the disease process by reducing clearance of noxious agents from the epithelium [215]. Several genetic polymorphisms and mutations predispose to IPF but how the fibrotic process is initiated and sustained remains unclear, partly because no good mouse model of the disease exist, leading to poor preclinical to clinical translation and extensive clinical trial failures [216]. Patient-specific in vitro models of the human alveolar epithelium are urgently needed to help elucidate the pathogenesis and role of genetic predisposition and environmental exposure in determining disease penetrance [214,217]. Specifically, on-chip model comprising the different cell types of the alveolar epithelium, ideally from the same patient, and recapitulating the cross talk between alveolar epithelial cells and the surrounding fibroblasts could reveal unknown mechanisms of pathogenesis and lead to new therapeutic targets. In addition, the ability to test the influence of physiological mechanical stretch on alveolar cells in vitro is another attractive feature of Lung-on-Chip for modeling IPF.
8.1.5. Lung cancer
Lung cancer is the leading cause of cancer mortality and causes more than 160,000 deaths per year in the United States [218]. Development of drug-resistant tumor cells and eventually fatal metastases is common despite aggressive treatment with surgery, radiation and chemotherapy, and long-term survival rates of lung cancer patients remain low [219]. Clearly, new treatment strategies are needed. Approaches that elicit anti-tumor immune responses such as antibody-based immune checkpoint receptor inhibitor therapy [220] or enhanced T-cell activation using gene editing [220] have raised great hopes; however, in many cases preclinical efficacy does not translate to human trials. For example, in some cases, immunologic response was not correlated with survival [221]. Perhaps more concerning, in recent phase II/III clinical trials, nearly 80% of all the studies failed [28] due to elevated levels of circulating immunosuppressive cytokines and various immunological checkpoints in humans that may not be present in animal models [29]. Indeed, there is a lack of human-relevant model systems where tumor biology in response to treatment and immune responses can be observed and quantified. Conventional subcutaneous implants of tumor tissue in mouse models do not mimic organ-specific differences in cancer growth or responses to drugs observed in the clinic [222]. Notably, human tumor xenografts implanted in mice at the corresponding organ site from which the tumors were derived are better at recapitulating physiological tumor growth and metastasis [223,224]. These results indicate the importance of the organ-specific microenvironment in determining tumor biology and drug response; however, the contributions and spatiotemporal dynamics of the tumor-organ interactions have remained a “black box” because of the difficulty to visualize and probe tumor development in vivo. Conventional 2D in vitro cancer cultures, on the other hand, are amenable to real time imaging and investigation and have been the workhorse for studying molecular mechanisms of cancer, providing high throughput and direct comparison of experimental results with a wealth of literature and online data [225,226]. However, there is evidence that the 3D architecture of real tumors might lead to different growth profiles and drug responses, and this has driven the development of 3D cancer cultures [227]. Extending these approaches to include the organ-specific microenvironment and heterogeneous cell populations, Organ-on-Chip models of human lung cancer provide an alternative approach by recapitulating the organ-specific 3D microenvironment, tissue-tissue interfaces, mechano-chemical cues, vascular perfusion and potential interactions with immune cells while also enabling continuous monitoring of tumor evolution and tumor-organ interactions [124]. Combining these approaches with stem-cell derived tumor tissues could improve translation of efficacy data from preclinical testing to the clinical and aid in the development of patient-specific therapies and treatment strategies overcoming tumor drug-resistance [228].
8.1.6. Pulmonary infectious disease
Respiratory infections of the lung from a viral or bacterial source are a major cause of human mortality worldwide. Third only to stroke and cardiovascular disease, lower respiratory infections constitute the most deadly communicable diseases, causing 3.2 million deaths in 2015 [2]. Tuberculosis and other bacterial diseases, together with influenza, as well as diseases caused by respiratory syncytial viruses, human metapneumovirus, parainfluenza virus, adenoviruses and coronaviruses, can cause life threatening conditions in healthy people. Even otherwise harmless infections can cause complications in patients with pre-existing lung diseases such as asthma, COPD, bronchiectasis, cystic fibrosis, or primary immunodeficiencies, and other conditions that alter immunologic mechanisms against microbial invasion [229]. Susceptibility to infection as well as onset, duration and severity of symptoms is controlled by dynamic and patient-specific host-microbe interactions which, in addition to the rise in antibiotic resistance, has contributed to the difficulty of developing effective treatments [[230], [231], [232]]. Lung homeostasis relies on a finely regulated balance between immune tolerance towards innocuous exogenous particles that continuously enter the lungs on the one hand, and defense mechanisms against infectious agents on the other hand. An imbalance in immune homeostasis of the lung can lead to exaggerated inflammatory responses such as allergies or a “cytokine storm” following viral infections which can lead to extensive tissue damage [233]. Respiratory infection pathogenesis is largely mediated by the interaction between the infected tissue and the host immune system, including lung resident and circulating immune cells. While most in vitro systems enable the study of isolated immune cells or epithelial cells in static conditions, human Lung-on-Chip models are dynamic fluidic systems that offer the unprecedented possibility to investigate interaction of lung endothelium and epithelium with the ability to flow immune cells in a dynamic fashion through lung capillaries. This unique feature can be leveraged to dissect individual roles of infiltrating immune cells in lung infection, to probe underlying disease mechanisms and to identify new therapeutic targets or facilitate vaccines development. Disease phenotypes are also modulated by more subtle interactions with the patient's resident immune cells. For example, pathogenesis of the tuberculosis causing bacterium Mycobacterium tuberculosis involves modification of core cell signaling pathways in immune cells and differences in patient-specific immune cell responses may explain why responses to infection are highly variable between patients [234,235]. Hence, both the development of novel, host-directed therapies as well as understanding the mechanisms of disease onset and progression in tuberculosis would benefit from patient-specific in vitro models of the dynamic interactions between bacteria, lung cells, and the immune system [[236], [237], [238]].
8.2. Pulmonary drug and toxicity testing
Preclinical drug discovery and development process involves testing of drug's safety, efficacy, pharmacodynamics, and pharmacokinetics (including drug absorption, distribution, metabolism, and excretion), as well as potential drug–drug interactions. Lung toxicity can also arise from inhalation of airborne substances or from prenatal exposure to blood borne chemicals. For the same reasons previously addressed, animal models, and particularly rodents, are limited in their ability to predict human responses, pointing to the opportunity of using stem-cell based Lung-on-Chips to advance the field.
8.2.1. Safety of pulmonary drugs and lung toxicity
Drug candidates can induce specific adverse effects in the lungs, and the lungs may be affected by general cytotoxicity too. Several hundred medications, delivered by various routes, are known to cause drug-induced adverse responses and respiratory complications [239]. Because the clinical, and histological symptoms are often non-specific, it can be difficult to diagnose lung-related adverse drug reactions, especially if the patient is taking multiple drugs, and hence pulmonary toxicity, estimated at approximately 3%, is likely under-diagnosed [240,241]. Adverse effects can be idiosyncratic or due to a toxic reaction of the drug or one of its metabolites, since the lungs metabolize certain substances. In addition, secondary mechanisms such as immune system responses can cause further damage. For example, drug-induced interstitial lung disease (DILD) is a common complication which can cause acute respiratory distress leading to pulmonary fibrosis (characterized by proliferation of fibroblasts and extensive ECM deposition) [242,243]. DILD is thought to be caused by direct, dose-dependent cytotoxicity to bronchial epithelial cells or the alveolar capillary endothelium, but also by a T-cell mediated immune response. While age, sex, existing lung disease, drug interactions, and drug dose are risk factors known to correlate with phenotype of DILD and other drug-induced respiratory diseases, a mechanistic understanding of factors influencing disease onset and progression is lacking. Human Lung-on-Chip models could help isolate and probe genetic predispositions of specific patient populations, elucidate interdependencies of risk factors, predict safety risks, and identify safe drug dosing regimens.
In addition to probing drug safety, Lung-on-Chip models are also an attractive route towards improved in vitro models of inhalation toxicology to probe adverse effects of inhaled substances originating from environmental, occupational or other external sources [244]. In particular, exposure to cigarette smoke, airborne nanoparticles and ultrafine particulates has become an emerging health concern and is associated, among other adverse health effects, with inflammation, exacerbation of preexisting respiratory disease, lung cancer, and particle-induced lung fibrosis. Furthermore, inhalation of toxic gaseous airborne substances remains an important cause of lung injury through occupational exposure and, in some parts of the world, chemical warfare. For many toxic gases, mechanistic understanding and specific treatment options for acute and chronic injury are lacking, and human in vitro inhalation models of gas toxicity would address an unmet need [245]. The in vitro modeling of inhalation toxicity is particularly challenging because no standardized procedure for delivering airborne toxins and gases to an air-liquid interphase exist, let alone for incorporating the effects of breathing flow dynamics. Mode of delivery and flow dynamics, however, can greatly affect the relative exposure times and effective dose of toxicants and safety margins. Hence, the lack of standardized procedures also prevents standardized toxicity and dosing assays for inhaled agents [246]. Lung-on-Chip models present the opportunity to address this challenge and deliver airborne toxic particles and gases in a spatiotemporally controlled manner, enabling the study of toxicity by physiological inhalation and also, in the future, providing a means to standardize safety testing for inhaled substances [126].
Lastly, stem-cell based Lung-on-Chip models could also provide a system for studying the ability of chemicals to disrupt neonatal development of the lung. For example, it is known from animal studies that maternal exposure to nicotine, which passes through the placenta and can accumulate in amniotic fluid, causes alterations in postnatal lung function, such as overexpression of nicotinic acetylcholine receptors and collagens [[247], [248], [249]]. Studies in human embryonic stem cells have demonstrated the direct modulation of differentiation pathways in lung fibroblasts by nicotine, supporting the hypothesis that lung matrix deposition and hence connective tissue mechanics could be altered in postnatal lungs [250]. A Lung-on-Chip model recapitulating some steps of airway development could enable the real-time study of how complex multi-cell type interactions and deposition of ECM are affected by exposure to nicotine.
8.2.2. Efficacy of pulmonary drugs
For most chronic lung diseases the available treatments are aimed at providing symptomatic relief rather than curing underlying disease mechanisms [251]. Further, many new drugs being developed are based on disease targets that were identified many years ago with clinical trials plagued by high failure rates. A number of recent trials have shown promise only to fail at later stages [252] which underlies the need for better understanding of patient phenotypes and preclinical models with greater relevance, reproducibility and predictability. For carefully designed scenarios and disease stages, Lung-on-Chip models in particular could help identify preclinical endpoints that can be translated to the clinic, stratify diagnosis and therapeutic care based on molecular mechanisms, tailor treatments to patient subgroups, and design trial end-points that better predict drug efficacy. Indeed, in 2014, the Respiratory Expert Council (REC), a global team of research and clinical experts, presented recommendations for improving drug research and development for airway diseases [253], three of which human Lung-on-Chip models are poised to facilitate.
First, to capture and understand the heterogeneity of airway disease pathophysiology and clinical presentation, airway diseases need to be redefined based on molecular mechanisms and systems-biology-based strategies, particularly network analysis, to characterize variability due to patient to patient differences and due to progression of the disease through various stages [254]. Categorizing the disease in this way could facilitate the design of patient-specific and stage-specific therapies and advance personalized respiratory medicine [255]. In the simplest application, systems-biology-based disease definitions can help address questions such as whether a new drug should be developed and tested in a broad disease category, such as COPD, or focus on specific disease phenotypes. In particular, even if patients exhibit similar symptoms on the clinical level, the molecular factors driving disease progression can be different, and vice versa [256]. Stem-cell based models can provide a platform to reveal genotype-phenotype relationships affecting drug responses in different patient populations. These pharmacogenomic insights could inform the design of clinical trials and determine the inclusion and exclusion criteria of patients to assure optimal drug targeting.
Second, preclinical studies also need to take into account systemic manifestations of airway diseases, such as inflammation and immune system responses [257] and link them both to molecular and tissue-level disease symptoms as well as clinical endpoints. Vascularized Lung-on-Chip models provide the dynamic environment allowing for controlled and experimentally accessible interaction between local lung tissue and systemic factors, such as immune system components, and even other organ tissues.
Finally, new biomarkers of lung function are needed that can be translated between experimental models and the clinic. Of particular interest is the development of imaging markers and biomarkers that are sensitive to different stages of airway disease, demarcate early-stage lung disease, and group patients into distinct phenotype or endotype populations. Towards these goals, Lung-on-Chip models provide a system to identify dynamic morphological, soluble, genetic and ‘omics’ markers that could be integrated with clinical readouts from blood tests, biopsies, imaging, patient-reported outcomes and clinical endpoints, such as forced expiratory volume in 1 s (FEV1). Such multi-dimensional disease signatures would then support the design of end-points that can be measured both in vitro and in the clinic which are directly related to disease progression and drug efficacy [258,259].
8.2.3. Pulmonary drug delivery
Inhalation is an attractive route for drug delivery because its large surface area (70–140 m2 in adults), good vascularization, and thin alveolar epithelium offer the potential for rapid absorption of pharmaceuticals either for local deposition or for systemic delivery [[260], [261], [262]]. There is also great interest in the development of inhaled drugs for localized treatment of lung diseases, including pneumonia [263]. However, accurate pulmonary drug delivery is not a trivial task because of the complex mechanical, chemical, and biological pharmacokinetics (PK) mechanisms at play, including drug deposition, absorption, distribution, metabolism, and various clearance mechanisms. The first step, drug deposition, depends mostly on the fluid-mechanics of lung morphology, breathing pattern, aerosol velocity, particle size and density. Only part of the dose will reach the target site in the lung whereas a significant fraction can remain in the inhalation device or be deposited in the mouth and throat. Due to the large spatial scales involved, understanding and predicting initial drug deposition relies on computational models and perfused animal lungs [264,265]. Organ-on-Chip models have great potential, however, to aid modeling of many aspects of drug fate following deposition in different regions of the lung; this is in addition to providing insight into region-specific pharmacodynamic activity due to varying target expression and density throughout the lung. Specifically, for pulmonary absorption into tissue and blood circulation to occur, deposited drug particles have to (i) dissolve in the surface lining fluids, (ii) overcome various defense mechanisms by which the lung clears inhaled particles, including mechanical (i.e., mucociliary clearance), chemical (e.g., surfactant lipids, antiproteases), and immunological (e.g., phagocytosis) responses, (iii) pass the transepithelial air-blood barrier by passive diffusion or carrier-mediated active transport via paracellular or transcellular pathway, and (iv) withstand (or be activated by) the lung's metabolic activity [264]. The relative role of these mechanisms in drug absorption varies depending on airway region. For example, the conducting airways (nasal cavity, sinuses, nasopharynx, oropharynx, larynx, trachea, bronchi, and bronchioles) are lined by ciliated cells and mucus-secreting cells, which together enable the mucociliary clearance of about 90% of inhaled particles, greatly reducing bioavailability of drug compounds [266]. In the respiratory regions, on the other hand, which comprise about 95% of the total surface area and consist of respiratory bronchioles, alveolar ducts, and alveolar sacs, drug bioavailability is limited by absorption, transport across the pulmonary air-blood barrier, and metabolic and macrophage clearance [267]. Because of the complexity of the lung morphology, the quantitative characterization of the in vivo interplay of all contributing factors either with in vitro assays or with in silico methods is an ongoing challenge. For example, rodents are generally exposed intra-tracheally or via nose-only inhalation, and hence the resulting plasma pharmacokinetics are a result of absorption from different regions of the lung [268]. Computational modeling approaches attempt to derive PK parameters mechanistically based on the physico-chemical characteristics of the drug (e.g., aqueous solubility) and the anatomical and physiological tissue properties, or, alternatively, from clinical PK data by means of statistical correlation; these approaches are limited by preexisting knowledge and data. Recently, there have also been microfluidic approaches to studying the dynamics and deposition of particles in true-scale, but cell-free, physical models of the pulmonary acinus which can help to isolate purely mechanical mechanisms [269]. Stem-cell based Lung-on-Chip models could complement these approaches and enable the empirical measurement of PK parameters of drug absorption at the cellular and tissue level. For example, local drug dissolution could be measured, which, especially for lipophilic drugs, can represent the rate-limiting process for pulmonary absorption. Further, temporal dynamics and patient-specific conditions can be revealed by recapitulating organ-specific barrier functions such as endothelial cell junctions and electrical resistance, presence of specific membrane transporters and metabolic enzymes, genotypic variation, disease conditions, and drug-drug interactions. Also, the effect of elimination processes such as mucociliary clearance on pulmonary bioavailability could be measured [270]. In addition, the flow environment of Organs-on-Chips enables the realistic test of time-dependent effects such as built-up of drug residue or drug carriers that can cause local toxicity [271].
8.3. Practical considerations and challenges of Lung-on-Chips engineering
The two principal challenges involved in the design and engineering of Lung-on-Chip models suitable for reproducible and scalable research are (i) to build the biology, i.e., to decide on the cells, ECM elements and the dynamic microenvironment that together enact the desired structure-function relationships of the organ [272], and (ii) to build the chip hardware, i.e. to identify materials and geometries supporting study and maintenance of tissue biology that are at the same time amenable to reliable batch fabrication and application (Fig. 6 ).
Fig. 6.
Practical biological and engineering approaches for building a Lung-on-Chip.
8.3.1. Biology
As discussed in great detail before, the main components determining the biology of the system are cell types, sources and architecture, ECM composition and distribution, and biomechanical forces such as flow shear and tissue strain. As with all model systems, tissue components and their mechanical properties have to be chosen carefully to achieve the structure-function relationships of interest without overly complexifying the system, which could mask important mechanisms, hinder reproducibility and make the results harder to interpret [273].
8.3.2. Hardware/materials
Complementary to the biological components, chip geometry and materials need to be chosen to provide appropriate structural support, implement the desired flow dynamics and mechanical forces, and support instrumentation for tissue maintenance and experimental readouts. Ideally, materials need to be non-toxic, optically clear, affordable, moldable or machinable, non-degrading under cell culture conditions, flexible to allow stretching, and not altering the chemistry of the fluid environment. Together, these criteria are difficult to achieve. For example, as discussed earlier, PDMS has been the material of choice for many proof-of-concept Organs-on-Chips designs; however, despite all its advantages for rapid prototyping, its use is sometimes limited by its tendency to absorb specific small hydrophobic pharmaceutical compounds, encouraging the development of new, non-absorbing Organs-on-Chips materials [274]. Similarly, chip geometry constitutes a major factor in providing a physiologically-relevant environment amenable to sampling. While most Organs-on-Chips currently published feature rather simple flow circuits serving mostly perfusion and gas exchange, more sophisticated scenarios containing in-flow sensors and drug dispensers could be designed with the help of design and simulation software, as is routinely done for microfluidics applications [275]. However, introducing complex fluidic microchannel networks may increase bubble formation which could in turn damage cultures through shear forces and cells drying out when bubbles block medium perfusion.
8.3.3. Fabrication, integration and scaling
Eventually, however, the success of Lung-on-Chip models in achieving significant scientific impact will depend as much on the scientific merits as on its ease of use and fabrication, which directly relate to reproducibility of results across samples, experiments, and users, as well as scalability of experimental work to provide sufficient capacity. For this reason, efforts need to be directed towards achieving the same streamlining of chip manufacture as has happened in non-biological microfluidics [276] and towards developing user-friendly platforms that can support large-scale experimental work [277]. Considering all these aspects together when designing a chip seems a daunting prospect. However, other branches of industry dealing with complex micro-engineered systems, such as mechatronics, have developed a range of computational and simulation tools for optimizing design and fabrication based on desired usage goals and past experiences. These same tools have started to emerge in the Organs-on-Chips community, promising yet another technology transfer from electronical to biological micro-engineering [278].
9. Conclusion and outlook
The recent advances in the development of Organs-on-Chips and stem cell based organoids make their combination particularly attractive for studying dynamic processes in lung development, regeneration, and pathogenesis, as well as for personalized medicine. However, nowadays a multitude of models are available, and it needs to be noted that each approach has its merits. Therefore, it is essential that researchers adapt their choice of a model based on the research questions and logistic considerations. Thus far, Lung-on-Chip models have been developed and mostly employed in academic research to generate proof-of-concept data. Broader accessibility of the technology and increased experimental throughput will enable the translation of this technology to pharmaceutical industry and clinical decision making. To meet the high expectations of Lung-on-Chip models, however, there is a need for a careful balance between continued increases in biological relevancy and complexity of the chip design on the one hand, and optimization for implementation in industry that requires greater reproducibility, capacity, and ease of use on the other hand. Meanwhile, patient-specific cells are already being used to predict the efficacy of drugs in the clinic; for example, in the Netherlands, the health care system covers the evaluation of cystic fibrosis drugs in patient-specific human organoids to assess whether a patient would benefit from the medication [183,279]. Merging the two technologies to engineer patient-specific, stem-cell based Lung-on-Chip models will amplify their respective advantages and enable the design of human models for predicting efficacy and safety, drug-drug interactions, clinical trial applications and precision medicine.
The demand for such models is expected to grow continuously, given that the current ban on animal testing for development of consumer products will likely expand from Europe (European Union Directive 76/768/EEC) to North America and worldwide, and given the pressing need for more predictive preclinical models for drug discovery and development. To enable such applications, however, the new Lung-on-Chip models need to be held to stringent and uniform quality control criteria [280]. This is a formidable challenge as many different chip designs are currently being developed in academic laboratories, highlighting the need for creating standards and user guidelines, in addition to harnessing the inherent variability of the cell sources.
One challenge is to balance the trade-off between increasing the biological complexity of the chip on one hand, and maintaining scalability of fabrication and ease of use on the other hand. In general, a greater degree of biological complexity will extend the range of functions that can be studied and are physiologically relevant, but might conversely impair economic factors, such as number of fabrication steps per sample, reproducibility of results, ease of use, throughput and volume of cells and reagents needed, and therefore slow down general adoption. To address this hurdle, US American funding agencies have started an initiative towards consolidating development of Organs-on-Chips and achieving robustness and replicability. The National Institutes of Health (NIH), in partnership with the Defense Advanced Research Projects Agency (DARPA), the FDA, and more recently the pharmaceutical industry, has invested in academic teams and companies to develop Organs-on-Chips technology suitable for industry applications.
These and other initiatives promise to address the challenges discussed in this review. If the current pace of innovation continues, we might soon witness the emergence of an “ideal human lung model” that can provide both large-scale screens of molecular level analysis of cell-cell interactions, and at the same time provide a platform to study or treat lung disorders with a personalized medicine approach.
Acknowledgments
Acknowledgements
SvR was supported by a grant from the Lung Foundation Netherlands (grant # 6.1.14.010).
Competing interests
J.C.N., R.B., D.C. and R.V. are employees at Emulate, Inc. and hold equity in it.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “Human stem cell based organ-on-a-chip models for drug discovery and development”.
References
- 1.Green G.M., Jakab G.J., Low R.B., Davis G.S. Defense mechanisms of the respiratory membrane. Am. Rev. Respir. Dis. 1977;115:479–514. doi: 10.1164/arrd.1977.115.3.479. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization The top 10 causes of death. 2017. http://www.who.int/mediacentre/factsheets/fs310/en/ (available at)
- 3.World Health Organization Global alliance against chronic respiratory disease. 2005. www.who.int/gard/news_events/1-3.%0AGARD-06-07-K1.pdf (available at)
- 4.European Respiratory Society European Lung White Book. 2012. https://www.erswhitebook.org/chapters/the-economic-burden-of-lung-disease/
- 5.Forum of International Respiratory Societies Respiratory diseases in the world realities of today – opportunities for tomorrow. 2013. https://www.ersnet.org/pdf/publications/firs-world-report.pdf
- 6.Landrigan P.J., Fuller R., Acosta N.J.R., Adeyi O., Arnold R., Basu N., Baldé A.B., Bertollini R., Bose-O'Reilly S., Boufford J.I., Breysse P.N., Chiles T., Mahidol C., Coll-Seck A.M., Cropper M.L., Fobil J., Fuster V., Greenstone M., Haines A., Hanrahan D., Hunter D., Khare M., Krupnick A., Lanphear B., Lohani B., Martin K., Mathiasen K.V., McTeer M.A., Murray C.J.L., Ndahimananjara J.D., Perera F., Potočnik J., Preker A.S., Ramesh J., Rockström J., Salinas C., Samson L.D., Sandilya K., Sly P.D., Smith K.R., Steiner A., Stewart R.B., Suk W.A., van Schayck O.C.P., Yadama G.N., Yumkella K., Zhong M. The Lancet Commission on pollution and health. Lancet. 2017;6736 doi: 10.1016/S0140-6736(17)32345-0. [DOI] [PubMed] [Google Scholar]
- 7.Snoeck H.-W. Modeling human lung development and disease using pluripotent stem cells. Development. 2015;142:13–16. doi: 10.1242/dev.115469. [DOI] [PubMed] [Google Scholar]
- 8.Persson C.G.A. Con: mice are not a good model of human airway disease. Am. J. Respir. Crit. Care Med. 2002;166(6–7) doi: 10.1164/rccm.2204001. (discussion 8) [DOI] [PubMed] [Google Scholar]
- 9.Metzger R.J., Klein O.D., Martin G.R., Krasnow M.A. The branching program of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driskell R.R., Engelhardt J.F. Handbook of Stem Cells. Elsevier; 2004. pp. 547–554. in. [Google Scholar]
- 11.B.R., Zander D.S., Popper H.H., Jagirdar J., Haque A.K., Cagle P.T. vol. 1. Springer; New York, NY, Ney York, NY: 2008. Molecular Pathology of Lung Diseases. Molecular Pathology Library.http://link.springer.com/10.1007/978-0-387-72430-0_14 [Google Scholar]
- 12.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 13.Hawkins F., Kotton D.N. Embryonic and induced pluripotent stem cells for lung regeneration. Ann. Am. Thorac. Soc. 2015;12(Suppl. 1):S50–S53. doi: 10.1513/AnnalsATS.201410-457MG. [DOI] [PubMed] [Google Scholar]
- 14.Franks T.J., Colby T.V., Travis W.D., Tuder R.M., Reynolds H.Y., Brody A.R., Cardoso W.V., Crystal R.G., Drake C.J., Engelhardt J., Frid M., Herzog E., Mason R., Phan S.H., Randell S.H., Rose M.C., Stevens T., Serge J., Sunday M.E., Voynow J.A., Weinstein B.M., Whitsett J., Williams M.C. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc. Am. Thorac. Soc. 2008;5:763–766. doi: 10.1513/pats.200803-025HR. [DOI] [PubMed] [Google Scholar]
- 15.Evans M.J., Van Winkle L.S., Fanucchi M.V., Plopper C.G. The attenuated fibroblast sheath of the respiratory tract epithelial-mesenchymal trophic unit. Am. J. Respir. Cell Mol. Biol. 1999;21:655–657. doi: 10.1165/ajrcmb.21.6.3807. [DOI] [PubMed] [Google Scholar]
- 16.Boers J.E., Ambergen A.W., Thunnissen F.B. Number and proliferation of Clara cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 1999;159:1585–1591. doi: 10.1164/ajrccm.159.5.9806044. [DOI] [PubMed] [Google Scholar]
- 17.Ordoñez C.L., Khashayar R., Wong H.H., Ferrando R., Wu R., Hyde D.M., Hotchkiss J.A., Zhang Y., Novikov A., Dolganov G., Fahy J.V. Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am. J. Respir. Crit. Care Med. 2001;163:517–523. doi: 10.1164/ajrccm.163.2.2004039. [DOI] [PubMed] [Google Scholar]
- 18.Saetta M., Turato G., Baraldo S., Zanin A., Braccioni F., Mapp C.E., Maestrelli P., Cavallesco G., Papi A., Fabbri L.M. Goblet cell hyperplasia and epithelial inflammation in peripheral airways of smokers with both symptoms of chronic bronchitis and chronic airflow limitation. Am. J. Respir. Crit. Care Med. 2000;161:1016–1021. doi: 10.1164/ajrccm.161.3.9907080. [DOI] [PubMed] [Google Scholar]
- 19.H.B., Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwasaki A., Foxman E.F., Molony R.D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 2016;17:7–20. doi: 10.1038/nri.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirst S.J. Airway smooth muscle as a target in asthma. Clin. Exp. Allergy. 2000;30(Suppl. 1):54–59. [PubMed] [Google Scholar]
- 22.Ramos C., Montaño M., García-Alvarez J., Ruiz V., Uhal B.D., Selman M., Pardo A. Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am. J. Respir. Cell Mol. Biol. 2001;24:591–598. doi: 10.1165/ajrcmb.24.5.4333. [DOI] [PubMed] [Google Scholar]
- 23.Doeing D.C., Solway J. Airway smooth muscle in the pathophysiology and treatment of asthma. J. Appl. Physiol. 2013;114:834–843. doi: 10.1152/japplphysiol.00950.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kendall R.T., Feghali-Bostwick C.A. Fibroblasts in fibrosis: novel roles and mediators. Front. Pharmacol. 2014;5(May):1–13. doi: 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herriges M., Morrisey E.E. Lung development: orchestrating the generation and regeneration of a complex organ. Development. 2014;141:502–513. doi: 10.1242/dev.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith L.G., Swartz M.A. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 27.Leist M., Hartung T. Inflammatory findings on species extrapolations: humans are definitely no 70-kg mice. ALTEX. 2013;30:227–230. doi: 10.14573/altex.2013.2.227. [DOI] [PubMed] [Google Scholar]
- 28.Ogi C., Aruga A. Immunological monitoring of anticancer vaccines in clinical trials. Oncoimmunology. 2013;2 doi: 10.4161/onci.26012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaddanapudi K., Mitchell R.A., Eaton J.W., Ogi C., Aruga A. Cancer vaccines: looking to the future. Oncoimmunology. 2013;2 doi: 10.4161/onci.23403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullane K. The increasing challenge of discovering asthma drugs. Biochem. Pharmacol. 2011;82:586–599. doi: 10.1016/j.bcp.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 31.Pilewski J.M., Frizzell R.A. Role of CFTR in airway disease. Physiol. Rev. 1999;79:S215–S255. doi: 10.1152/physrev.1999.79.1.S215. [DOI] [PubMed] [Google Scholar]
- 32.Bella J., Rossmann M.G. Review: rhinoviruses and their ICAM receptors. J. Struct. Biol. 1999;128:69–74. doi: 10.1006/jsbi.1999.4143. [DOI] [PubMed] [Google Scholar]
- 33.Bartlett N.W., Walton R.P., Edwards M.R., Aniscenko J., Caramori G., Zhu J., Glanville N., Choy K.J., Jourdan P., Burnet J., Tuthill T.J., Pedrick M.S., Hurle M.J., Plumpton C., Sharp N.A., Bussell J.N., Swallow D.M., Schwarze J., Guy B., Almond J.W., Jeffery P.K., Lloyd C.M., Papi A., Killington R.A., Rowlands D.J., Blair E.D., Clarke N.J., Johnston S.L. Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nat. Med. 2008;14:199–204. doi: 10.1038/nm1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen A.L., Racaniello V.R. Selection of rhinovirus 1A variants adapted for growth in mouse lung epithelial cells. Virology. 2011;420:82–88. doi: 10.1016/j.virol.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byrd L.G., Prince G.A. Animal models of respiratory syncytial virus infection. Clin. Infect. Dis. 1997;25:1363–1368. doi: 10.1086/516152. [DOI] [PubMed] [Google Scholar]
- 36.Holmes A.M., Solari R., Holgate S.T. Animal models of asthma: value, limitations and opportunities for alternative approaches. Drug Discov. Today. 2011;16:659–670. doi: 10.1016/j.drudis.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345 doi: 10.1126/science.1247125. (1247125-1247125, 80-.) [DOI] [PubMed] [Google Scholar]
- 38.Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., Hogan B.L.M. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tata P.R., Mou H., Pardo-Saganta A., Zhao R., Prabhu M., Law B.M., Vinarsky V., Cho J.L., Breton S., Sahay A., Medoff B.D., Rajagopal J. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y.-W., Huang S.X., de Carvalho A.L.R.T., Ho S.-H., Islam M.N., Volpi S., Notarangelo L.D., Ciancanelli M., Casanova J.-L., Bhattacharya J., Liang A.F., Palermo L.M., Porotto M., Moscona A., Snoeck H.-W. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017;19:542–549. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 42.Dvorak A., Tilley A.E., Shaykhiev R., Wang R., Crystal R.G. Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am. J. Respir. Cell Mol. Biol. 2011;44:465–473. doi: 10.1165/rcmb.2009-0453OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross A.J., Dailey L.A., Brighton L.E., Devlin R.B. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2007;37(2):169–185. doi: 10.1165/rcmb.2006-0466OC. [DOI] [PubMed] [Google Scholar]
- 44.Gray T.E., Guzman K., Davis C.W., Abdullah L.H., Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- 45.Fulcher M.L., Gabriel S., Burns K.A., Yankaskas J.R., Randell S.H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 46.Crystal R.G., Randell S.H., Engelhardt J.F., Voynow J., Sunday M.E. Airway epithelial cells: current concepts and challenges. Proc. Am. Thorac. Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livraghi A., Randell S.H. Cystic fibrosis and other respiratory diseases of impaired mucus clearance. Toxicol. Pathol. 2007;35:116–129. doi: 10.1080/01926230601060025. [DOI] [PubMed] [Google Scholar]
- 48.Malavia N.K., Raub C.B., Mahon S.B., Brenner M., Panettieri R.A., George S.C. Airway epithelium stimulates smooth muscle proliferation. Am. J. Respir. Cell Mol. Biol. 2009;41:297–304. doi: 10.1165/rcmb.2008-0358OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matrosovich M.N., Matrosovich T.Y., Gray T., Roberts N.A., Klenk H.D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Hsin H.Y., Ingber D.E. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huh D., Hamilton G.A., Ingber D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benam K.H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H.-H., Alves S.E., Salmon M., Ferrante T.C., Weaver J.C., Bahinski A., Hamilton G.A., Ingber D.E. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 53.Dunsmore S.E., Rannels D.E. Extracellular matrix biology in the lung. Am. J. Phys. 1996;270:L3–27. doi: 10.1152/ajplung.1996.270.1.L3. [DOI] [PubMed] [Google Scholar]
- 54.Bottaro D.P., Liebmann-Vinson A., Heidaran M.A. Molecular signaling in bioengineered tissue microenvironments. Ann. N. Y. Acad. Sci. 2002;961:143–153. doi: 10.1111/j.1749-6632.2002.tb03068.x. [DOI] [PubMed] [Google Scholar]
- 55.Alford A.I., Rannels D.E. Extracellular matrix fibronectin alters connexin43 expression by alveolar epithelial cells. Am. J. Phys. Lung Cell. Mol. Phys. 2001;280:L680–L688. doi: 10.1152/ajplung.2001.280.4.L680. [DOI] [PubMed] [Google Scholar]
- 56.Clark R.A., Mason R.J., Folkvord J.M., McDonald J.A. Fibronectin mediates adherence of rat alveolar type II epithelial cells via the fibroblastic cell-attachment domain. J. Clin. Invest. 1986;77:1831–1840. doi: 10.1172/JCI112509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeQuach J.A., Mezzano V., Miglani A., Lange S., Keller G.M., Sheikh F., Christman K.L. Simple and high yielding method for preparing tissue specific extracellular matrix coatings for cell culture. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serini G., Bochaton-Piallat M.L., Ropraz P., Geinoz A., Borsi L., Zardi L., Gabbiani G. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J. Cell Biol. 1998;142(873–81) doi: 10.1083/jcb.142.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu F., Mih J.D., Shea B.S., Kho A.T., Sharif A.S., Tager A.M., Tschumperlin D.J. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J. Cell Biol. 2010;190:693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mercer R.R., Crapo J.D. Spatial distribution of collagen and elastin fibers in the lungs. J. Appl. Physiol. 1990;69:756–765. doi: 10.1152/jappl.1990.69.2.756. [DOI] [PubMed] [Google Scholar]
- 61.Balestrini J.L., Niklason L.E. Extracellular matrix as a driver for lung regeneration. Ann. Biomed. Eng. 2015;43:568–576. doi: 10.1007/s10439-014-1167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomos I.P., Tzouvelekis A., Aidinis V., Manali E.D., Bouros E., Bouros D., Papiris S.A. Extracellular matrix remodeling in idiopathic pulmonary fibrosis. It is the “bed” that counts and not “the sleepers”. Expert Rev. Respir. Med. 2017;11:299–309. doi: 10.1080/17476348.2017.1300533. [DOI] [PubMed] [Google Scholar]
- 63.Sakai N., Tager A.M. Fibrosis of two: epithelial cell-fibroblast interactions in pulmonary fibrosis. Biochim. Biophys. Acta Mol. basis Dis. 2013;1832:911–921. doi: 10.1016/j.bbadis.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 65.Teijaro J.R., Walsh K.B., Cahalan S., Fremgen D.M., Roberts E., Scott F., Martinborough E., Peach R., Oldstone M.B.A., Rosen H. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee J., Bhang D.H., Beede A., Huang T.L., Stripp B.R., Bloch K.D., Wagers A.J., Tseng Y., Ryeom S., Kim C.F. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huh D., Kim H.J., Fraser J.P., Shea D.E., Khan M., Bahinski A., Hamilton G.A., Ingber DE. Microfabrication of human organs-on-chips. Nat Protoc. 2013;8(11):2135–2157. doi: 10.1038/nprot.2013.137. https://www.ncbi.nlm.nih.gov/pubmed/24113786 [DOI] [PubMed] [Google Scholar]
- 68.Waters C.M., Roan E., Navajas D. Mechanobiology in lung epithelial cells: measurements, perturbations, and responses. Compr. Physiol. 2012;2:1–29. doi: 10.1002/cphy.c100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Forrest J. The effect of changes in lung volume on the size and shape of alveoli. J. Physiol. 1970;210:533–547. doi: 10.1113/jphysiol.1970.sp009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fredberg J.J., Kamm R.D. Stress transmission in the lung: pathways from organ to molecule. Annu. Rev. Physiol. 2006;68:507–541. doi: 10.1146/annurev.physiol.68.072304.114110. [DOI] [PubMed] [Google Scholar]
- 71.Fredberg J.J., Inouye D., Miller B., Nathan M., Jafari S., Helioui Raboudi S., Butler J.P., Shore S.A. Airway smooth muscle, tidal stretches, and dynamically determined contractile states. Am. J. Respir. Crit. Care Med. 1997;156:1752–1759. doi: 10.1164/ajrccm.156.6.9611016. [DOI] [PubMed] [Google Scholar]
- 72.Grainge C.L., Lau L.C.K., Ward J.A., Dulay V., Lahiff G., Wilson S., Holgate S., Davies D.E., Howarth P.H. Effect of bronchoconstriction on airway remodeling in asthma. N. Engl. J. Med. 2011;364:2006–2015. doi: 10.1056/NEJMoa1014350. [DOI] [PubMed] [Google Scholar]
- 73.Sanchez-Esteban J., Cicchiello L.A., Wang Y., Tsai S.W., Williams L.K., Torday J.S., Rubin L.P. Mechanical stretch promotes alveolar epithelial type II cell differentiation. J. Appl. Physiol. 2001;91:589–595. doi: 10.1152/jappl.2001.91.2.589. [DOI] [PubMed] [Google Scholar]
- 74.Edwards Y.S. Stretch stimulation: its effects on alveolar type II cell function in the lung. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001;129:245–260. doi: 10.1016/s1095-6433(01)00321-x. [DOI] [PubMed] [Google Scholar]
- 75.Arold S.P., Bartolák-Suki E., Suki B. Variable stretch pattern enhances surfactant secretion in alveolar type II cells in culture. Am. J. Phys. Lung Cell. Mol. Phys. 2009;296:L574–L581. doi: 10.1152/ajplung.90454.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu M., Tanswell A.K., Post M. Mechanical force-induced signal transduction in lung cells. Am. J. Phys. 1999;277:L667–L683. doi: 10.1152/ajplung.1999.277.4.L667. [DOI] [PubMed] [Google Scholar]
- 77.Davies P.F., Dewey C.F., Bussolari S.R., Gordon E.J., Gimbrone M.A. Influence of hemodynamic forces on vascular endothelial function. In vitro studies of shear stress and pinocytosis in bovine aortic cells. J. Clin. Invest. 1984;73:1121–1129. doi: 10.1172/JCI111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levesque M.J., Nerem R.M., Sprague E.A. Vascular endothelial cell proliferation in culture and the influence of flow. Biomaterials. 1990;11:702–707. doi: 10.1016/0142-9612(90)90031-k. [DOI] [PubMed] [Google Scholar]
- 79.Davies P.F. Flow-mediated endothelial mechanotransduction. Physiol. Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stucki A.O., Stucki J.D., Hall S.R.R., Felder M., Mermoud Y., Schmid R.A., Geiser T., Guenat O.T. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip. 2015;15:1302–1310. doi: 10.1039/c4lc01252f. [DOI] [PubMed] [Google Scholar]
- 81.Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A., Thorneloe K.S., McAlexander M.A., Ingber D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jain A., Barrile R., van der Meer A.D., Mammoto A., Mammoto T., De Ceunynck K., Aisiku O., Otieno M.A., Louden C.S., Hamilton G.A., Flaumenhaft R., Ingber D.E. Primary human lung alveolus-on-a-chip model of intravascular thrombosis for assessment of therapeutics. Clin. Pharmacol. Ther. 2017 doi: 10.1002/cpt.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCulley D., Wienhold M., Sun X. The pulmonary mesenchyme directs lung development. Curr. Opin. Genet. Dev. 2015;32:98–105. doi: 10.1016/j.gde.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Whitsett J.A., Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 2014;16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fisher A.B. Normal and pathologic biochemistry of the lung. Environ. Health Perspect. 1976;16:3–9. doi: 10.1289/ehp.76163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hukkanen J., Pelkonen O., Hakkola J., Raunio H. Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit. Rev. Toxicol. 2002;32:391–411. doi: 10.1080/20024091064273. [DOI] [PubMed] [Google Scholar]
- 87.Boei J.J.W.A., Vermeulen S., Klein B., Hiemstra P.S., Verhoosel R.M., Jennen D.G.J., Lahoz A., Gmuender H., Vrieling H. Xenobiotic metabolism in differentiated human bronchial epithelial cells. Arch. Toxicol. 2017;91:2093–2105. doi: 10.1007/s00204-016-1868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kopf M., Schneider C., Nobs S.P. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 2015;16:36–44. doi: 10.1038/ni.3052. [DOI] [PubMed] [Google Scholar]
- 89.Seo J., Conegliano D., Farrell M., Cho M., Ding X., Seykora T., Qing D., Mangalmurti N.S., Huh D. A microengineered model of RBC transfusion-induced pulmonary vascular injury. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-03597-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jain A., van der Meer A.D., Papa A.-L., Barrile R., Lai A., Schlechter B.L., Otieno M.A., Louden C.S., Hamilton G.A., Michelson A.D., Frelinger A.L., Ingber D.E. Assessment of whole blood thrombosis in a microfluidic device lined by fixed human endothelium. Biomed. Microdevices. 2016;18(73) doi: 10.1007/s10544-016-0095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Manz A., Widmers H.M., Graber N. Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sensors Actuators B Chem. 1990;1:244–248. [Google Scholar]
- 92.van den Berg A., Bergveld P. 1995. Proceedings of the μTAS; p. 1994. [Google Scholar]
- 93.Kopp M.U., De Mello A.J., Manz A. Chemical amplification: continuous flow PCR on a chip. Science. 1998;280:1046–1048. doi: 10.1126/science.280.5366.1046. (80-.) [DOI] [PubMed] [Google Scholar]
- 94.Daw R., Finkelstein J. Lab on a chip. Nature. 2006;442:367-367. [Google Scholar]
- 95.Duffy D.C., McDonald J.C., Schueller O.J.A., Whitesides G.M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 96.Folch A., Toner M. Microengineering of cellular interactions. Annu. Rev. Biomed. Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 97.Folch A., Ayon A., Hurtado O., Schmidt M.A., Toner M. Molding of deep polydimethylsiloxane microstructures for microfluidics and biological applications. J. Biomech. Eng. 1999;121:28–34. doi: 10.1115/1.2798038. [DOI] [PubMed] [Google Scholar]
- 98.Bhatia S.N., Balis U.J., Yarmush M.L., Toner M. Effect of cell–cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 99.Bhatia S.N., Yarmush M.L., Toner M. Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts. J. Biomed. Mater. Res. 1997;34:189–199. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 100.El-Ali J., Sorger P.K., Jensen K.F. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 101.Beebe D.J., Moore J.S., Bauer J.M., Yu Q., Liu R.H., Devadoss C., Jo B.-H. Functional hydrogel structures for autonomous flow control inside microfluidic channels: abstract: nature. Nature. 2000;404:588–590. doi: 10.1038/35007047. [DOI] [PubMed] [Google Scholar]
- 102.Whitesides G.M., Ostuni E., Takayama S., Jiang X., Ingber D.E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 103.Quake S.R. From micro- to nanofabrication with soft materials. Science. 2000;290:1536–1540. doi: 10.1126/science.290.5496.1536. (80-.) [DOI] [PubMed] [Google Scholar]
- 104.McDonald J.C., Whitesides G.M. Poly (dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 105.Langer R., Vacanti J.P. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. (80-.) [DOI] [PubMed] [Google Scholar]
- 106.Khademhosseini A., Bong G.C. 2009 IEEE/NIH Life Sci. Syst. Appl. Work. LiSSA 2009. Vol. 103. 2009. Microscale technologies for tissue engineering; pp. 56–57. [Google Scholar]
- 107.Borenstein J.T., Terai H., King K.R., Weinberg E.J., Kaazempur-Mofrad M.R., Vacanti J.P. Microfabrication technology for vascularized tissue engineering. Biomed. Microdevices. 2002;4:167–175. [Google Scholar]
- 108.Kaihara S., Borenstein J., Koka R., Lalan S., Ochoa E.R., Ravens M., Pien H., Cunningham B., Vacanti J.P. Silicon micromachining to tissue engineer branched vascular channels for liver fabrication. Tissue Eng. 2000;6:105–117. doi: 10.1089/107632700320739. [DOI] [PubMed] [Google Scholar]
- 109.Powers M.J., Domansky K., Kaazempur-Mofrad M.R., Kalezi A., Capitano A., Upadhyaya A., Kurzawski P., Wack K.E., Stolz D.B., Kamm R., Griffith L.G. A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol. Bioeng. 2002;78:257–269. doi: 10.1002/bit.10143. [DOI] [PubMed] [Google Scholar]
- 110.Leclerc E., Sakai Y., Fujii T. Cell culture in 3-dimensional microfluidic structure of PDMS (polydimenthylsiloxane) Biomed. Microdevices. 2003;5:109–114. [Google Scholar]
- 111.Lee P.J., Hung P.J., Lee L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 112.Park J., Berthiaume F., Toner M., Yarmush M.L., Tilles A.W. Microfabricated grooved substrates as platforms for bioartificial liver reactors. Biotechnol. Bioeng. 2005;90:632–644. doi: 10.1002/bit.20463. [DOI] [PubMed] [Google Scholar]
- 113.Lam M.T., Huang Y.-C., Birla R.K., Takayama S. Microfeature guided skeletal muscle tissue engineering for highly organized 3-dimensional free-standing constructs. Biomaterials. 2009;30:1150–1155. doi: 10.1016/j.biomaterials.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 114.Jang K., Sato K., Igawa K., Chung U., Kitamori T. Development of an osteoblast-based 3D continuous-perfusion microfluidic system for drug screening. Anal. Bioanal. Chem. 2008;390:825–832. doi: 10.1007/s00216-007-1752-7. [DOI] [PubMed] [Google Scholar]
- 115.Park J.W., Vahidi B., Taylor A.M., Rhee S.W., Jeon N.L. Microfluidic culture platform for neuroscience research. Nat. Protoc. 2006;1:2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 116.Mahler G.J., Esch M.B., Glahn R.P., Shuler M.L. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- 117.Baudoin R., Griscom L., Monge M., Legallais C., Leclerc E. Development of a renal microchip for in vitro distal tubule models. Biotechnol. Prog. 2007;23:0-0. doi: 10.1021/bp0603513. [DOI] [PubMed] [Google Scholar]
- 118.Jang K.-J., Suh K.-Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip. 2010;10:36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 119.Sin A., Chin K.C., Jamil M.F., Kostov Y., Rao G., Shuler M.L. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol. Prog. 2004;20:338–345. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- 120.Sung J.H., Srinivasan B., Esch M.B., McLamb W.T., Bernabini C., Shuler M.L., Hickman J.J. Using PBPK guided “body on a Chip” systems to predict mammalian response to drug and chemical exposure. Exp. Biol. Med. 2014;239:1225–1239. doi: 10.1177/1535370214529397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Douville N.J., Zamankhan P., Tung Y.-C., Li R., Vaughan B.L., Tai C.-F., White J., Christensen P.J., Grotberg J.B., Takayama S. Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab Chip. 2011;11:609–619. doi: 10.1039/c0lc00251h. [DOI] [PubMed] [Google Scholar]
- 122.Huh D., Fujioka H., Tung Y.-C., Futai N., Paine R., Grotberg J.B., Takayama S. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc. Natl. Acad. Sci. 2007;104:18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nalayanda D.D., Puleo C.M., Fulton W.B., Wang T.H., Abdullah F. Characterization of pulmonary cell growth parameters in a continuous perfusion microfluidic environment. Exp. Lung Res. 2007;33:321–335. doi: 10.1080/01902140701557754. [DOI] [PubMed] [Google Scholar]
- 124.Hassell B.A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C.S., Ingber D.E. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–516. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 125.Villenave R., Lucchesi C., Lee H.-H., Nguyen J., Varone A., Karalis K., Alves S., Salmon M., Hamilton G. European Respiratory Society; 2017. Respiratory Infections; p. PA4136. [Google Scholar]
- 126.Benam K.H., Novak R., Nawroth J., Villenave R., Hirano-Kobayashi M., Lucchesi C., Ferrante T.C., Choe Y.-J., Prantil-Braun R., Weaver J.C., Parker K.K., Bahinski A., Ingber D.E. Matched-comparative modeling of normal and COPD human airway responses to inhaled smoke in vitro. Cell Syst. 2016;3:456–466. doi: 10.1016/j.cels.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 127.Nesmith A.P., Agarwal A., McCain M.L., Parker K.K. Human airway musculature on a chip: an in vitro model of allergic asthmatic bronchoconstriction and bronchodilation. Lab Chip. 2014;14:3925–3936. doi: 10.1039/c4lc00688g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sellgren K.L., Butala E.J., Gilmour B.P., Randell S.H., Grego S. A biomimetic multicellular model of the airways using primary human cells. Lab Chip. 2014:3349–3358. doi: 10.1039/c4lc00552j. [DOI] [PubMed] [Google Scholar]
- 129.Wanner A., Salathé M., O'Riordan T.G. Mucociliary clearance in the airways. Am. J. Respir. Crit. Care Med. 1996;154:1868–1902. doi: 10.1164/ajrccm.154.6.8970383. [DOI] [PubMed] [Google Scholar]
- 130.Mercer R.R., Russell M.L., Roggli V.L., Crapo J.D. Cell number and distribution in human and rat airways. Am. J. Respir. Cell Mol. Biol. 1994;10:613–624. doi: 10.1165/ajrcmb.10.6.8003339. [DOI] [PubMed] [Google Scholar]
- 131.Barkal L.J., Procknow C.L., Álvarez-García Y.R., Niu M., Jiménez-Torres J.A., Brockman-Schneider R.A., Gern J.E., Denlinger L.C., Theberge A.B., Keller N.P., Berthier E., Beebe D.J. Microbial volatile communication in human organotypic lung models. Nat. Commun. 2017;8(1770) doi: 10.1038/s41467-017-01985-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hughes P., Marshall D., Reid Y., Parkes H., Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques. 2007;575(43):577–578. doi: 10.2144/000112598. (581–2 passim) [DOI] [PubMed] [Google Scholar]
- 133.Butler C.R., Hynds R.E., Gowers K.H.C., Lee D.D.H., Brown J.M., Crowley C., Teixeira V.H., Smith C.M., Urbani L., Hamilton N.J., Thakrar R.M., Booth H.L., Birchall M.A., De Coppi P., Giangreco A., O'Callaghan C., Janes S.M. Rapid expansion of human epithelial stem cells suitable for airway tissue engineering. Am. J. Respir. Crit. Care Med. 2016;194:156–168. doi: 10.1164/rccm.201507-1414OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mou H., Vinarsky V., Tata P.R., Brazauskas K., Choi S.H., Crooke A.K., Zhang B., Solomon G.M., Turner B., Bihler H., Harrington J., Lapey A., Channick C., Keyes C., Freund A., Artandi S., Mense M., Rowe S., Engelhardt J.F., Hsu Y.C., Rajagopal J. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell. 2016;19:217–231. doi: 10.1016/j.stem.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kadzik R.S., Morrisey E.E. Directing lung endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2012;10:355–361. doi: 10.1016/j.stem.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.D'Amour K.A., Bang A.G., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., Baetge E.E. Production of pancreatic hormone–expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 137.Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., Shroyer N.F., Wells J.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cai J., Zhao Y., Liu Y., Ye F., Song Z., Qin H., Meng S., Chen Y., Zhou R., Song X., Guo Y., Ding M., Deng H. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 139.Green M.D., Chen A., Nostro M.-C., D'Souza S.L., Schaniel C., Lemischka I.R., Gouon-Evans V., Keller G., Snoeck H.-W. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 141.Morrisey E.E., Hogan B.L.M. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kimura S., Hara Y., Pineau T., Fernandez-Salguero P., Fox C.H., Ward J.M., Gonzalez F.J. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 143.Xu Y., Mizuno T., Sridharan A., Du Y., Guo M., Tang J., Wikenheiser-Brokamp K.A., Perl A.-K.T., Funari V.A., Gokey J.J., Stripp B.R., Whitsett J.A. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:1–18. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gonzales L.W., Guttentag S.H., Wade K.C., Postle A.D., Ballard P.L. Differentiation of human pulmonary type II cells in vitro by glucocorticoid plus cAMP. Am. J. Phys. Lung Cell. Mol. Phys. 2002;283:L940–L951. doi: 10.1152/ajplung.00127.2002. [DOI] [PubMed] [Google Scholar]
- 145.Wade K.C., Guttentag S.H., Gonzales L.W., Maschhoff K.L., Gonzales J., Kolla V., Singhal S., Ballard P.L. Gene induction during differentiation of human pulmonary type II cells in vitro. Am. J. Respir. Cell Mol. Biol. 2006;34:727–737. doi: 10.1165/rcmb.2004-0389OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Andreeva A.V., Kutuzov M.A., Voyno-Yasenetskaya T.A. Regulation of surfactant secretion in alveolar type II cells. Am. J. Phys. Lung Cell. Mol. Phys. 2007;293:L259–L271. doi: 10.1152/ajplung.00112.2007. [DOI] [PubMed] [Google Scholar]
- 147.Huang S.X.L., Green M.D., de Carvalho A.T., Mumau M., Chen Y.-W., D'Souza S.L., Snoeck H.-W. The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc. 2015;10:413–425. doi: 10.1038/nprot.2015.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gotoh S., Ito I., Nagasaki T., Yamamoto Y., Konishi S., Korogi Y., Matsumoto H., Muro S., Hirai T., Funato M., Mae S.I., Toyoda T., Sato-Otsubo A., Ogawa S., Osafune K., Mishima M. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Rep. 2014;3:394–403. doi: 10.1016/j.stemcr.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jacob A., Morley M., Hawkins F., McCauley K.B., Jean J.C., Heins H., Na C.-L., Weaver T.E., Vedaie M., Hurley K., Hinds A., Russo S.J., Kook S., Zacharias W., Ochs M., Traber K., Quinton L.J., Crane A., Davis B.R., White F.V., Wambach J., Whitsett J.A., Cole F.S., Morrisey E.E., Guttentag S.H., Beers M.F., Kotton D.N. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell. 2017;21:472–488.e10. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yamamoto Y., Gotoh S., Korogi Y., Seki M., Konishi S., Ikeo S., Sone N., Nagasaki T., Matsumoto H., Muro S., Ito I., Hirai T., Kohno T., Suzuki Y., Mishima M. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat. Methods. 2017;14:1097–1106. doi: 10.1038/nmeth.4448. [DOI] [PubMed] [Google Scholar]
- 151.Konishi S., Gotoh S., Tateishi K., Yamamoto Y., Korogi Y., Nagasaki T., Matsumoto H., Muro S., Hirai T., Ito I., Tsukita S., Mishima M. Directed induction of functional multi-ciliated cells in proximal airway epithelial spheroids from human pluripotent stem cells. Stem Cell Rep. 2016;6:18–25. doi: 10.1016/j.stemcr.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell. 2017;20:844–857.e6. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wong A.A.P., Bear C.C.E., Chin S., Pasceri P., Thompson T.T.O., Huan L.-J.L.-J., Ratjen F., Ellis J., Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Firth A.L., Dargitz C.T., Qualls S.J., Menon T., Wright R., Singer O., Gage F.H., Khanna A., Verma I.M. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. 2014;111:E1723–E1730. doi: 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wong A.P., Chin S., Xia S., Garner J., Bear C.E., Rossant J. Efficient generation of functional CFTR-expressing airway epithelial cells from human pluripotent stem cells. Nat. Protoc. 2015;10:363–381. doi: 10.1038/nprot.2015.021. [DOI] [PubMed] [Google Scholar]
- 156.Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.-H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D., White E.S., Deutsch G.H., Spence J.R. In vitro generation of human pluripotent stem cell derived lung organoids. elife. 2015;4:1–25. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dye B.R., Dedhia P.H., Miller A.J., Nagy M.S., White E.S., Shea L.D., Spence J.R. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. elife. 2016;5:1–18. doi: 10.7554/eLife.19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nikolić M.Z., Caritg O., Jeng Q., Johnson J.-A., Sun D., Howell K.J., Brady J.L., Laresgoiti U., Allen G., Butler R., Zilbauer M., Giangreco A., Rawlins E.L. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. elife. 2017;6:1–33. doi: 10.7554/eLife.26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huang S.X.L., Islam M.N., O'Neill J., Hu Z., Yang Y.-G., Chen Y.-W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., Snoeck H.-W. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Serra M., Alysandratos K.-D., Hawkins F., McCauley K.B., Jacob A., Choi J., Caballero I.S., Vedaie M., Kurmann A.A., Ikonomou L., Hollenberg A.N., Shannon J.M., Kotton D.N. Pluripotent stem cell differentiation reveals distinct developmental pathways regulating lung- versus thyroid-lineage specification. Development. 2017;144:3879–3893. doi: 10.1242/dev.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hawkins F., Kramer P., Jacob A., Driver I., Thomas D.C., McCauley K.B., Skvir N., Crane A.M., Kurmann A.A., Hollenberg A.N., Nguyen S., Wong B.G., Khalil A.S., Huang S.X.L., Guttentag S., Rock J.R., Shannon J.M., Davis B.R., Kotton D.N. Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells. J. Clin. Invest. 2017;127:2277–2294. doi: 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Beers M.F., Moodley Y. When is an alveolar type 2 cell an alveolar type 2 cell? A conundrum for lung stem cell biology and regenerative medicine. Am. J. Respir. Cell Mol. Biol. 2017;57:1–31. doi: 10.1165/rcmb.2016-0426PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ghaedi M., Calle E.A., Mendez J.J., Gard A.L., Balestrini J., Booth A., Bove P.F., Gui L., White E.S., Niklason L.E. Technical advance human iPS cell – derived alveolar epithelium repopulates lung extracellular matrix. J. Clin. Densitom. 2013;123:4950. doi: 10.1172/JCI68793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Barkauskas C.E., Chung M.-I., Fioret B., Gao X., Katsura H., Hogan B.L.M. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vining K.H., Mooney D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Robinton D.A., Daley G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Somers A., Jean J.-C., Sommer C.A., Omari A., Ford C.C., Mills J.A., Ying L., Sommer A.G., Jean J.M., Smith B.W., Lafyatis R., Demierre M.-F., Weiss D.J., French D.L., Gadue P., Murphy G.J., Mostoslavsky G., Kotton D.N. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bellin M., Marchetto M.C., Gage F.H., Mummery C.L. Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- 169.Spagnolo P., du Bois R.M., Cottin V. Rare lung disease and orphan drug development. Lancet Respir. Med. 2013;1:479–487. doi: 10.1016/S2213-2600(13)70085-7. [DOI] [PubMed] [Google Scholar]
- 170.Elborn J.S. Cystic fibrosis. Lancet (London, England) 2016;388:2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- 171.Jaffé A., Bush A. Cystic fibrosis: review of the decade. Monaldi Arch. Chest Dis. 2001;56:240–247. (= Arch. Monaldi per le Mal. del torace) [PubMed] [Google Scholar]
- 172.Griesenbach U., Boyd A.C. UK cystic fibrosis gene therapy consortium, pre-clinical and clinical endpoint assays for cystic fibrosis gene therapy. J. Cyst. Fibros. 2005;4:89–100. doi: 10.1016/j.jcf.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 173.Pier G.B. The challenges and promises of new therapies for cystic fibrosis. J. Exp. Med. 2012;209:1235–1239. doi: 10.1084/jem.20121248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ratjen F., Döring G. Cystic fibrosis. Lancet (London, England) 2003;361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 175.Rozmahel R., Wilschanski M., Matin A., Plyte S., Oliver M., Auerbach W., Moore A., Forstner J., Durie P., Nadeau J., Bear C., Tsui L.C. Modulation of disease severity in cystic fibrosis transmembrane conductance regulator deficient mice by a secondary genetic factor. Nat. Genet. 1996;12:280–287. doi: 10.1038/ng0396-280. [DOI] [PubMed] [Google Scholar]
- 176.Lavelle G.M., White M.M., Browne N., McElvaney N.G., Reeves E.P. Animal models of cystic fibrosis pathology: phenotypic parallels and divergences. Biomed. Res. Int. 2016;2016:1–14. doi: 10.1155/2016/5258727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration. 2000;67:117–133. doi: 10.1159/000029497. [DOI] [PubMed] [Google Scholar]
- 178.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat. Rev. Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Konar D., Devarasetty M., Yildiz D.V., Atala A., Murphy S.V. Lung-On-A-Chip technologies for disease modeling and drug development. Biomed. Eng. Comput. Biol. 2016;7:17–27. doi: 10.4137/BECB.S34252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Huh D. (Dan) A human breathing Lung-on-a-Chip. Ann. Am. Thorac. Soc. 2015;12:S42–S44. doi: 10.1513/AnnalsATS.201410-442MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rada B. Interactions between neutrophils and Pseudomonas aeruginosa in cystic fibrosis. Pathog. (Basel, Switzerland) 2017;6 doi: 10.3390/pathogens6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.De Boeck K., Kent L., Davies J., Derichs N., Amaral M., Rowe S.M., Middleton P., de Jonge H., Bronsveld I., Wilschanski M., Melotti P., Danner-Boucher I., Boerner S., Fajac I., Southern K., de Nooijer R.A., Bot A., de Rijke Y., de Wachter E., Leal T., Vermeulen F., Hug M.J., Rault G., Nguyen-Khoa T., Barreto C., Proesmans M., Sermet-Gaudelus I. European cystic fibrosis society clinical trial network standardisation committee, CFTR biomarkers: time for promotion to surrogate end-point. Eur. Respir. J. 2013;41:203–216. doi: 10.1183/09031936.00057512. [DOI] [PubMed] [Google Scholar]
- 183.Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W.M., Bijvelds M.J.C., Scholte B.J., Nieuwenhuis E.E.S., van den Brink S., Clevers H., van der Ent C.K., Middendorp S., Beekman J.M. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 184.Ahmadi S., Bozoky Z., Di Paola M., Xia S., Li C., Wong A.P., Wellhauser L., Molinski S.V., Ip W., Ouyang H., Avolio J., Forman-Kay J.D., Ratjen F., Hirota J.A., Rommens J., Rossant J., Gonska T., Moraes T.J., Bear C.E. Phenotypic profiling of CFTR modulators in patient-derived respiratory epithelia. NPJ Genomic Med. 2017;2(12) doi: 10.1038/s41525-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.S. M. Rowe, J. P. Clancy, M. Wilschanski, (in) (2011), pp. 69–86. [DOI] [PMC free article] [PubMed]
- 186.Schöni M.H., Nikolaizik W.H. Aerosol therapy in patients with cystic fibrosis. Schweiz. Med. Wochenschr. 1997;127:158–164. [PubMed] [Google Scholar]
- 187.Quon B.S., Rowe S.M. New and emerging targeted therapies for cystic fibrosis. BMJ. 2016:i859. doi: 10.1136/bmj.i859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Maiuri L., Raia V., Kroemer G. Strategies for the etiological therapy of cystic fibrosis. Cell Death Differ. 2017;24:1825–1844. doi: 10.1038/cdd.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K., Nieuwenhuis E.E.S., Beekman J.M., Clevers H. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 190.Montiel-Gonzalez M.F., Vallecillo-Viejo I., Yudowski G.A., Rosenthal J.J.C. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18285–18290. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Whitsett J.A., Wert S.E., Weaver T.E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 2010;61:105–119. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.GBD 2015 Chronic Respiratory Disease Collaborators, Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir. Med. 2017;5:691–706. doi: 10.1016/S2213-2600(17)30293-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Balkissoon R., Lommatzsch S., Carolan B., Make B. Chronic obstructive pulmonary disease: a concise review. Med. Clin. N. Am. 2011;95:1125–1141. doi: 10.1016/j.mcna.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 194.Olin J.T., Wechsler M.E. Asthma: pathogenesis and novel drugs for treatment. BMJ. 2014;349:g5517. doi: 10.1136/bmj.g5517. [DOI] [PubMed] [Google Scholar]
- 195.Barnes P.J., Burney P.G.J., Silverman E.K., Celli B.R., Vestbo J., Wedzicha J.A., Wouters E.F.M. Chronic obstructive pulmonary disease. Nat. Rev. Dis. Prim. 2015;11:507–513. doi: 10.1038/nrdp.2015.76. [DOI] [PubMed] [Google Scholar]
- 196.McCracken J.L., Veeranki S.P., Ameredes B.T., Calhoun W.J. Diagnosis and management of asthma in adults: a review. JAMA. 2017;318:279–290. doi: 10.1001/jama.2017.8372. [DOI] [PubMed] [Google Scholar]
- 197.Blume C., Davies D.E. In vitro and ex vivo models of human asthma. Eur. J. Pharm. Biopharm. 2013;84:394–400. doi: 10.1016/j.ejpb.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 198.Epstein M.M. Do mouse models of allergic asthma mimic clinical disease? Int. Arch. Allergy Immunol. 2004;133:84–100. doi: 10.1159/000076131. [DOI] [PubMed] [Google Scholar]
- 199.Shin Y.S., Takeda K., Gelfand E.W. Understanding asthma using animal models. Allergy, Asthma Immunol. Res. 2009;1:10–18. doi: 10.4168/aair.2009.1.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bates J.H.T., Rincon M., Irvin C.G. Animal models of asthma. Am. J. Phys. Lung Cell. Mol. Phys. 2009;297:L401–L410. doi: 10.1152/ajplung.00027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Canning B.J., Wright J.L. Animal models of asthma and chronic obstructive pulmonary disease. Pulm. Pharmacol. Ther. 2008;21:695. doi: 10.1016/j.pupt.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 202.Fehrenbach H. Animal models of pulmonary emphysema: a stereologist's perspective. Eur. Respir. Rev. 2006;15:136–147. [Google Scholar]
- 203.Martin J.G., Ramos-Barbón D. Airway smooth muscle growth from the perspective of animal models. Respir. Physiol. Neurobiol. 2003;137:251–261. doi: 10.1016/s1569-9048(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 204.Guseh J.S., Bores S.A., Stanger B.Z., Zhou Q., Anderson W.J., Melton D.A., Rajagopal J. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development. 2009;136:1751–1759. doi: 10.1242/dev.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bayram H., Devalia J.L., Khair O.A., Abdelaziz M.M., Sapsford R.J., Sagai M., Davies R.J. Comparison of ciliary activity and inflammatory mediator release from bronchial epithelial cells of nonatopic nonasthmatic subjects and atopic asthmatic patients and the effect of diesel exhaust particles in vitro. J. Allergy Clin. Immunol. 1998;102:771–782. doi: 10.1016/s0091-6749(98)70017-x. [DOI] [PubMed] [Google Scholar]
- 206.Woodruff P.G., Boushey H.A., Dolganov G.M., Barker C.S., Yang Y.H., Donnelly S., Ellwanger A., Sidhu S.S., Dao-Pick T.P., Pantoja C., Erle D.J., Yamamoto K.R., Fahy J.V. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Holtzman M.J., Byers D.E., Alexander-Brett J., Wang X. The role of airway epithelial cells and innate immune cells in chronic respiratory disease. Nat. Rev. Immunol. 2014;14:686–698. doi: 10.1038/nri3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sharma S., Chhabra D., Kho A.T., Hayden L.P., Tantisira K.G., Weiss S.T. The genomic origins of asthma. Thorax. 2014;69:481–487. doi: 10.1136/thoraxjnl-2014-205166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stiemsma L.T., Turvey S.E. Asthma and the microbiome: defining the critical window in early life. Allergy, Asthma Clin. Immunol. 2017;13(3) doi: 10.1186/s13223-016-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Parker J., Sarlang S., Thavagnanam S., Williamson G., O'Donoghue D., Villenave R., Power U., Shields M., Heaney L., Skibinski G. A 3-D well-differentiated model of pediatric bronchial epithelium demonstrates unstimulated morphological differences between asthmatic and nonasthmatic cells. Pediatr. Res. 2010;67:17–22. doi: 10.1203/PDR.0b013e3181c0b200. [DOI] [PubMed] [Google Scholar]
- 211.Wolters P.J., Collard H.R., Jones K.D. Pathogenesis of idiopathic pulmonary fibrosis. Annu. Rev. Pathol.: Mech. Dis. 2014;9:157–179. doi: 10.1146/annurev-pathol-012513-104706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hutchinson J.P., McKeever T.M., Fogarty A.W., Navaratnam V., Hubbard R.B. Increasing global mortality from idiopathic pulmonary fibrosis in the twenty-first century. Ann. Am. Thorac. Soc. 2014;11:1176–1185. doi: 10.1513/AnnalsATS.201404-145OC. [DOI] [PubMed] [Google Scholar]
- 213.Martinez F.J., Collard H.R., Pardo A., Raghu G., Richeldi L., Selman M., Swigris J.J., Taniguchi H., Wells A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Prim. 2017;3 doi: 10.1038/nrdp.2017.74. [DOI] [PubMed] [Google Scholar]
- 214.Ryu J.H., Moua T., Daniels C.E., Hartman T.E., Yi E.S., Utz J.P., Limper A.H. Idiopathic pulmonary fibrosis: evolving concepts. Mayo Clin. Proc. 2014;89:1130–1142. doi: 10.1016/j.mayocp.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 215.Renzoni E., Srihari V., Sestini P. F1000Prime Rep. Vol. 6. 2014. Pathogenesis of idiopathic pulmonary fibrosis: review of recent findings; p. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Varone F., Montemurro G., Macagno F., Calvello M., Conte E., Intini E., Iovene B., Leone P.M., Mari P.-V., Richeldi L. Investigational drugs for idiopathic pulmonary fibrosis. Expert Opin. Investig. Drugs. 2017;26:1019–1031. doi: 10.1080/13543784.2017.1364361. [DOI] [PubMed] [Google Scholar]
- 217.Noble P.W., Barkauskas C.E., Jiang D. Pulmonary fibrosis: patterns and perpetrators. J. Clin. Invest. 2012;122:2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Jemal A., Murray T., Ward E., Samuels A., Tiwari R.C., Ghafoor A., Feuer E.J., Thun M.J. Cancer statistics. CA Cancer J. Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 219.Shanker M., Willcutts D., Roth J.A., Ramesh R. Drug resistance in lung cancer. Lung Cancer (Auckland, N.Z.) 2010;1:23–36. [PMC free article] [PubMed] [Google Scholar]
- 220.Raez L.E., Fein S., Podack E.R. Lung cancer immunotherapy. Clin. Med. Res. 2005;3:221–228. doi: 10.3121/cmr.3.4.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nemunaitis J., Sterman D., Jablons D., Smith J.W., Fox B., Maples P., Hamilton S., Borellini F., Lin A., Morali S., Hege K. Granulocyte-macrophage colony-stimulating factor gene-modified autologous tumor vaccines in non-small-cell lung cancer. J. Natl. Cancer Inst. 2004;96:326–331. doi: 10.1093/jnci/djh028. [DOI] [PubMed] [Google Scholar]
- 222.Fidler I.J., Wilmanns C., Staroselsky A., Radinsky R., Dong Z., Fan D. Modulation of tumor cell response to chemotherapy by the organ environment. Cancer Metastasis Rev. 1994;13:209–222. doi: 10.1007/BF00689637. [DOI] [PubMed] [Google Scholar]
- 223.Killion J.J., Radinsky R., Fidler I.J. Orthotopic models are necessary to predict therapy of transplantable tumors in mice. Cancer Metastasis Rev. 1998;17:279–284. doi: 10.1023/a:1006140513233. [DOI] [PubMed] [Google Scholar]
- 224.Gould S.E., Junttila M.R., de Sauvage F.J. Translational value of mouse models in oncology drug development. Nat. Med. 2015;21:431–439. doi: 10.1038/nm.3853. [DOI] [PubMed] [Google Scholar]
- 225.Holliday D.L., Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13(215) doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Greshock J., Nathanson K., Martin A.-M., Zhang L., Coukos G., Weber B.L., Zaks T.Z. Cancer cell lines as genetic models of their parent histology: analyses based on array comparative genomic hybridization. Cancer Res. 2007;67:3594–3600. doi: 10.1158/0008-5472.CAN-06-3674. [DOI] [PubMed] [Google Scholar]
- 227.Katt M.E., Placone A.L., Wong A.D., Xu Z.S., Searson P.C. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016;4(12) doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Weeber F., Ooft S.N., Dijkstra K.K., Voest E.E. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell. Chem. Biol. 2017;24:1092–1100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 229.Alvarez B., Arcos J., Fernández-Guerrero M.L. Pulmonary infectious diseases in patients with primary immunodeficiency and those treated with biologic immunomodulating agents. Curr. Opin. Pulm. Med. 2011;17:172–179. doi: 10.1097/MCP.0b013e3283455c0b. [DOI] [PubMed] [Google Scholar]
- 230.Bhavnani S.M., Rubino C.M., Hammel J.P., Forrest A., Dartois N., Cooper C.A., Korth-Bradley J., Ambrose P.G. Pharmacological and patient-specific response determinants in patients with hospital-acquired pneumonia treated with tigecycline. Antimicrob. Agents Chemother. 2012;56:1065–1072. doi: 10.1128/AAC.01615-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pirofski L., Casadevall A. The damage-response framework of microbial pathogenesis and infectious diseases. Adv. Exp. Med. Biol. 2008;635:135–146. doi: 10.1007/978-0-387-09550-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kling H.M., Nau G.J., Ross T.M., Evans T.G., Chakraborty K., Empey K.M., Flynn J.L. Challenges and future in vaccines, drug development, and immunomodulatory therapy. Ann. Am. Thorac. Soc. 2014;11:S201–S210. doi: 10.1513/AnnalsATS.201401-036PL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chiu C., Openshaw P.J. Antiviral B cell and T cell immunity in the lungs. Nat. Immunol. 2015;16:18–26. doi: 10.1038/ni.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mahon R.N., Hafner R. Immune cell regulatory pathways unexplored as host-directed therapeutic targets for mycobacterium tuberculosis: an opportunity to apply precision medicine innovations to infectious diseases. Clin. Infect. Dis. 2015;61(Suppl. 3):S200–S216. doi: 10.1093/cid/civ621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hawn T.R., Matheson A.I., Maley S.N., Vandal O. Host-directed therapeutics for tuberculosis: can we harness the host? Microbiol. Mol. Biol. Rev. 2013;77:608–627. doi: 10.1128/MMBR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wallis R.S., Hafner R. Advancing host-directed therapy for tuberculosis. Nat. Rev. Immunol. 2015;15:255–263. doi: 10.1038/nri3813. [DOI] [PubMed] [Google Scholar]
- 237.Fonseca K.L., Rodrigues P.N.S., Olsson I.A.S., Saraiva M. Experimental study of tuberculosis: from animal models to complex cell systems and organoids. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lerner T.R., Borel S., Gutierrez M.G. The innate immune response in human tuberculosis. Cell. Microbiol. 2015;17:1277–1285. doi: 10.1111/cmi.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Flieder D.B., Travis W.D. Pathologic characteristics of drug-induced lung disease. Clin. Chest Med. 2004;25:37–45. doi: 10.1016/S0272-5231(03)00138-2. [DOI] [PubMed] [Google Scholar]
- 240.Coultas D.B., Zumwalt R.E., Black W.C., Sobonya R.E. The epidemiology of interstitial lung diseases. Am. J. Respir. Crit. Care Med. 1994;150:967–972. doi: 10.1164/ajrccm.150.4.7921471. [DOI] [PubMed] [Google Scholar]
- 241.Nemery B., Bast A., Behr J., Borm P.J., Bourke S.J., Camus P.H., De Vuyst P., Jansen H.M., Kinnula V.L., Lison D., Pelkonen O., Saltini C. Interstitial lung disease induced by exogenous agents: factors governing susceptibility. Eur. Respir. J. Suppl. 2001;32:30s–42s. [PubMed] [Google Scholar]
- 242.Matsuno O. Drug-induced interstitial lung disease: mechanisms and best diagnostic approaches. Respir. Res. 2012;13:39. doi: 10.1186/1465-9921-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Schwaiblmair M. Drug induced interstitial lung disease. Open Respir. Med. J. 2012;6:63–74. doi: 10.2174/1874306401206010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hiemstra P.S., Grootaers G., van der Does A.M., Krul C.A.M., Kooter I.M. Human lung epithelial cell cultures for analysis of inhaled toxicants: lessons learned and future directions. Toxicol. in Vitro. 2018;47:137–146. doi: 10.1016/j.tiv.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 245.White C.W., Martin J.G. Chlorine gas inhalation: human clinical evidence of toxicity and experience in animal models. Proc. Am. Thorac. Soc. 2010;7:257–263. doi: 10.1513/pats.201001-008SM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Thorne D., Adamson J. A review of in vitro cigarette smoke exposure systems. Exp. Toxicol. Pathol. 2013;65:1183–1193. doi: 10.1016/j.etp.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 247.Sandberg K., Poole S.D., Hamdan A., Arbogast P., Sundell H.W. Altered lung development after prenatal nicotine exposure in young lambs. Pediatr. Res. 2004;56:432–439. doi: 10.1203/01.PDR.0000136276.52104.61. [DOI] [PubMed] [Google Scholar]
- 248.Sekhon H.S., Jia Y., Raab R., Kuryatov A., Pankow J.F., Whitsett J.A., Lindstrom J., Spindel E.R. Prenatal nicotine increases pulmonary alpha7 nicotinic receptor expression and alters fetal lung development in monkeys. J. Clin. Invest. 1999;103:637–647. doi: 10.1172/JCI5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Sekhon H.S., Proskocil B.J., Clark J.A., Spindel E.R. Prenatal nicotine exposure increases connective tissue expression in foetal monkey pulmonary vessels. Eur. Respir. J. 2004;23:906–915. doi: 10.1183/09031936.04.00069604. [DOI] [PubMed] [Google Scholar]
- 250.Ben-Yehudah A., Campanaro B.M., Wakefield L.M., Kinney T.N., Brekosky J., Eisinger V.M., Castro C.A., Carlisle D.L. Nicotine exposure during differentiation causes inhibition of N-myc expression. Respir. Res. 2013;14:119. doi: 10.1186/1465-9921-14-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ask K., Kolb M.R.J. Drug development for chronic lung disease–mission impossible? Respirology. 2015;20:13–14. doi: 10.1111/resp.12445. [DOI] [PubMed] [Google Scholar]
- 252.Hanania N.A., Korenblat P., Chapman K.R., Bateman E.D., Kopecky P., Paggiaro P., Yokoyama A., Olsson J., Gray S., Holweg C.T.J., Eisner M., Asare C., Fischer S.K., Peng K., Putnam W.S., Matthews J.G. Efficacy and safety of lebrikizumab in patients with uncontrolled asthma (LAVOLTA I and LAVOLTA II): replicate, phase 3, randomised, double-blind, placebo-controlled trials. Lancet Respir. Med. 2016;4:781–796. doi: 10.1016/S2213-2600(16)30265-X. [DOI] [PubMed] [Google Scholar]
- 253.Holgate S., Agusti A., Strieter R.M., Anderson G.P., Fogel R., Bel E., Martin T.R., Reiss T.F. Drug development for airway diseases: looking forward. Nat. Rev. Drug Discov. 2015;14:367–368. doi: 10.1038/nrd4645. [DOI] [PubMed] [Google Scholar]
- 254.Diez D., Agustí A., Wheelock C.E. Network analysis in the investigation of chronic respiratory diseases. From basics to application. Am. J. Respir. Crit. Care Med. 2014;190:981–988. doi: 10.1164/rccm.201403-0421PP. [DOI] [PubMed] [Google Scholar]
- 255.Agustí A., Antó J.M., Auffray C., Barbé F., Barreiro E., Dorca J., Escarrabill J., Faner R., Furlong L.I., Garcia-Aymerich J., Gea J., Lindmark B., Monsó E., Plaza V., Puhan M.A., Roca J., Ruiz-Manzano J., Sampietro-Colom L., Sanz F., Serrano L., Sharpe J., Sibila O., Silverman E.K., Sterk P.J., Sznajder J.I. Personalized respiratory medicine: exploring the horizon, addressing the issues. Summary of a BRN-AJRCCM workshop held in Barcelona on June 12, 2014. Am. J. Respir. Crit. Care Med. 2015;191:391–401. doi: 10.1164/rccm.201410-1935PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Vanfleteren L.E.G.W., Kocks J.W.H., Stone I.S., Breyer-Kohansal R., Greulich T., Lacedonia D., Buhl R., Fabbri L.M., Pavord I.D., Barnes N., Wouters E.F.M., Agusti A. Moving from the Oslerian paradigm to the post-genomic era: are asthma and COPD outdated terms? Thorax. 2014;69:72–79. doi: 10.1136/thoraxjnl-2013-203602. [DOI] [PubMed] [Google Scholar]
- 257.Wouters E.F.M., Reynaert N.L., Dentener M.A., Vernooy J.H.J. Systemic and local inflammation in asthma and chronic obstructive pulmonary disease: is there a connection? Proc. Am. Thorac. Soc. 2009;6:638–647. doi: 10.1513/pats.200907-073DP. [DOI] [PubMed] [Google Scholar]
- 258.Raghu G., Collard H.R., Anstrom K.J., Flaherty K.R., Fleming T.R., King T.E., Martinez F.J., Brown K.K. Idiopathic pulmonary fibrosis: clinically meaningful primary endpoints in phase 3 clinical trials. Am. J. Respir. Crit. Care Med. 2012;185:1044–1048. doi: 10.1164/rccm.201201-0006PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Nathan S.D., Meyer K.C. IPF clinical trial design and endpoints. Curr. Opin. Pulm. Med. 2014;20:463–471. doi: 10.1097/MCP.0000000000000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Gonda I. Systemic delivery of drugs to humans via inhalation. J. Aerosol Med. 2006;19:47–53. doi: 10.1089/jam.2006.19.47. [DOI] [PubMed] [Google Scholar]
- 261.Gonda I. The ascent of pulmonary drug delivery. J. Pharm. Sci. 2000;89:940–945. doi: 10.1002/1520-6017(200007)89:7<940::AID-JPS11>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 262.Patton J.S., Fishburn C.S., Weers J.G. The lungs as a portal of entry for systemic drug delivery. Proc. Am. Thorac. Soc. 2004;1:338–344. doi: 10.1513/pats.200409-049TA. [DOI] [PubMed] [Google Scholar]
- 263.Safdar A., Shelburne S.A., Evans S.E., Dickey B.F. Inhaled therapeutics for prevention and treatment of pneumonia. Expert Opin. Drug Saf. 2009;8:435–449. doi: 10.1517/14740330903036083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Liu X., Jin L., Upham J.W., Roberts M.S. The development of models for the evaluation of pulmonary drug disposition. Expert Opin. Drug Metab. Toxicol. 2013;9:487–505. doi: 10.1517/17425255.2013.754009. [DOI] [PubMed] [Google Scholar]
- 265.Tsuda A., Henry F.S., Butler J.P. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2013. Comprehensive Physiology; pp. 1437–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Gizurarson S. The effect of cilia and the mucociliary clearance on successful drug delivery. Biol. Pharm. Bull. 2015;38:497–506. doi: 10.1248/bpb.b14-00398. [DOI] [PubMed] [Google Scholar]
- 267.Hastings R.H., Folkesson H.G., Matthay M.A. Mechanisms of alveolar protein clearance in the intact lung. Am. J. Phys. Lung Cell. Mol. Phys. 2004;286:L679–L689. doi: 10.1152/ajplung.00205.2003. [DOI] [PubMed] [Google Scholar]
- 268.Forbes B., Asgharian B., Dailey L.A., Ferguson D., Gerde P., Gumbleton M., Gustavsson L., Hardy C., Hassall D., Jones R., Lock R., Maas J., McGovern T., Pitcairn G.R., Somers G., Wolff R.K. Challenges in inhaled product development and opportunities for open innovation. Adv. Drug Deliv. Rev. 2011;63:69–87. doi: 10.1016/j.addr.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 269.Fishler R., Hofemeier P., Etzion Y., Dubowski Y., Sznitman J. Particle dynamics and deposition in true-scale pulmonary acinar models. Sci. Rep. 2015;5(14071) doi: 10.1038/srep14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Borghardt J.M., Weber B., Staab A., Kloft C. Pharmacometric models for characterizing the pharmacokinetics of orally inhaled drugs. AAPS J. 2015;17:853–870. doi: 10.1208/s12248-015-9760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hickey A.J., Garcia-Contreras L. Immunological and toxicological implications of short-term studies in animals of pharmaceutical aerosol delivery to the lungs: relevance to humans. Crit. Rev. Ther. Drug Carrier Syst. 2001;18:387–431. [PubMed] [Google Scholar]
- 272.Capulli A.K., Tian K., Mehandru N., Bukhta A., Choudhury S.F., Suchyta M., Parker K.K. Approaching the in vitro clinical trial: engineering organs on chips. Lab Chip. 2014;14:3181. doi: 10.1039/c4lc00276h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Nawroth J.C., Parker K.K. Design standards for engineered tissues. Biotechnol. Adv. 2013;31:632–637. doi: 10.1016/j.biotechadv.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Shirure V.S., George S.C. Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab Chip. 2017;17:681–690. doi: 10.1039/c6lc01401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bedekar A., Wang Y., Siddhaye S.S., Krishnamoorthy S., Malin S.F. Design software for application-specific microfluidic devices. Clin. Chem. 2007;53:2023–2026. doi: 10.1373/clinchem.2007.090498. [DOI] [PubMed] [Google Scholar]
- 276.Hu C., Lin S., Li W., Sun H., Chen Y., Chan C.-W., Leung C.-H., Ma D.-L., Wu H., Ren K. A one-step strategy for ultra-fast and low-cost mass production of plastic membrane microfluidic chips. Lab Chip. 2016;16:3909–3918. doi: 10.1039/c6lc00957c. [DOI] [PubMed] [Google Scholar]
- 277.Wikswo J.P., Block F.E., Cliffel D.E., Goodwin C.R., Marasco C.C., Markov D.A., McLean D.L., McLean J.A., McKenzie J.R., Reiserer R.S., Samson P.C., Schaffer D.K., Seale K.T., Sherrod S.D. Engineering challenges for instrumenting and controlling integrated Organ-on-Chip systems. IEEE Trans. Biomed. Eng. 2013;60:682–690. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Christoffersson J., van Noort D., Mandenius C.-F. Developing organ-on-a-chip concepts using bio-mechatronic design methodology. Biofabrication. 2017;9(25023) doi: 10.1088/1758-5090/aa71ca. [DOI] [PubMed] [Google Scholar]
- 279.Dekkers J.F., Berkers G., Kruisselbrink E., Vonk A., de Jonge H.R., Janssens H.M., Bronsveld I., van de Graaf E.A., Nieuwenhuis E.E.S., Houwen R.H.J., Vleggaar F.P., Escher J.C., de Rijke Y.B., Majoor C.J., Heijerman H.G.M., De Winter-De Groot K.M., Clevers H., van der Ent C.K., Beekman J.M. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aad8278. (344ra84-344ra84) [DOI] [PubMed] [Google Scholar]
- 280.Food U.S. 2017. Drug administration, FDA researchers to evaluate “Organs-on-Chips”. (available at) https://www.fda.gov/Food/NewsEvents/ConstituentUpdates/ucm551503.htm. [Google Scholar]