Abstract
Oat blue dwarf virus (OBDV) is a member of the genus Marafivirus whose genome encodes a 227 kDa polyprotein (p227) ostensibly processed post-translationally into its functional components. Encoded near the 3' terminus and coterminal with the p227 ORF are ORFs specifying major and minor capsid proteins (CP). Since the CP expression strategy of marafiviruses has not been thoroughly investigated, we produced a series of point mutants in the OBDV CP encoding gene and examined expression in protoplasts. Results support a model in which the 21 kDa major CP is the product of direct translation of a sgRNA, while the 24 kDa minor CP is a cleavage product derived from both the polyprotein and a larger ~26 kDa precursor translated directly from the sgRNA. Cleavage occurs at an LXG[G/A] motif conserved in many viruses that use papain-like proteases for polyprotein processing and protection against degradation via the ubiquitin-proteasome system.
Keywords: Marafivirus, Tymoviridae, OBDV
Highlights
-
•
The 21 kDa major CP is the product of direct translation of a sgRNA.
-
•
The 24 kDa minor CP is a cleavage product derived in two ways.
-
•
Cleavage occurs at an LXG[G/A] motif.
-
•
Relevant to viruses using papain-like proteases for polyprotein processing.
-
•
Relevant to protection against degradation via the ubiquitin-proteasome system.
Introduction
Members of the genus Marafivirus are alpha-like plant viruses (Goldbach et al., 1991, Rozanov et al., 1992) belonging to the family Tymoviridae in the order Tymovirales. These small, isometric, positive stranded RNA viruses have approximately 6.3–6.8 kb genomes that are similar in organization to those of the tymoviruses and encode large polyproteins with methyltransferase, helicase, and polymerase motifs (reviewed in Dreher et al., 2011). While the tymovirus coat protein (CP) is encoded in a separate open reading frame (ORF) just downstream of the polyprotein ORF, marafivirus CP-encoding sequences are nested within, and are 3' co-terminal with, the ORF encoding the polyprotein. The tymovirus genome expression strategy has been shown to employ a papain-like protease to process the large precursor polyprotein, while the CP is translated directly from a subgenomic RNA (Dreher et al., 2011). A highly conserved core sequence known as the tymobox serves as a promoter for transcription of this sgRNA (Ding et al., 1990).
Precise details of marafivirus genome expression have not been demonstrated, although Edwards et al. (1997) proposed a model for expression of the oat blue dwarf virus (OBDV) genome based in part on its genomic similarities with tymoviruses. Marafiviruses possess a sequence analogous to the highly conserved tymobox subgenomic promoter sequence, known as the marafibox (Izadpanah et al., 2002), which is presumed to have a similar function. Virions of marafiviruses, however, contain a major CP of 21 kDa and a minor CP of 24 kDa that differ only by an amino terminal extension present in the minor CP, whereas tymoviral particles contain a single CP. Because these CPs likely have a role in transmissibility of leafhopper-borne marafiviruses and the fact that there are structural differences between marafivirus and tymovirus genomes, the CP expression strategy of marafiviruses is an important feature to characterize. We now report the use of mutation analysis with an infectious OBDV clone to dissect and analyze CP expression of OBDV as a first step toward a better understanding of marafivirus gene expression. We also map the initial nucleotide at the 5'-terminus of the sgRNA encoding the CPs and show that it is consistent in position relative to the tymobox/marafibox with that of sgRNA termini of the tymoviruses.
Results and discussion
The OBDV genomic region targeted to investigate CP expression and the mutants used in this investigation are shown in Fig. 1. Our strategy employed mutation to create premature stop codons, disrupt proposed initiation codons and a potential protease cleavage site, as well as to disrupt the putative sgRNA promoter sequence. Inoculation of oat leaf protoplasts with OBDV wild type transcripts (OBDV-2r, GenBank #GU396990) resulted in the accumulation after 24 h of readily detectable amounts of viral CP and gRNA+sgRNA on western and northern blots, respectively. Infection of protoplasts by OBDV was confirmed using fluorescence microscopy to visualize a variant in which the entire major CP was replaced by the enhanced green fluorescent protein gene (not shown).
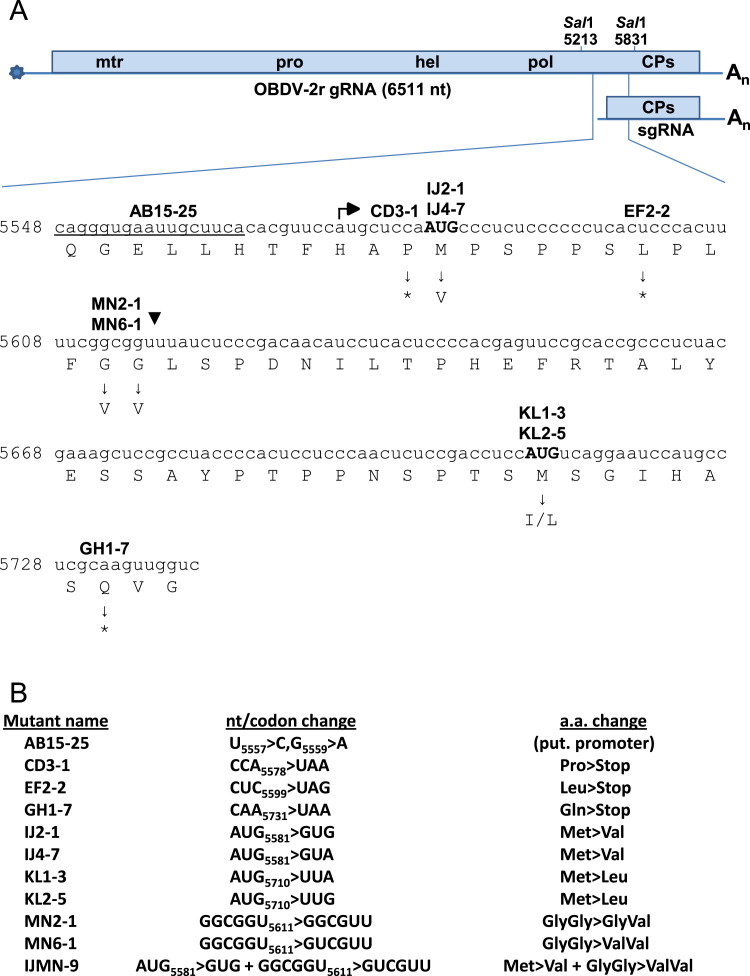
Fig. 1.
Illustration of the OBDV genome, and the genomic region and mutants used in the investigation of coat protein (CP) expression. (A) Diagram of the OBDV genome and the nucleotide sequence in the region encoding the CPs. The genomic RNA possesses a 5' cap structure (star) and a 3'-poly A tail (An). The single, large ORF encodes a polyprotein with domains specifying methyltransferase (mtr), protease (pro), helicase/NTpase (hel), and polymerase (pol) activities fused to the sequence encoding the CPs. The major and minor coat proteins map to the same CP sequence and size differences between them are potentially determined by translation initiation at two different start codons (minor, AUG5581 and major, AUG5710). Whereas the major CP is postulated to be translated primarily from sgRNA, the minor CP could be produced by translation directly from the sgRNA, by cleavage after Gly–Gly(▼) of either or both the large polyprotein or a protein initiated at AUG5581, or by both direct translation and cleavage. The putative promoter for sgRNA synthesis, the marafibox, is underlined and the start site for sgRNA synthesis (this work) is indicated by the bent arrow. Sites where premature termination codons were introduced are indicated by asterisks. Mutant names are shown above the corresponding sequence locations, and amino acid changes in potential initiation and protease cleavage site codons are shown under the sequence. (B) Mutant description and nomenclature. The mutant name is followed by indication of nucleotide/codon substitutions and the cognate amino acid change. Mutant IJMN-9 represents a double mutant altering both the putative minor CP initiation codon and the potential cleavage site.
Initiation point for sgRNA transcription and role of sgRNA in CP production
Alignment of various tymo/marafibox sequences (a hallmark of the Tymoviridae and putative promoter for sgRNA synthesis of tymo- and marafiviruses) reveals conservation of an adenine nucleotide ~10 nt downstream from the 3'-edge of the core sequence ( Fig. 2). This is the known 5'-end nucleotide for the sgRNAs of turnip yellow mosaic virus (TYMV), ononis yellow mosaic virus (OYMV), and kennedya yellow mosaic virus (KYMV) (Ding et al., 1990, Guilley and Briand, 1978). To determine whether the conserved A in that position in OBDV (A5573) is the 5'-terminal nucleotide of the OBDV sgRNA, 5'-RACE was performed on size-fractionated RNA extracted from infected protoplasts and the resulting amplicons were sequenced (not shown). Sequence data showed that A5573 represents the 5'-end of the OBDV sgRNA encoding the viral CPs and is the likely start site of transcription for this 939 nt sgRNA (excluding the poly A tail). OBDV is the first marafivirus for which this has been determined, thus providing the first experimental evidence that this highly conserved adenine nucleotide likely represents the 5'-terminus of other marafivirus sgRNAs as well. These results are consistent with the evolutionary homology of the tymobox/marafibox sequence and its putative role as promoter of sgRNA synthesis.
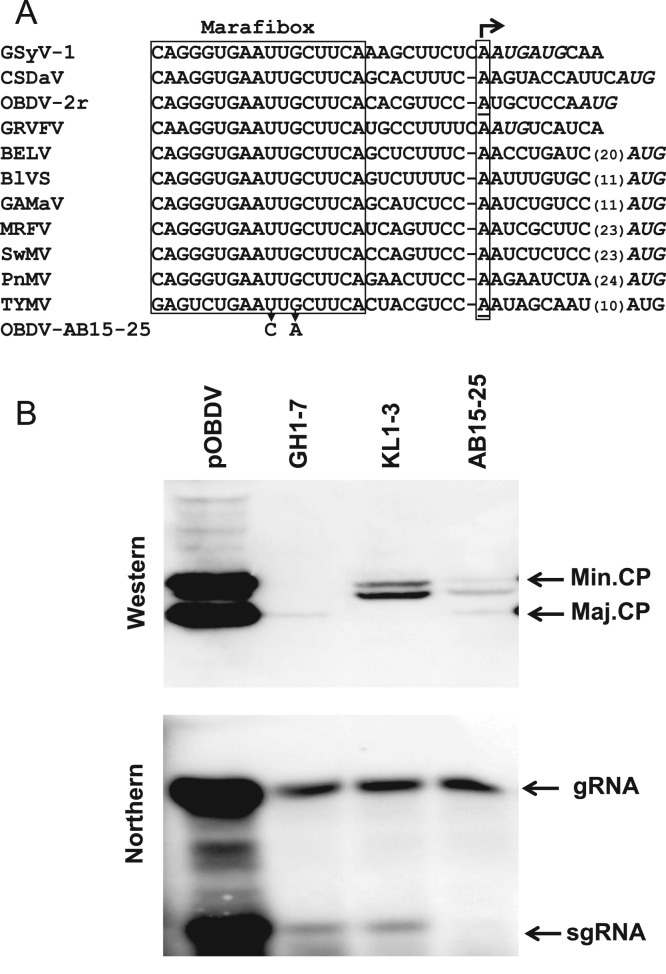
Fig. 2.
Effects of marafibox and major coat protein (CP) gene mutations on accumulation of viral CPs, and genomic and sgRNAs. (A) Nucleotide sequences from known (OBDV, MRFV, BELV, CSDaV) and proposed (SwMV, BlVS, GAMaV, GRVFV, GSyV-1) marafiviruses aligned with those of PnMV (unassigned) and TYMV (type member of the genus Tymovirus). Silent substitutions (U>C, G>A) introduced to produce mutant OBDV-AB15-25 are shown and the adenine position representing the proposed start site of sgRNA synthesis is indicated (bent arrow); underlined adenine residues below the horizontal arrow have been experimentally confirmed as sgRNA 5'-ends. (B) Accumulation of marafibox and major CP mutants in oat protoplasts. Oat protoplasts were inoculated with capped transcripts of wild type clone pOBDV, major CP mutants GH1-7 (premature termination mutant) and KL1-3 (initiation codon mutant), and AB15-25 (marafibox mutant) and incubated for 40 h. Extracted viral RNA and protein were detected on northern and western blots using a 3'-end dsDNA probe and anti-OBDV antiserum, respectively. The locations of major CP (Maj. CP), minor CP (Min. CP), genomic (gRNA), and subgenomic RNA (sgRNA), on the blots are shown.
To confirm this role, silent mutations (in relation to the polyprotein) were made in the marafibox in positions analogous to those known to severely reduce production of sgRNA in TYMV (Schirawski et al., 2000). Mutation of two key nucleotides (mutant AB15-25) reduced transcription of the sgRNA to an undetectable level even after 40 h of incubation, with a concomitant large reduction in accumulation of both forms of CP (Fig. 2). Production of neither CP was completely eliminated, suggesting that either sgRNA production was not completely shut down or that a low level of translation from gRNA occurred. An extraneous band migrating slightly faster than the minor CP appeared in some instances on western blots, perhaps representing a conformational variant (Edwards et al., 1997). Interestingly, gRNA accumulation was also significantly reduced relative to that of wild type. This reduction appeared to be due to the loss of the major CP and not to the marafibox mutations, a view supported by similar reductions in gRNA accumulation for two other mutants whose ability to produce major CP was severely compromised (GH1-7 premature termination mutant and KL1-3 initiation codon mutant; Fig. 2).
The predicted initiation codons for the major and minor CPs are functional in vivo
As we noted previously (Edwards et al., 1997), two initiation codons (AUG5581 and AUG5710) exist with the potential to encode the minor and major CPs, respectively. The possible roles of these codons in the production of the OBDV CPs were investigated using a combination of site-directed mutants designed to prevent translation either through introduction of premature stop codons or elimination of initiation codons AUG5581 and AUG5710.
Results with point mutants generating premature stop codons (CD3-1, EF2-2, and GH1-7) confirmed and expanded our previous model for the origins of the major and minor CPs of OBDV. When a stop codon (UAA5731) was placed downstream of the major CP initiation codon (mutant GH1-7), neither CP was detectable after 24 h ( Fig. 3) and gRNA accumulation was greatly reduced (Fig. 2, Fig. 3). However, a trace amount of major CP was detectable after 40 h (Fig. 2), possibly due to UAA suppression from this highly translated ORF (Lao et al., 2009). With the stop codon (UAG5599) placed upstream of the major CP initiation codon (mutant EF2-2), abundant major CP was produced without detectable minor CP (Fig. 3). That the accumulation of gRNA and major CP was restored to near wild type levels in mutant EF2-2 suggests that the reduction of gRNA accumulation in mutant GH1-7 was linked to the absence of major CP expression. Mutant CD3-1 (UAA5578) positions the stop codon upstream of both the candidate start codon for the minor CP as well as the start codon for the major CP and thus precludes translation of either CP as a part of the polyprotein; both a minor and major CP are produced by this mutant (not shown). Thus, complete translation of the entire polyprotein ORF is not an absolute requirement for the production of either CP species, and AUG5581 is the probable initiation codon for the production of at least some fraction of the minor CP pool despite its close proximity to the 5' terminus of the sgRNA. Although such a short leader (8 nt for OBDV, as close as 1 nt for other marafiviruses) is unusual, initiation of translation at AUG codons positioned very close to the 5' end has been observed in other biological systems (Li and Wang, 2004, Elfakess and Dikstein, 2008).
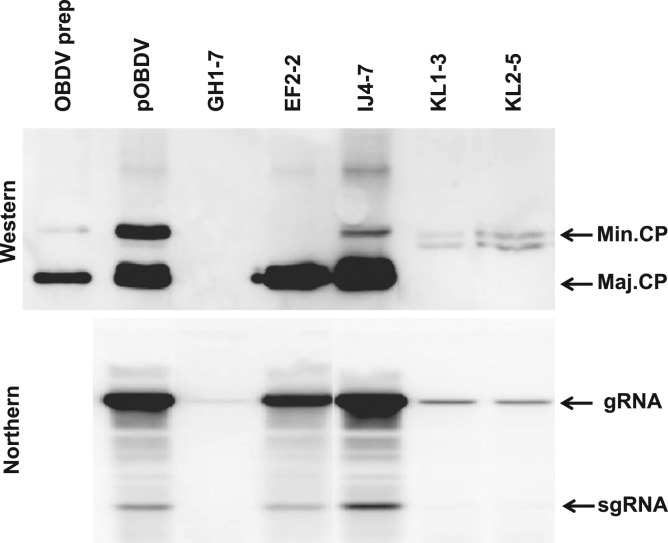
Fig. 3.
Accumulation of viral coat proteins (CPs) and RNA in oat protoplasts inoculated with premature termination and initiation codon mutants of the CP gene. Oat protoplasts were inoculated with capped transcripts of wild type clone pOBDV, premature termination mutants GH1-7 and EF2-2, and initiation codon mutants IJ4-7 (minor CP), and KL1-3 and KL2-5 (major CP) and incubated for 24 h. Methods for detection of viral protein and RNA were as described in the legend for Fig. 2. The locations of major CP (Maj. CP), minor CP (Min. CP), genomic (gRNA), and subgenomic RNA (sgRNA) on the blots are shown. Partially purified OBDV (OBDV prep) was included on the western blot for size comparison.
Additional mutants were made to confirm the use of AUG5581 and AUG5710 as initiation codons in the production of minor CP and major CP, respectively. Mutation of AUG5710 to UUA (KL1-3) or UUG (KL2-5) abolished accumulation of major CP in protoplasts, as expected, but also greatly reduced accumulation of viral RNA (Fig. 3). However, near wild-type levels of viral RNA and the major CP, along with reduced levels of the minor CP, accumulated in protoplasts inoculated with mutants IJ4-7 and IJ2-1, in which AUG5581 was replaced by GUA or GUG, respectively (Fig. 3, IJ2-1 not shown). This supports the notion that the minor CP of OBDV can be produced via proteolytic cleavage from the large polyprotein.
Proteolytic cleavage plays a role in formation of the minor CP species
Previous peptide sequence evidence indicated the presence of a potential papain-like protease cleavage site located immediately following Gly1834 of the OBDV polyprotein (Edwards et al., 1997). Taken together with the experimental evidence above, it is thus conceivable that functional minor CP might be produced via proteolytic cleavage from the large polyprotein precursor. To further investigate this possibility, the candidate cleavage site was changed from Gly–Gly1834 to Val–Val1834 (mutant MN6-1; Fig. 4). Typical accumulation of major CP was observed for this mutant, but the apparent size of the minor CP increased slightly, suggesting production exclusively from translation initiating at AUG5581 (Fig. 4). A similar result was obtained when Gly–Gly1834 was changed to Gly–Val1834 in mutant MN2-1 (not shown). When both the putative cleavage and initiation sites were knocked out by changing AUG5581 to GUA in the MN6-1 background (double mutants IJMN9A and B), only major CP was detected in infected protoplasts. When combined with the results from premature stop codon mutant CD3-1 and initiation codon mutant IJ4-7, it is clear the minor CP can be produced by cleavage of either or both the polyprotein and the minor CP originating from sgRNA translation.
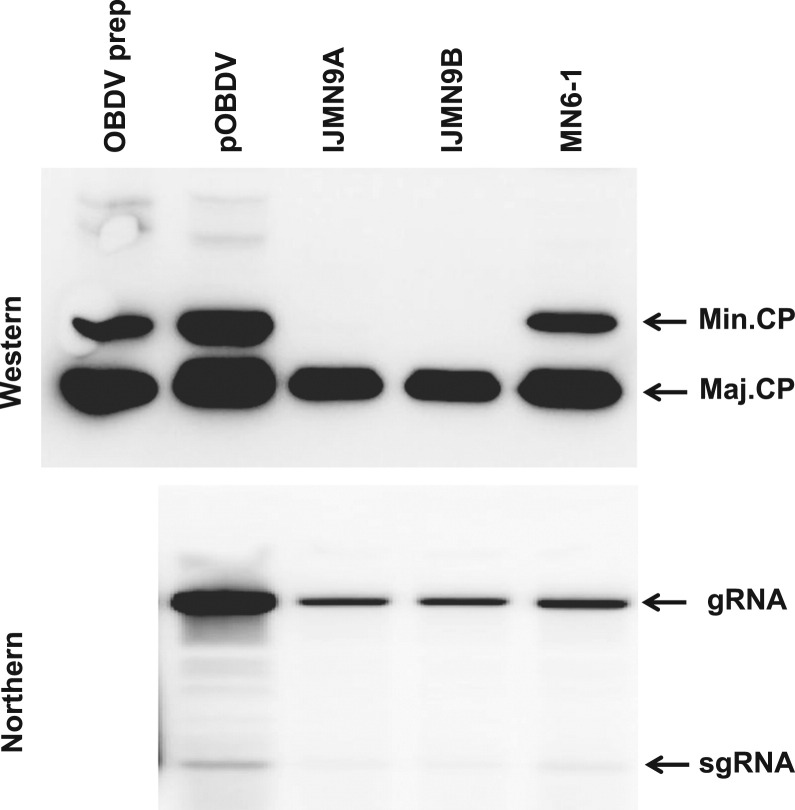
Fig. 4.
Accumulation of viral coat proteins (CPs) and RNA in oat protoplasts inoculated with putative polymerase/CP cleavage site mutants. Oat protoplasts were inoculated with capped transcripts of wild type clone pOBDV, putative cleavage site mutant MN6-1, and double mutant IJMN-9 (minor CP initiation codon mutation combined with putative cleavage site mutation; IJMN-9A and B represent two different inoculations) and incubated for 24 h. Methods for detection of viral protein and RNA were as described in the legend for Fig. 2. The locations of major CP (Maj. CP), minor CP (Min. CP), genomic (gRNA), and subgenomic RNA (sgRNA) on the blots are shown. Partially purified OBDV (OBDV prep) was included on the western blot for size comparison.
A model for OBDV CP expression and its applicability to other marafiviruses
The empirical evidence offered here, taken together with previous sequence analysis by Edwards et al. (1997), supports a model for OBDV CP expression in which the major CP is translated directly from the 939 nt sgRNA (excluding the polyA tail), while the minor CP is cleaved from both the polyprotein and a minor CP precursor translated from the sgRNA. The underlying reason for the existence of multiple means of expression of the minor CP is not evident. While cleavage from the replicase polyprotein provides a source of minor CP, it may be that production of minor CP exclusively through this mechanism doesn't provide stoichiometric amounts necessary for virion assembly. It is also possible that viruses such as OBDV are in an evolutionary transition toward CP production solely via sgRNAs and that readthrough of the larger replicase polyprotein is vestigial. In any case, production of the minor CP through a cleavage mechanism provides a regulatory feature with probable functional significance for both replication and encapsidation. Ultimately, the complexity of CP expression relative to that of tymoviruses may relate in some way to the infection by OBDV of both plant and insect hosts.
To consider this model in the larger context of CP expression in the marafiviruses as a group, amino acid sequences of known and proposed marafiviruses were aligned and compared ( Fig. 5). In all cases, putative initiator methionine residues for both minor and major CPs are evident. The putative initiator Met for the minor CP is 5–15 amino acids downstream of the highly conserved GELL motif that is encoded by the core sequence of the marafibox. With the exception of maize rayado fino virus (MRFV), switchgrass mosaic virus (SwMV), and Bermudagrass etched-line virus (BELV), a conserved motif comprised of LXGG is present 7–8 amino acids downstream of the initiator Met. Since this incorporates the demonstrated cleavage site that creates the amino terminus of the OBDV minor CP, it is reasonable to speculate that production of these other marafivirus minor CPs also involves cleavage at this site. A similar motif is present immediately upstream of the putative initiator Met in the polyproteins of MRFV, SwMV, and BELV. The SwMV sequence encodes an alanine at the putative cleavage site, but that is consistent with the previous observations of Kadaré et al. (1995) and Sulea et al. (2006), who noted the presence of G or A residues in the P1 and P2 positions of confirmed and predicted cleavage site sequences of tymo- and coronaviruses, respectively. Thus, we propose the consensus sequence at the predicted cleavage site associated with minor CP expression in marafiviruses is LXG[G/A]. Intriguingly, to counter cellular antiviral strategies that utilize the ubiquitin-proteasome system, the protease of the closely-related TYMV cleaves ubiquitin conjugates from its RdRp at the same consensus sequence in addition to its role in polyprotein processing (Chenon et al., 2012). Moreover, this consensus sequence is targeted by other viral cysteine proteases with both deubiquitinating and polyprotein processing activities (Barretto et al., 2005, Clementz et al., 2010, Karpe and Lole, 2011, Wang et al., 2011).
Fig. 5.
Amino acid sequence alignment of the encoded polyproteins for known and proposed marafiviruses and PnMV in the region surrounding the coat protein (CP) amino termini. In (A) the proposed initiator methionine residues for the minor (left) and major (right) marafiviral CPs are boxed, as are invariant amino acids encoded by the marafibox. Underlined amino acids indicate conserved sequence around the proposed LXG[G/A] cleavage recognition site for the encoded papain-like protease. (B) Re-alignment of sequences to highlight the proposed conserved protease recognition sequence (boxed). The conserved LXG[G/A] motif resembles that targeted by cellular deubiquitinating enzymes (Ubiquitin). Proposed initiator methionine residues for the minor and major marafiviral CPs are underlined. Although GSyV and GVQ are the same species, the reported sequences for each isolate are not identical and therefore both are included here.
The model for expression of OBDV CPs appears directly applicable to the majority of accepted and proposed marafiviruses, while an interesting variation of this strategy likely occurs with SwMV, MRFV, and BELV. For the latter, the predicted cleavage sites lie upstream of the predicted minor CP initiation codons, necessitating further studies to determine the precise CP expression strategies of these viruses. The location of the predicted cleavage site downstream from the initiator methionine of the minor CP would provide a consistent amino terminus for minor CP produced by cleavage of both the replicase polyprotein and a minor CP precursor translated directly from sgRNA. Irrespective of the role of protein cleavage in the formation of the minor CP N-terminus, an important function of the cleavage site may be to liberate the polymerase from the CP. The location of a putative protease cleavage site upstream of the initiator methionine for the minor CPs of SwMV, MRFV, and BELV is consistent with this view. Furthermore, the single CP of the closely-related but unclassified Poinsettia mosaic virus (PnMV) is fused to the PnMV replicase polyprotein, and the amino acid sequence between the marafibox and the CP also harbors a predicted LXGA cleavage site sequence (Fig. 5). Thus, the LXG[G/A] motif is well conserved among viruses of the Tymoviridae that possess a replicase-CP fusion, regardless of the precise location of the motif relative to the CP translation initiation codon.
This is the first investigation of a marafivirus CP expression strategy and a step toward a better understanding of overall gene expression in marafiviruses. The sequence conservation and similarity with other viruses that utilize papain-like proteases as a means to process polyproteins and defend against protein degradation via the ubiquitin-proteasome system suggests that similar mechanisms are used by marafiviruses. With the existence of tymoviral and marafiviral infectious clones and techniques for their transmission, the tools are now available to investigate the roles of CPs and other factors in the infection process within whole plants and their potential roles in marafivirus transmission by leafhoppers (Edwards and Weiland, 2010, Spetz et al., 2008, Weiland and Dreher, 1989, Weiland and Edwards, 2011).
Materials and methods
Parental and mutant clone construction
A full-length cDNA clone of OBDV from which infectious transcripts can be derived was generated previously (Edwards and Weiland, 2010). This clone (pOBDV-2r; GenBank #GU396990) served as the parent plasmid in the generation of mutants of OBDV with alterations in the CP gene(s). General amplification primers OBD5140fwd and OBD5901rev were used in conjunction with mutagenic primers (Fig. 1, Table 1) to produce overlapping amplicons. PCR reactions included 1 ng of linearized plasmid as template and conditions were as described in Higuchi (1990). Typical reactions contained 1X Platinum Taq buffer, 0.1 mM each d(GATC)TP, 30 ng of each oligonucleotide primer, and 0.5 units of Platinum Taq High-Fidelity DNA polymerase (Life Technologies, Grand Island, NY USA) in a volume of 30 µl. Thermocycling parameters were 95 °C, 30 s; 55 °C, 30 s; and 72 °C, 2 min per cycle over 30 cycles total. Paired amplicons were mixed and subjected to a second round of amplification using only primers OBD5140fwd and OBD5901rev (Higuchi, 1990). Digestion of the resulting amplicon with Sal 1 (located at nts 5213 and 5831) released a 618 bp fragment that was used to replace the homologous region in pOBDV-13B2 (a derivative of pOBDV-2r in which the MCS was altered to remove the Sal 1 site). Capped and polyadenylated transcripts were synthesized as previously described (Edwards and Weiland, 2010). The wild type pOBDV-13B2 is designated pOBDV herein.
Table 1.
Mutagenic and amplification oligonucleotide primers for the generation of OBDV-CP mutants.
| Primer name a | Primer type b | Primer sequence c |
|---|---|---|
| OBD5140fwd | Amplification | CGGTCTGTATGCCACCTTCT5140 |
| OBD5901rev | Amplification | TCAGGTGAGAGGATCCTGAG5901 |
| Tymobox (+) | Mutagenic/promoter | GGTGAACTACTTCACACGTTC5571 |
| Tymobox (−) | Mutagenic/promoter | GAACGTGTGAAGTAGTTCACC5554 |
| Term1-221 K(+) | Mutagenic/termination | TCCATGCTTAAATGCCCTCTCC5591 |
| Term1-221 K(−) | Mutagenic/termination | GGAGAGGGCATTTAAGCATGGA5570 |
| Term2-221 K(+) | Mutagenic/termination | CCCTCATAGCCACTTTTCG5611 |
| Term2-221 K(−) | Mutagenic/termination | CGAAAAGTGGCTATGAGGG5593 |
| Term3-221 K(+) | Mutagenic/termination | ATGCCTCGTAAGTTGGTCCG5742 |
| Term3-221 K(−) | Mutagenic/termination | CGGACCAACTTACGAGGCAT5723 |
| CPinit1(+) | Mutagenic/initiation | TCCATGCTCCAKTRCCCTCTCC5591 |
| CPinit1(−) | Mutagenic/initiation | GGAGAGGGYAMTGGAGCATGGA5570 |
| CPinit2(+) | Mutagenic/initiation | GACCTCCKTRTCAGGAATC5721 |
| CPinit2(−) | Mutagenic/initiation | GATTCCTGAYAMGGAGGTC5702 |
| CleavSite1(+) | Mutagenic/cleavage | CACTTTTCGKCGKTTTATCTCC5625 |
| Cleavsite1(−) | Mutagenic/cleavage | GGAGATAAAMCGMCGAAAAGTG5603 |
Designation of (+) and (−) denotes complementary pair of primers where (+) is of “sense” orientation with respect to the genomic RNA.
Changes to ablate putative transcriptional promoter, translational initiation, and protease cleavage sites or produce premature termination of translation.
Standard nucleotide designation in 5'->3' polarity with degeneracies where K=T+G, M=A+C, Y=T+C, and R=A+G. The last nucleotide in each primer is numbered relative to its position in the sequence of OBDV-2r (GenBank Accession #GU396990). Nucleotides changed from the OBDV-2r sequence are underlined.
Protoplast inoculation
Protoplasts were produced from oat (Avena sativa L. cv ‘Rodney’) as previously described (Weiland and Edwards, 1994). The procedure routinely produced 106 protoplasts per gram fresh weight tissue from 7 day-old oat seedlings. A modification of the procedure by Matsuda and Dreher (2005) was used to transfect protoplasts with capped transcript RNA. Isolated protoplasts prepared from 3 g fresh weight of oat leaves were collected from the surface of a sucrose pad after centrifugation and diluted in 0.55 M mannitol/0.1% MES pH 5.6 (MM) containing 5 mM CaCl2 and 40 mM KCl (MMCK). Protoplasts were pelleted by centrifugation at 115×g for 8 min. and resuspended in 4 ml MMCK. Approx. 5×105 protoplasts (per inoculation) were transferred to a microfuge tube, spun to collect the protoplasts, and the cells resuspended in 0.7 ml MMCK. Tubes containing protoplasts were incubated on ice for 30 min prior to inoculation.
Transcript RNA was adjusted to a concentration of 0.2 µg/µl, and 5 µl was transferred to a 1.5 ml microfuge tube and placed on ice. A 0.8 ml aliquot of incubation medium (IM) consisting of MM supplemented with micronutrients as previously described (Weiland and Edwards, 1994) was chilled on ice in a microfuge tube along with a 0.4 cm gap electroporation cuvette. Chilled protoplasts were transferred to the tube containing transcript RNA and the mixture was rapidly pipetted into the pre-chilled cuvette. Contents of the cuvette were subjected to an exponential decay pulse of 100 V, 2 mF, and 480 Ω, yielding a pulse duration of ~110 ms (delivered by a BTX ECM-600 electroporation system, Harvard Apparatus, Holliston, MA, USA). Following transfer of the contents of the cuvette to 0.8 ml of chilled IM, samples were incubated on ice for 10 min. Protoplasts were collected by centrifugation, resuspended in 1 ml fresh IM containing 100 µg/ml cefotaxime, and incubated for 24 h or 40 h at 22 °C under continuous fluorescent light. Each mutant was tested in protoplast inoculations at least 4 times and included parallel mock and pOBDV inoculations within each experimental replication.
Northern and western blot analysis
Samples were harvested and processed for the analysis of viral RNA and protein as previously described (Weiland and Edwards, 1994). Total protein (for Western blots) and nucleic acids (for Northern blots) from 105 protoplasts were separated on 12% sodium dodecylsulfate polyacrylamide gels and 1% denaturing agarose gels, respectively, as indicated (Edwards and Weiland, 2010). Proteins were electroblotted from polyacrylamide gels to nitrocellulose membranes (Schleicher and Schuell [Keene, N.H. USA], BA-85, 0.2 µ), which subsequently were incubated in a 1:1000 dilution of rabbit anti-OBDV, followed by incubation in a 1:2000 dilution of alkaline phosphatase-conjugated goat anti-rabbit IgG (Product #A0418, Sigma-Aldrich, St. Louis, MO USA). Protein complexes were detected on a Kodak Image Station 2000 MM following treatment of the blot with LumiPhos WB (Thermo Scientific, Lafayette, CO USA). Nucleic acids transferred to positively-charged nylon membranes by capillary blotting (Roche Applied Science, Indianapolis, IN USA) were probed with a digoxigenin-labeled, denatured dsDNA probe representing nucleotides 5929–6508 of OBDV-2r. After incubating blots with alkaline phosphatase-conjugated anti-digoxigenin IgG, CDP-Star (Roche Applied Science, Indianapolis, IN USA) was added and viral RNAs were detected by chemiluminescence.
Mapping of the 5' end of the sgRNA
Total nucleic acids were prepared from OBDV-infected oat protoplasts as previously described (Edwards and Weiland, 2010). Due to substrate competition with the genomic RNA in the cDNA generation step of the 5'-RACE procedure, gel-purified RNA was used as template in the mapping of the 5'-end of the sgRNA. Thus, RNAs were size-fractionated by agarose gel electrophoresis (1% GTG agarose [Cambrex Bioscience, Rockland, ME, USA], 1X Tris-borate-EDTA buffer), and RNA migrating a distance consistent with the estimated size of OBDV sgRNA (~1.0 kb) was extracted and purified using the Zymoclean™ Gel RNA Recovery Kit (ZymoResearch, Irvine, CA USA). Following the manufacturer's recommendations, primer OBD5901rev (Table 1) was used to prime cDNA synthesis in the presence of SmartScribe reverse transcriptase and anchor primers contained in the SmartRACE kit (Clontech, Mountain View, CA USA). Final amplification employed virus-specific primer Term2-221 K(−) and a SmartRACE nested universal anchor primer and the products were cloned and sequenced (MWG Operon, Huntsville, AL, USA). CLUSTAL-W (Larkin et al., 2007) was used to align the resulting nucleotide sequences and the sgRNA 5'-end determined; initial protein sequence alignments used to illustrate conservation of amino acid residues around the CP amino termini employed the same software.
Virus names, acronyms, and sequences used
Acronyms and GenBank accession numbers for viruses discussed in the text and figures are: Bermuda grass etched-line virus (BELV, AY040531), Blackberry virus S (BlVS, FJ915122), Citrus sudden death-associated virus (CSDaV, NC_006950), Grapevine asteroid mosaic-associated virus (GAMaV, AJ249357), Grapevine rupestris vein feathering virus (GRVFV, AY706994), Grapevine Syrah virus-1 (GSyV-1, NC_012484), Grapevine virus Q (GVQ, an isolate of GSyV-1, FJ977041); Maize rayado fino virus (MRFV, NC_002786), Oat blue dwarf virus isolate 2r (OBDV-2r, GU396990), Poinsettia mosaic virus (PnMV, NC_002164), Switchgrass mosaic virus (SwMV, NC_015522), Turnip yellow mosaic virus (TYMV, X16378).
Acknowledgments
Funding for this work was provided by USDA-ARS CRIS project number 5442-22000-048-00D.
Footnotes
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
References
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenon M., Camborde L., Cheminant S., Jupin I. A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity. EMBO J. 2012;31:741–753. doi: 10.1038/emboj.2011.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K., Li K., Mesecar A.D., Baker S.C. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Howe J., Keese P., Mackenzie A., Meek D., Osorio-Keese M., Skotnicki M., Srifah P., Torronen M., Gibbs A. The tymobox, a sequence shared by most tymoviruses: its use in molecular studies of tymoviruses. Nucleic Acids Res. 1990;18:1181–1187. doi: 10.1093/nar/18.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T.W., Edwards M.C., Gibbs A.J., Haenni A.-L., Hammond R.W., Jupin I., Koenig R., Sabanadzovic S., Martelli G.P. Tymoviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; Oxford: 2011. pp. 944–952. [Google Scholar]
- Edwards M.C., Weiland J.J. First infectious clone of the propagatively transmitted Oat blue dwarf virus. Arch. Virol. 2010;155:463–470. doi: 10.1007/s00705-010-0603-6. [DOI] [PubMed] [Google Scholar]
- Edwards M.C., Zhang Z., Weiland J.J. Oat blue dwarf marafivirus resembles the tymoviruses in sequence, genome organization, and expression strategy. Virology. 1997;232:217–229. doi: 10.1006/viro.1997.8555. [DOI] [PubMed] [Google Scholar]
- Elfakess R., Dikstein R. A translation initiation element specific to mRNAs with very short 5'UTR that also regulates transcription. PLoS One. 2008;3:e3094. doi: 10.1371/journal.pone.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R., Le Gall O., Wellink J. Alpha-like viruses in plants. Semin. Virol. 1991;2:19–25. [Google Scholar]
- Guilley H., Briand J.P. Nucleotide sequence of turnip yellow mosaic virus coat protein mRNA. Cell. 1978;15:113–122. doi: 10.1016/0092-8674(78)90087-9. [DOI] [PubMed] [Google Scholar]
- Higuchi R. Recombinant PCR. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA: 1990. pp. 171–183. [Google Scholar]
- Izadpanah K., Zhang Y.P., Daubert S., Masumi M., Rowhani A. Sequence of the coat protein gene of Bermuda grass etched-line virus, and of the adjacent ‘marafibox’ motif. Virus Genes. 2002;24:131–134. doi: 10.1023/a:1014516515454. [DOI] [PubMed] [Google Scholar]
- Kadaré G., Rozanov M., Haenni A-L. Expression of the turnip yellow mosaic virus proteinase in Escherichia coli and determination of the cleavage site within the 206 kDa protein. J. Gen. Virol. 1995;76:2853–2857. doi: 10.1099/0022-1317-76-11-2853. [DOI] [PubMed] [Google Scholar]
- Karpe Y.A., Lole K.S. Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J. Gen. Virol. 2011;92:2088–2092. doi: 10.1099/vir.0.033738-0. [DOI] [PubMed] [Google Scholar]
- Lao N.T., Maloney A.P., Atkins J.F., Kavanagh T.A. Versatile dual reporter gene systems for investigating stop codon readthrough in plants. PLoS One. 2009;4:e7354. doi: 10.1371/journal.pone.0007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li L., Wang C.C. Capped mRNA with a single nucleotide leader is optimally translated in a primitive eukaryote, Giardia lamblia. J. Biol. Chem. 2004;279:14656–14664. doi: 10.1074/jbc.M309879200. [DOI] [PubMed] [Google Scholar]
- Matsuda D., Dreher T.W. In vivo translation studies of plant viral RNAs using reporter genes. Curr. Protoc. Microbiol. 2005;2:11. doi: 10.1002/9780471729259.mc16k02s00. (00:16K.2.1–16K) [DOI] [PubMed] [Google Scholar]
- Rozanov M.N., Koonin E.V., Gorbalenya A.E. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- Schirawski J., Voyatzakis A., Zaccomer B., Bernardi F., Haenni A.-L. Identification and functional analysis of the turnip yellow mosaic tymovirus subgenomic promoter. J. Virol. 2000;74:11073–11080. doi: 10.1128/jvi.74.23.11073-11080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetz C., Moe R., Blystad D.-R. Symptomless infectious cDNA clone of a Norwegian isolate of Poinsettia mosaic virus. Arch. Virol. 2008;153:1347–1351. doi: 10.1007/s00705-008-0109-7. [DOI] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Purisima E.O., Ménard R. Binding site-based classification of coronaviral papain-like proteases. Proteins: Struct., Funct., Bioinf. 2006;62:760–775. doi: 10.1002/prot.20802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q., Luo R., Liu X., Li K., Chen H., Chen Z., Xiao S. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 2011;85:3758–3766. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland J.J., Dreher T.W. Infectious TYMV RNA from cloned cDNA: effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 1989;17:4675–4687. doi: 10.1093/nar/17.12.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland J.J., Edwards M.C. Evidence that the αa gene of barley stripe mosaic virus encodes determinants of pathogenicity to oat (Avena sativa) Virology. 1994;201:116–126. doi: 10.1006/viro.1994.1271. [DOI] [PubMed] [Google Scholar]
- Weiland J.J., Edwards M.C. Linear-motion tattoo machine and prefabricated needle sets for the delivery of plant viruses by vascular puncture inoculation. Eur. J. Plant Pathol. 2011;131:553–558. [Google Scholar]