Abstract
Background
Rapid tests have had a significant impact on influenza diagnosis, but more accurate tests are needed for hospitalized patients who test negative by rapid methods.
Objective
We sought to determine the increased yield obtained from influenza RT-PCR in hospitalized patients compared to two rapid methods.
Study design
Binax NOW, cytospin-enhanced direct immunofluoroescence (DFA), and influenza A and B multiplex TaqMan RT-PCR were performed on 237 clinical samples.
Results
Binax NOW detected 70 (53.0%), cytospin-DFA detected 127 (96.2%), and TaqMan RT-PCR detected 132 (100%) influenza-positive samples. The difference in sensitivity was significant between RT-PCR and Binax NOW (p < 0.0001), but not between RT-PCR and cytospin-DFA (p = 0.0736). Two samples testing positive for influenza B by all three methods, tested falsely positive for influenza A by Binax. Eight true positive samples did not become reactive by Binax until 30 min, and thus were counted as negative.
Conclusions
The accuracy of real-time RT-PCR should greatly improve the diagnosis of influenza in hospitals using simple rapid flu tests, but may have a more modest impact in hospitals with expertise in cytospin-DFA. Further studies are needed to determine the effect of influenza RT-PCR on patient management and costs in hospitalized patients.
Keywords: Influenza, TaqMan, RT-PCR, Binax NOW, Rapid influenza tests, Immunofluorescence, Cytospin DFA
The impact of influenza on morbidity and mortality of young children as well as on elderly adults has been increasingly recognized (CDC, 2004). Early diagnosis can impact therapy and reduce unnecessary tests and treatments (Barenfanger et al., 2000, Bonner et al., 2003).
In the past, laboratory diagnosis of influenza was largely confined to culture in specialized laboratories. In recent years, rapid and simple influenza tests have become widely implemented, both in general laboratories and at the point of care (Hurt et al., 2007). With the introduction of real-time molecular methods and commercial kits, polymerase chain reaction (PCR) is more accessible to hospital laboratories and is becoming an option to replace culture (Boivin et al., 2004).
In our hospital, cytospin-enhanced direct immunofluorescence (DFA) is performed on respiratory samples when Virology is open, and a rapid influenza test, Binax NOW, is used in the Core Laboratory when Virology is closed (Landry and Ferguson, 2000, Landry et al., 2004). Culture is reserved for hospitalized patients.
Due to the shorter assay time and greater sensitivity of PCR over conventional culture, a multiplex real-time RT-PCR procedure for influenza A and B was validated for clinical use in our laboratory (Ward et al., 2004). As part of the validation process, a prospective study was performed on a subset of clinical samples to determine the increased yield obtained from PCR in hospitalized patients.
1. Methods
In total, 237 nasopharyngeal swabs from 234 patients collected in viral transport medium (M4, Remel, Lenexa, KS) and submitted to the Clinical Virology Laboratory between January and April 2006 for respiratory virus DFA were utilized. Patients ranged in age from 4 months to 93 years; 107 were ≤18 years and 130 were over 18 years of age. Specimens were transported to the laboratory within 1 h of collection and were entered into the study if adequate sample was available to test by all three methods. Since all DFA-negative samples could not be tested, only DFA-negative or inadequate samples (less than 25 respiratory epithelial cells) from inpatients were included. Samples were tested on receipt by DFA and Binax NOW. An aliquot was frozen in lysis buffer at −70 °C and batch tested within 2 weeks by RT-PCR.
During the study period, reflex viral cultures were ordered by physicians on another 683 DFA-negative non-study samples from hospitalized patients. Samples were inoculated into primary rhesus monkey kidney (RhMK), MRC-5 and A549 cells in roller tubes, incubated at 35 °C, and examined for CPE and hemadsorption for 10 days. The influenza detection rate from cultured samples was compared to study samples tested by PCR.
Cytospin enhanced DFA using SimulFluor Respiratory Screen reagents (Chemicon, Temecula, CA) and Binax NOW (Binax Inc., Portland, ME) were performed as previously described (Landry and Ferguson, 2000). Binax results were read at 15 min according to manufacturer's instructions. Nucleic acids from 200 μL of sample were extracted using Nuclisens EasyMag (bioMerieux, Durham, NC). A multiplex one-step influenza A and B TaqMan RT-PCR assay was performed using previously published primers and probes (Ward et al., 2004), Universal Master Mix with UNG and Multiscribe (Applied Biosystems), 900 nM of each primer, 200 nM of each probe, and 5 μL of nucleic acid extract in a 50-μL reaction. The amplification protocol consisted of 48 °C for 30 min, one cycle for 10 min at 95 °C, followed by 45 cycles of 15 s at 95 °C and 1 min at 60 °C. A cycle threshold of <38 was considered a true positive. DFA-negative samples with a C T of ≥38 were re-extracted and tested in duplicate. If both replicates were positive, the sample was called positive. RT-PCR sensitivity was ≤0.01 TCID50/mL for both influenza A and B. RT-PCR was considered the reference standard. A subset of samples was monitored for inhibitors, and none were found.
2. Results
The results for Binax NOW, cytospin-DFA and RT-PCR are shown in Table 1 . One hundred thirty-two samples were positive by RT-PCR, 127 by cytospin-DFA and 70 by Binax NOW. The difference in sensitivity between Binax NOW and DFA, or Binax NOW and RT-PCR, was significant (p < 0.0001). The difference between DFA and RT-PCR was not (p = 0.0736) (McNemar's test). The results for detection of influenza A and B were similar and the impact of age on the sensitivity of Binax NOW and DFA was minimal (Table 2 ).
Table 1.
Comparison of Binax NOW, cytospin-enhanced DFA and TaqMan RT-PCR for detection influenza positives
| True positives detected (%)a |
|||
|---|---|---|---|
| Binax NOWb | Cytopsin DFA | TaqMan RT-PCR | |
| Influenza A | 35 (52.2%) | 62 (92.5%) | 67 (100%) |
| Influenza B | 35 (53.8%) | 65 (100%) | 65 (100%) |
| Total | 70 (53.0%) | 127 (96.2%) | 132 (100%) |
Both PCR and DFA were more sensitive than Binax (p < 0.0001). PCR and DFA sensitivities were not significantly different (p = 0.0736); McNemar's test.
Two samples testing positive for influenza B by all three methods, also tested falsely positive for influenza A. Eight true positive samples (two influenza A and six influenza B) were not reactive until ≥30 min of incubation and were called negative.
Table 2.
Effect of age on sensitivity of Binax NOW and cytospin-DFA
| Method | No. positive by test/No. positive by RT-PCR (%) |
|
|---|---|---|
| Pediatrica | Adult | |
| Binax NOW | 41/78 (52.6%)b | 29/54 (53.7%) |
| Cytospin DFA | 77/78 (98.7%) | 50/54 (92.6%) |
≤18 years of age.
14 of 23 samples (60.9%) from children ≤2 years of age were Binax positive; all were DFA positive.
Two samples positive for influenza B by all three methods tested falsely positive for influenza A by Binax. For one of these, the false reactivity for influenza A was stronger than the true reactivity for influenza B. Eight true positive samples (two influenza A and six influenza B) became reactive by Binax after 30 min of incubation. According to kit instructions, results must be read at 15 min to avoid erroneous results, thus these were considered negative. Sixty-two Binax NOW negative samples were RT-PCR positive.
The specificity of cytospin-enhanced Respiratory Screen DFA was very high, with 127 of 128 DFA positives confirmed by PCR. The one sample that failed to confirm by PCR had only one DFA positive cell. While this could have been a true positive, for the purposes of the study it was deemed false positive.
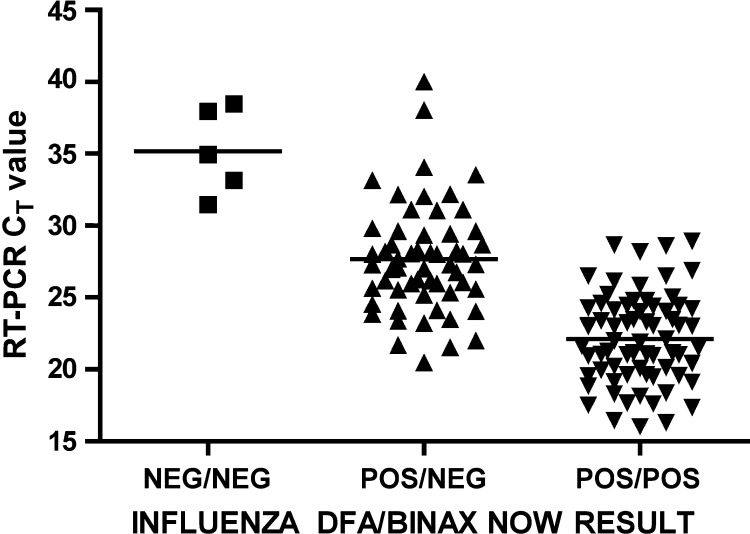
The C T values of samples positive by both DFA and Binax NOW ranged from 16.00 to 28.92 (median 21.93). C T values of samples positive by DFA, but negative by Binax, ranged from 20.49 to 40.02 (median 27.30). The eight samples with delayed reactivity by Binax, and thus classified as negative, had C T values of 23.21–33.53. The relation of C T values to DFA and Binax results is given in Fig. 1 . C T results for influenza A and B were not significantly different and have been combined.
Fig. 1.
CT values according to influenza DFA and Binax NOW test results for 132 samples positive by TaqMan RT-PCR. The five samples positive only by RT-PCR had a median (—) CT value of 34.93 (range 31.47–38.47). The 57 samples positive by cytopsin-DFA but negative by Binax had a median (—) CT value of 27.30 (range 20.49–40.02). The 70 samples positive by both cytospin-DFA and Binax NOW had a median (—) CT value of 21.93 (range 16.00–28.92).
Five DFA-negative samples were found to be influenza A positive by RT-PCR only (Table 3 ). C T values ranged from 31.47 to 38.47, with a median of 34.93. Four of the five were adults, and all had influenza-like illnesses for 2 days or more. One was from a child with underlying disease who had a prior positive sample. In a separate study, it was shown that culture-positive samples generally have C T values of <33 (data not shown). Thus, only one of these five samples would be expected to be culture positive.
Table 3.
Analysis of samples positive by RT-PCR only
| Age (years) | DFA result | Duration of flu-like illness at time of collection (days) | PCR (CT value) | Received in lab | Expected time to resulta (h) |
|---|---|---|---|---|---|
| 12 | Inadequate cells | 14b | 38.47 | Thursday PM | 25 |
| 25 | Negative | 2–3 | 34.93 | Thursday AM | 15 |
| 48 | Negative | 2 | 31.47 | Friday PM | 72 |
| 64 | Negative | 7 | 33.14 | Monday PM | 25 |
| 89 | Negative | 2–3 | 37.94 | Monday PM | 20 |
If PCR performed once a day, Monday to Friday.
Sample from this patient obtained 12 days prior was influenza A-positive by all three tests.
Since the study was performed as part of test validation prior to clinical use, the samples were batch tested by PCR. Currently, the influenza PCR assay is performed once a day, Monday through Friday. According to the day and time of receipt in the lab, results for these samples would have been reported in 15–72 h (median 25 h).
Compared to influenza A and B RT-PCR, the sensitivity, specificity, positive and negative predictive values were 53.0%, 98.1%, 97.2% and 62.9% for Binax NOW and 96.2%, 99.0%, 99.2% and 95.4% for cytospin-DFA.
From the 683 separate DFA-negative samples that were cultured for clinical purposes, only three influenza A positives were detected. Eight adenovirus, 16 rhinovirus, 1 parainfluenza type 4, 17 herpes simplex types 1 and 10 cytomegalovirus were also isolated.
3. Discussion
Rapid flu tests are simple and fast, require no equipment, and can be performed at the point of care. In most situations, these are the only tests used. For hospitalized patients, a more sensitive and specific diagnosis may be warranted and DFA, rapid cell culture, conventional cell culture, and PCR are all options. In this study we assessed the potential impact of influenza A and B RT-PCR on hospitalized patients. Selecting DFA negative and inadequate samples on hospitalized patients rather than random samples could have been introduced bias. However, we sought to evaluate the utility of PCR as it would be employed in our hospital.
PCR greatly enhanced the accuracy of influenza detection compared to the rapid flu test, detecting an additional 62 positives beyond the 70 detected by Binax NOW. Binax also had two false positive influenza A results. Of note, M4 transport media is used in our laboratory, since it allows multiple tests, including rapid flu, DFA, culture, and PCR, to be performed on one sample, However, it is possible that Binax NOW would have performed better with a different, low volume collection fluid and a more concentrated sample.
Until now, conventional culture has been the standard clinical test for DFA-negative samples in our facility. During the study period, reflex cultures were performed on 683 DFA-negative samples, but only three influenza A positives were detected. In contrast, PCR detected five additional influenza positives out of 109 samples (93 DFA-negative and 16 DFA inadequate). Thus, PCR provided a higher yield than culture, as expected from the analytical sensitivity of the PCR assay (≤0.01 TCID50/mL).
Ultimately, the impact of influenza diagnosis on management of hospitalized patients will depend not only on sensitivity, but also on time to result. Antiviral treatment should be given within 48 h of symptom onset, but by the time patient arrives at the hospital, as shown in Table 3, often 2 days or more has already passed. For infection control practices to be implemented, it is best to have results prior to bed assignment. Likewise to avert unnecessary use of antibiotics or diagnostic tests, results should as close to the time of admission as possible.
Of the more sensitive rapid test options, DFA results are available the fastest, namely within 2 h when Virology is open, and within 10–14 h if the sample is submitted when Virology is closed. However, DFA accuracy varies with the laboratory and cytospin preparation of slides is rarely employed. The newer R-mix cell cultures are within the capability of most laboratories, are highly sensitive for influenza, and results are reported at 1 and 2 days (Barenfanger et al., 2001, Dunn et al., 2004). In our institution, influenza A and B PCR is now performed once a day, 5 days a week. Thus time to result is 8–32 h weekdays, but up to 80 h or more on weekends. For the five samples detected by PCR only, results would have been reported in 15–72 h, and beyond the 48 h window recommended for antiviral therapy.
The utility of PCR would be greatly enhanced if multiplexed to include multiple viruses while retaining sensitivity greater than culture; if viruses not detected by DFA, such as coronaviruses and rhinoviruses, were included; and if performed at least once a day. There are now commercially available kits using conventional RT-PCR that detect multiple respiratory virus targets (Lee et al., 2007, Li et al., 2007, Mahony et al., 2007, Nolte et al., 2007). However the frequency with which these assays could be performed in the routine hospital laboratory remains uncertain.
It is important to note that multiplex PCR assays may have sensitivities of only 82.8–86.2% for influenza A and 63.3–73.3% for RSV, the two most important viral respiratory pathogens (Lee et al., 2007), or only 83% for influenza B (Mahony et al., 2007). Therefore it is critical that laboratories compare the newer methods in their own laboratories to validate improved sensitivity over current methods.
To date, the cost benefits of rapid influenza tests in children (Bonner et al., 2003) and respiratory virus cytospin DFA in adults and children (Barenfanger et al., 2000, Bonner et al., 2003) have been shown to have both clinical and cost benefits. Whether respiratory virus PCRs will also have favorable cost/benefit profiles is not known, and will require prospective, controlled studies stratified by the age of the patient, severity of disease, and the presence of co-morbid conditions.
In this study, RT-PCR for influenza A and B detected many additional positives not detected by Binax NOW, but only a small number of positives not detected by cytospin-DFA. While the accuracy of real-time RT-PCR should be of significant benefit to those hospitals using simple rapid influenza tests, it may have a more modest impact on hospitals with expertise in cytospin-DFA. Further studies are needed to assess the impact of influenza PCR, including highly multiplexed assays, on both patient outcomes and costs in the hospital setting.
Acknowledgement
We thank Gerri Russo for collecting and organizing the data.
References
- Barenfanger J., Drake C. Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J Clin Microbiol. 2000;38(8):2824–2828. doi: 10.1128/jcm.38.8.2824-2828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenfanger J., Drake C. R-mix cells are faster, at least as sensitive and marginally more costly than conventional cell lines for the detection of respiratory viruses. J Clin Virol. 2001;22(1):101–110. doi: 10.1016/s1386-6532(01)00171-8. [DOI] [PubMed] [Google Scholar]
- Boivin G., Cote S. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J Clin Microbiol. 2004;42(1):45–51. doi: 10.1128/JCM.42.1.45-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner A.B., Monroe K.W. Impact of the rapid diagnosis of influenza on physician decision-making and patient management in the pediatric emergency department: results of a randomized, prospective, controlled trial. Pediatrics. 2003;112(2):363–367. doi: 10.1542/peds.112.2.363. [DOI] [PubMed] [Google Scholar]
- CDC (2004). Update: influenza-associated deaths reported among children aged <18 years—United States, 2003–2004 influenza season. MMWR Morb Mortal Wkly Rep 52(53), 1286–8. [PubMed]
- Dunn J.J., Woolstenhulme R.D. Sensitivity of respiratory virus culture when screening with R-mix fresh cells. J Clin Microbiol. 2004;42(1):79–82. doi: 10.1128/JCM.42.1.79-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt A.C., Alexander R. Performance of six influenza rapid tests in detecting human influenza in clinical specimens. J Clin Virol. 2007;39(2):132–135. doi: 10.1016/j.jcv.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Landry M.L., Cohen S. Comparison of Binax NOW and Directigen for rapid detection of influenza A and B. J Clin Virol. 2004;31(2):113–115. doi: 10.1016/j.jcv.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Landry M.L., Ferguson D. SimulFluor respiratory screen for rapid detection of multiple respiratory viruses in clinical specimens by immunofluorescence staining. J Clin Microbiol. 2000;38(2):708–711. doi: 10.1128/jcm.38.2.708-711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.M., Grindle K. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J Clin Microbiol. 2007;45(8):2626–2634. doi: 10.1128/JCM.02501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., McCormac M.A. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J Clin Microbiol. 2007;45(7):2105–2109. doi: 10.1128/JCM.00210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J., Chong S. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007;45(9):2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte F.S., Marshall D.J. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J Clin Microbiol. 2007;45(9):2779–2786. doi: 10.1128/JCM.00669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.L., Dempsey M.H. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004;29(3):179–188. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]