Abstract
Macroautophagy (herein referred to as autophagy) is an evolutionary ancient mechanism that culminates with the lysosomal degradation of superfluous or potentially dangerous cytosolic entities. Over the past 2 decades, the molecular mechanisms underlying several variants of autophagy have been characterized in detail. Accumulating evidence suggests that most, if not all, components of the molecular machinery for autophagy also mediate autophagy-independent functions. Here, we discuss emerging data on the non-autophagic functions of autophagy-relevant proteins.
Keywords: ATG5, BECN1, LC3-associated phagocytosis, proliferation, regulated cell death, vesicular trafficking
Proteins that regulate macroautophagy mediate a number of non-autophagy roles in physiology and disease.
Main Text
Introduction
From an evolutionary perspective, the convergence of multiple, biochemically unrelated functions into a single protein constitutes a valuable strategy for organisms to economize genetic and metabolic resources (Storz, 2016). Nonetheless, there is a general tendency to attribute unique functions to specific proteins, often reflecting the earliest or most abundant literature on the topic. As an example, while caspase 3 (CASP3) is globally recognized as a key effector in apoptosis (Galluzzi et al., 2018a, Singh et al., 2019), its non-apoptotic role in the differentiation of multiple cell types (Nakajima and Kuranaga, 2017) is largely underappreciated. The same issue applies to hundreds other proteins that have been characterized mostly, if not only, in the context of a single cellular process, including multiple components of the molecular machinery for macroautophagy.
Macroautophagy is an evolutionarily conserved stress-responsive process that disposes of superfluous or potentially dangerous cytosolic entities (e.g., damaged mitochondria, invading pathogens) upon sequestration within double-membraned vesicles (autophagosomes) and delivery to lysosomes for degradation (Galluzzi et al., 2017a, Levine and Kroemer, 2019) (Figures 1A and 1B). Two other forms of autophagy have been described: (1) microautophagy, which involves the delivery of autophagy substrates to lysosomes upon invagination of the lysosomal membrane, and (2) chaperone-medicated autophagy (CMA), which relies on a specific splicing isoform of lysosomal-associated membrane protein 2 (LAMP2) as a translocase for KFERQ-containing cytosolic proteins into the lysosomal lumen (Kaushik and Cuervo, 2018, Li et al., 2012a). The molecular apparatus that underlies these multiple variants of autophagy has been characterized with increasing precision (Dikic and Elazar, 2018) (Box 1 ). The crosstalk between bona fide autophagic responses and multiple other cellular processes has also been intensively investigated. Thus, it has become clear that autophagy occupies a central position in the biology of most eukaryotes, interfacing with the regulation of core metabolism (He et al., 2012, Sousa et al., 2016), damage control (Fernández et al., 2018, Khaminets et al., 2015, Mathew et al., 2007), and cell death (Green et al., 2014, Wang et al., 2012, Wei et al., 2013). Until recently, however, relatively little attention has been given to the possibility that components of the autophagy apparatus could mediate non-autophagic functions (Levine and Kroemer, 2019).
Figure 1.
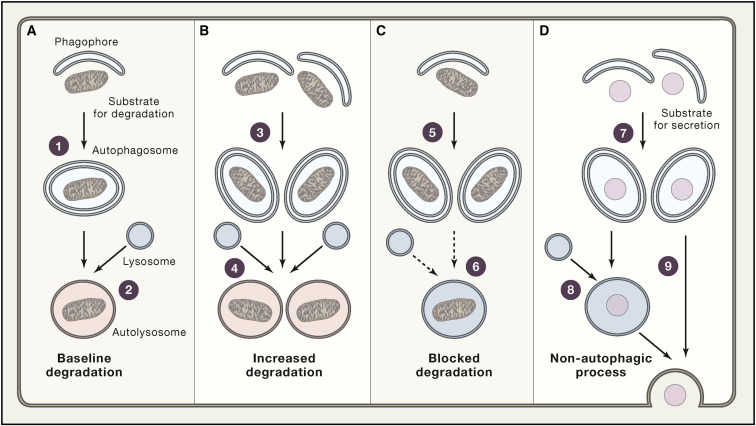
Degradative Autophagic Responses, Autophagy Blockade, and Non-autophagic Functions of Autophagy Machinery
(A) In physiological conditions, autophagosomes form (1) and successfully fuse with lysosomes (2) at baseline rates, underling the ability of autophagy to support normal cellular functions.
(B) In the presence of an autophagic stimulus such as nutrient deprivation, the rate of autophagosome formation (3), autophagosome-lysosome fusion, and lysosomal degradation increases (4), resulting in accelerated degradation of autophagic substrates.
(C) Autophagosomes also accumulate in the absence of an upstream autophagic stimulus (5) when lysosomal functions are inhibited (6), such as in the presence of lysosomotropic agents.
(D) Finally, the autophagosome compartment expands, driven by an upstream stimulus (7), when autophagosomal content is destined to secretion, either upon (8) or independent of (9) fusion with lysosomes in the absence of lysosomal degradation. Thus, widely employed assays only based on the maturation of LC3 not only are unable to determine whether an expansion of the autophagosomal compartment compared to baseline (A) reflects upstream autophagy activation coupled to efficient lysosomal degradation (B) or downstream inhibition of autophagosome-lysosome fusion or lysosomal acidification (C), but also they cannot identify situations in which activation of upstream autophagy-relevant signaling modules mediate non-autophagic effects (D).
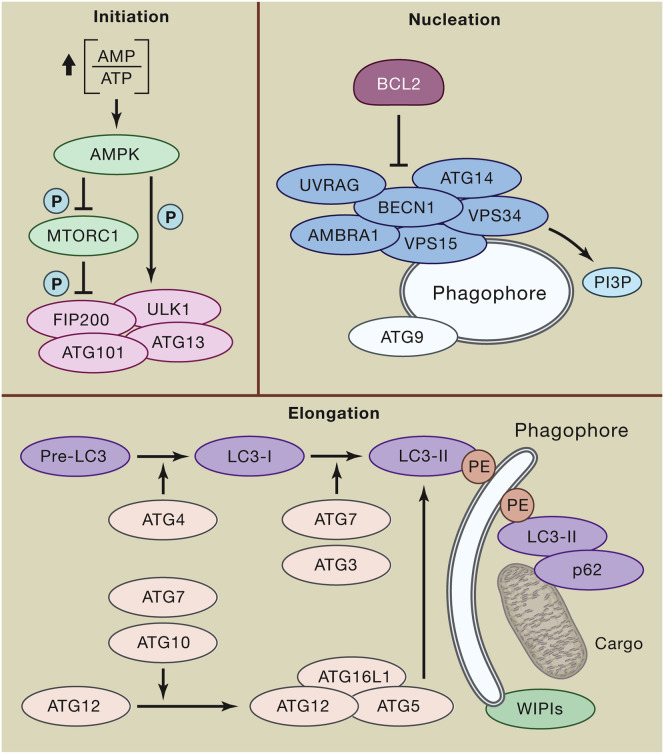
Box 1. Principles of Autophagy Regulation in Mammals.
Canonical autophagic responses, which are best exemplified by starvation-driven autophagy, can be schematically subdivided into five phases: (1) initiation, (2) phagophore nucleation, (3) phagophore expansion and substrate selection, (4) autophagosome-lysosome fusion, and (5) lysosomal substrate degradation (Galluzzi et al., 2017b). During initiation, the bioenergetic stress imposed by decreased nutrient availability manifests with increasing AMP levels, resulting in the activation of AMPK and consequent (1) inactivating phosphorylation of mechanistic target of rapamycin (MTOR) complex 1 (MTORC1) and (2) direct activating phosphorylation or indirect activating dephosphorylation (as a consequence of MTORC1 inhibition) of multiple proteins involved in initiation (e.g., ATG13, ULK1) and nucleation (e.g., ATG14, AMBRA1, UVRAG, BECN1). In this setting, ULK1 operates as part of a multiprotein complex containing ATG13, ATG101, and FIP200. ULK1-, AMPK-, and MTORC1-related phosphorylation/dephosphorylation events trigger phagophore nucleation (mostly at the ER) upon PI3P production by a supramolecular complex with class III PI3K activity consisting of VPS34 (the catalytic subunit), VPS15, BECN1, AMBRA1, and/or UVRAG, coupled to recruitment of ATG9-containing vesicles. The activity of this complex is under tonic inhibition by BCL2, reflecting the ability of BCL2 to engage in physical inhibitory interactions with BECN1. Phagophore elongation involves two ubiquitin-like conjugation systems. On one hand, ATG7 and ATG10 operate sequentially to catalyze the formation of ATG12-ATG5:ATG16L1 complexes. On the other hand, ATG4, ATG7, and ATG3 cooperate to cleave the precursors of LC3-like proteins into their mature forms, followed by conjugation to phosphatidylethanolamine (PE) and recruitment to autophagosomes forming with the support of WIPI proteins. LC3 and LC3 homologues enable autophagosomes with the ability to bind autophagic substrates and/or proteins that mediate cargo selectivity (including p62). Indeed, while autophagic responses to nutrient deprivation are relatively non-selective, multiple other variants of autophagy exhibit exquisite substrate specificity. On closure, autophagosomes fuse with lysosomes to generate autolysosomes, generally followed by luminal acidification and consequent activation of lysosomal hydrolases that mediate substrate degradation. This latter step, which appears to involve (at least to some extent) the conjugation systems responsible for elongation (Tsuboyama et al., 2016), is critical to discriminate bona fide autophagic responses from autophagy-independent functions of the autophagic machinery (Galluzzi et al., 2017a).
P, inorganic phosphate.
Along with methodological issues linked to widely employed approaches to measure degradative macroautophagy (from here onward referred to as autophagy) (Evans et al., 2018b), such under-appreciation of biological complexity may have considerably confounded the interpretation of hundreds of experiments investigating the impact of autophagy on several physiological and pathological states (Figures 1C and 1D). In particular, this may have led investigators to misattribute phenotypic or functional effects caused by the pharmacological or genetic perturbation of single autophagy-regulatory factors to autophagy as a process (Galluzzi et al., 2017b). Accumulating evidence suggests indeed that most, if not all, components of the molecular apparatus for autophagy mediate one or multiple effects that do not depend on lysosomal degradation of autophagy substrates (Table 1 ). In particular, autophagy-relevant proteins mediate non-autophagic effects that impinge on cellular functions linked to membrane biology, such as (1) endocytosis, phagocytosis, and intracellular vesicular trafficking; (2) conventional and non-conventional secretion; and (3) cytokinesis, as well as on (at least apparently) membrane-unrelated functions, such as (1) inflammatory and immune responses, (2) cell death, (3) genomic stability, and (4) cell proliferation (Table 2 ). Moreover, several pathogens acquired the ability to hijack non-autophagic functions of the autophagy machinery for their own benefit, suggesting that such functions may have appeared early in the course of host-pathogen co-evolution (Choi et al., 2018, Evans et al., 2018a).
Table 1.
Non-autophagic Functions of Core Components of the Autophagy Apparatus
ADCD, autophagy-dependent cell death; ER, endoplasmic reticulum; GA, Golgi apparatus; LAP, LC3-associated phagocytosis; PRR, pattern recognition receptor.
Referring to bacteria replicating in the cytoplasm of infected cells
Table 2.
Major cellular functions involving components of the autophagy machinery
| Functions | Protein | Autophagic Role | References |
|---|---|---|---|
| Related to membrane biology | |||
| Cytokine secretion | ATG7 | Elongation | Henault et al., 2012, Li et al., 2016 |
| ULK1 | Initiation | ||
| Cytokinesis | BECN1 | Nucleation | Thoresen et al., 2010, You et al., 2016 |
| BIF-1 | Nucleation | ||
| UVRAG | Initiation | ||
| VPS15 | Nucleation | ||
| VPS34 | Nucleation | ||
| ULK1 | Initiation | ||
| Endocytosis | BECN1 | Nucleation | Lee et al., 2011, Liang et al., 2008, McKnight et al., 2014, Pirooz et al., 2014, Rohatgi et al., 2015, Shravage et al., 2013, Thoresen et al., 2010 |
| BIF-1 | Nucleation | ||
| RAB7A | Fusion | ||
| UVRAG | Initiation | ||
| VPS15 | Nucleation | ||
| VPS34 | Nucleation | ||
| ER-to-GA anterograde transport | ULK1 | Initiation | Joo et al., 2016, Wang et al., 2018 |
| ULK2 | Initiation | ||
| Exosome secretion | ATG12 | Elongation | Guo et al., 2017, Jaé et al., 2015, Murrow et al., 2015, Shrivastava et al., 2015 |
| ATG16L1 | Elongation | ||
| ATG3 | Elongation | ||
| ATG5 | Elongation | ||
| ATG7 | Elongation | ||
| RAB7A | Fusion | ||
| GA-to-ER retrograde transport | VPS34 | Nucleation | He et al., 2013 |
| UVRAG | Initiation | ||
| Granule exocytosis | ATG4B | Elongation | DeSelm et al., 2011, Patel et al., 2013 |
| ATG5 | Elongation | ||
| ATG7 | Elongation | ||
| LC3 | Cargo selection | ||
| RAB7A | Fusion | ||
| LAP | ATG12 | Elongation | Martinez et al., 2015, Martinez et al., 2016, Sanjuan et al., 2007 |
| ATG16L1 | Elongation | ||
| ATG3 | Elongation | ||
| ATG4B | Elongation | ||
| ATG5 | Elongation | ||
| ATG7 | Elongation | ||
| BECN1 | Nucleation | ||
| LC3 | Cargo selection | ||
| RUBCN | Nucleation | ||
| UVRAG | Initiation | ||
| VPS34 | Nucleation | ||
| Melanogenesis | UVRAG | Initiation | Yang et al., 2018 |
| Non-canonical protein secretion | ATG5 | Elongation | Dupont et al., 2011, Kimura et al., 2017, Zhang et al., 2015 |
| RAB7A | Fusion | ||
| Pathogen controla | ATG12 | Elongation | Hwang et al., 2012, Kimmey et al., 2015, Mauthe et al., 2016, Selleck et al., 2015, Zhao et al., 2008 |
| ATG13 | Initiation | ||
| ATG16L1 | Elongation | ||
| ATG5 | Elongation | ||
| ATG7 | Elongation | ||
| FIP200 | Initiation | ||
| LC3 | Cargo selection | ||
| NDP52 | Cargo selection | ||
| p62 | Cargo selection | ||
| Pathogen replicationb and release | LC3 | Cargo selection | Al-Younes et al., 2011, Alirezaei et al., 2015, Sin et al., 2017, Wong et al., 2008 |
| Phagocytosis | ATG16L1 | Elongation | Tung et al., 2010, Xiong et al., 2015 |
| ATG9 | Initiation | ||
| Vision cycle | ATG5 | Elongation | Muniz-Feliciano et al., 2017, Kim et al., 2013 |
| Others | |||
| ADCD | AMBRA1 | Nucleation | Elgendy et al., 2014, Gao et al., 2011, Goodall et al., 2016, Haller et al., 2014, Imagawa et al., 2016, Lee et al., 2012, Rubinstein et al., 2011, Singh et al., 2010, Strappazzon et al., 2016 |
| ATG12 | Elongation | ||
| ATG13 | Initiation | ||
| ATG14 | Nucleation | ||
| ATG5 | Elongation | ||
| ATG7 | Elongation | ||
| ATG9 | Initiation | ||
| BECN1 | Nucleation | ||
| IRGM | Unclear | ||
| p62 | Cargo selection | ||
| VPS34 | Nucleation | ||
| Cell proliferation | ATG3 | Elongation | Afzal et al., 2015, O’Sullivan et al., 2016, Pei et al., 2015 |
| ATG5 | Elongation | ||
| ATG7 | Elongation | ||
| UVRAG | Initiation | ||
| Centrosome functions | BECN1 | Nucleation | Park et al., 2014 |
| DNA repair | UVRAG | Initiation | Yang et al., 2016, Zhao et al., 2012 |
| Immunological memory | ATG5 | Elongation | Puleston et al., 2014, Xu et al., 2014 |
| ATG7 | Elongation | ||
| PRR signaling | ATG7 | Elongation | Henault et al., 2012, Sorbara et al., 2013 |
| ATG16L1 | Elongation | ||
| BECN1 | Nucleation | ||
| LC3 | Cargo selection | ||
| RUBCN | Nucleation | ||
ADCD, autophagy-dependent cell death; ER, endoplasmic reticulum; GA, Golgi apparatus; LAP, LC3-associated phagocytosis; PRR, pattern recognition receptor.
Partially unrelated to membrane biology
Referring to the replication of pathogens in the cytoplasm of infected cells
Here, we discuss emerging data on non-autophagic functions of components of the molecular apparatus for autophagy.
Endocytosis and Phagocytosis
Autophagy relies on the formation, maturation, and subcellular relocalization of autophagosomes, ultimately resulting in their fusion with lysosomes (Galluzzi et al., 2017a). Not surprisingly, several components of the molecular machinery for autophagy regulate vesicular trafficking, including endocytosis and phagocytosis, often as signaling modules (Figure 2 ).
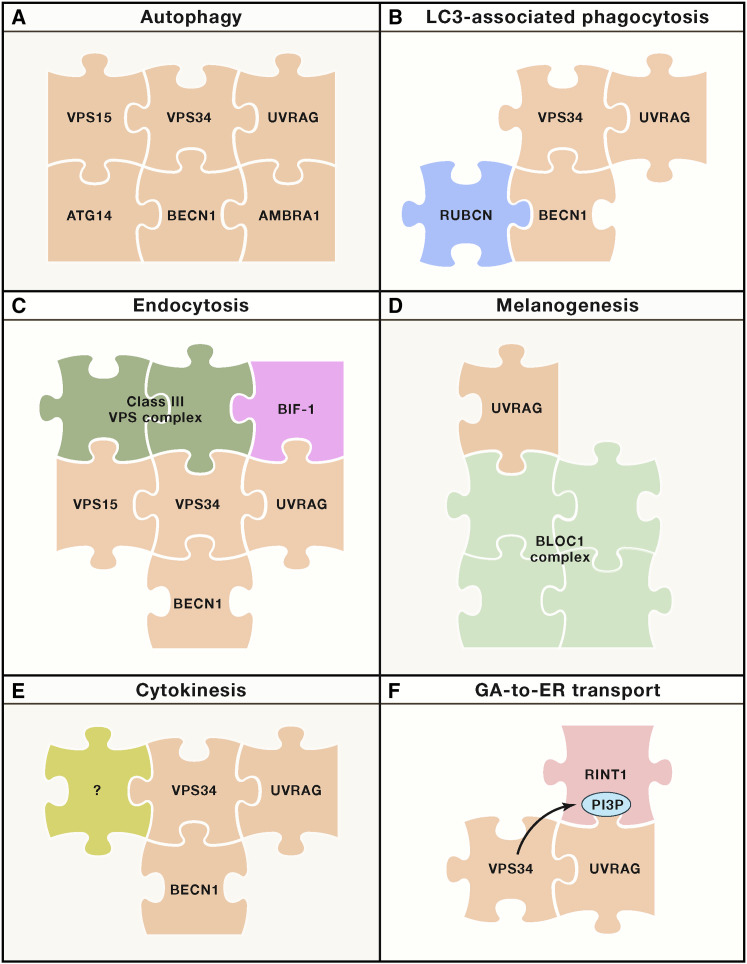
Figure 2.
Molecular Interface between Autophagy and Membrane Biology
Multiple components of the molecular machinery for autophagy mediate non-autophagic functions linked to the rearrangement and trafficking of intracellular membranes independently of bona fide autophagic responses. In this setting, different supramolecular entities can be assembled around components of the class III phosphatidylinositol 3-kinase complex that drives autophagy (A) to differentially regulate specific non-autophagic functions, including LC3-associated phagocytosis (C), endocytosis (C), melanogenesis (D), cytokinesis (E), and GA-to-ER transport (F).
ER, endoplasmic reticulum; GA, Golgi apparatus; PI3P, phosphatidylinositol 3-phosphate.
Independently of their pro-autophagic functions, phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3, best known as VPS34), phosphoinositide-3-kinase regulatory subunit 4 (PI3KR4, best known as VPS15), and beclin 1 (BECN1) promote endocytosis and endosome maturation. UV radiation resistance-associated (UVRAG), a pro-autophagic BECN1 activator, also favors endocytosis through interactions with the class C vacuolar protein-sorting tethering complex and consequent increase in RAB7A activity (Liang et al., 2008, Pirooz et al., 2014). Initially, such an effect, which involves accelerated endosomal maturation, was believed to be unrelated to BECN1 and the VPS34 complex (Liang et al., 2008). However, at least in some cell types, VPS34 and VPS34 interactors involved in the control of growing membranes, including BECN1, VPS15, and SH3 domain containing GRB2 like, endophilin B1 (SH3GLB1, best known as BIF-1), have been implicated in this process, with no apparent roles for other VPS34 regulators such as ATG14 and RUN and cysteine-rich domain containing beclin 1 interacting protein (RUBCN) (McKnight et al., 2014, Thoresen et al., 2010). Thus, while mice with a cerebellum-specific knockout of Atg7 exhibit a neurodegenerative disorder that develops over multiple months, the deletion of Becn1 from the same cellular compartment imposes rapid neurodegeneration (McKnight et al., 2014). Most likely, this difference reflects the endocytic defect imposed by the loss of BECN1, leading to alterations in growth factor receptor signaling (McKnight et al., 2014). Reinforcing this notion, downregulation of BECN1 (which is a common event in human tumors) (Tang et al., 2015, Yue et al., 2003) has been associated with increased AKT serine/threonine kinase 1 (AKT1) signaling in breast cancer cells (Rohatgi et al., 2015). Such an observation could be ascribed to defective endosomal maturation in BECN1-incompetent cells, resulting in increased residency of growth factor receptors at phosphatidylinositol 3-phosphate (PI3P)-negative signaling-competent endosomal compartments (Rohatgi et al., 2015). The function of BECN1 and UVRAG in endocytosis appears to be evolutionarily conserved (Lee et al., 2011, Shravage et al., 2013).
PI3P-bound UVRAG localizes to the endoplasmic reticulum (ER), where it interacts with RAD50 interactor 1 (RINT1) to constitute a tethering platform for vesicles coming in from the Golgi apparatus (GA) in the context of retrograde transport (He et al., 2013). Both downregulation of UVRAG and PI3P depletion cause defects in Golgi-to-ER retrograde transport that are not linked to the autophagic functions of UVRAG (He et al., 2013). However, autophagy induction by starvation and rapamycin results in the redistribution of UVRAG from RING1-containing complexes to BECN1-, VPS34- and BIF-1-containing complexes and consequent ATG9 recruitment at sites of autophagosome production (He et al., 2013). This scenario exemplifies the mutual regulation of two biological functions based on the limited availability of a common molecular player. Interestingly, Uvrag −/− mouse melanoma cells display significant whitening as compared to their wild-type counterparts, a phenotype that cannot be recapitulated by ATG5, ATG16L1 or BECN1 depletion, or pharmacological VPS34 inhibition (Yang et al., 2018). In this context, loss of UVRAG destabilizes a supramolecular entity involved in endosome-to-melanosome fusion commonly known as BLOC-1 complex, an effect that is not sensitive to lysosomal inhibition (Yang et al., 2018). At least in some cells, a BECN1- and UVRAG-containing, ATG14-independent PI3K complex has also been suggested to participate in the control of cytokinesis, the final stage of mitosis whereby daughter cells separate once their nuclei have divided (Thoresen et al., 2010, You et al., 2016). These latter observations reinforce the notion that multiple modules of the autophagy apparatus regulate the rearrangement of cellular membranes.
A specific form of phagocytosis engaged by several pathogens and dead cell corpses, which is known as LC3-associated phagocytosis (LAP), depends on several proteins involved in bona fide autophagic responses (Martinez et al., 2015, Martinez et al., 2016, Sanjuan et al., 2007). These include not only UVRAG, BECN1, and VPS34, which promote autophagy as a part of a single supramolecular entity, but also ATG3, ATG4, ATG5, ATG7, ATG12, ATG16L1, and multiple members of the microtubule-associated protein 1 light chain 3 (MAP1LC3, best known as LC3) family, which are all involved in the two major conjugation systems required for canonical autophagy (Box 1) (Martinez et al., 2015, Martinez et al., 2016, Sanjuan et al., 2007). Importantly, LAP differs from bona fide autophagy in that (1) it targets to lysosomal degradation extracellular entities that never acquire a cytosolic localization and (2) it relies on single-membraned vesicles (Galluzzi et al., 2017a). Moreover, LAP occurs independently of unc-51 like autophagy-activating kinase 1 (ULK1), autophagy and beclin 1 regulator 1 (AMBRA1), ATG14, and RB1-inducible coiled-coil 1 (RB1CC1, best known as FIP200), which are all involved in autophagy, but relies on RUBCN, NAPDH oxidase 2, and the WD domain of ATG16L1, which are all dispensable for autophagy (Fletcher et al., 2018, Martinez et al., 2015, Martinez et al., 2016, Rai et al., 2018). Thus, the deletion of Becn1, Atg5, or Atg7 (but not Rb1cc1) from myeloid cells drives an autoimmune disorder similar to human systemic lupus erythematosus (SLE), which can be recapitulated by the whole-body knockout of Rubcn but not by that of Ulk1 (Martinez et al., 2016). Similarly, the myeloid cell-specific deletion of Becn1, Pik3c3, Atg5, Atg7, or Atg16l1 (but not Rb1cc1 or Atg14) as well as the whole-body knockout of Rbcn or Cybb (coding for a subunit of NADPH oxidase 2), but not Ulk1, promotes anti-tumor T cell immunity as a consequence of improved inflammatory responses to dead cell corpses (Cunha et al., 2018). In the retinal epithelium, BECN1, ATG5, RUBCN, and LC3 are required for the phagocytosis of photoreceptor outer segments (POS), which is fundamental for normal vision (Kim et al., 2013, Muniz-Feliciano et al., 2017). This process, however, occurs independently of ULK1, ATG13, and FIP200, which are all required for conventional autophagic responses (Kim et al., 2013). Thus, at least some of the phenotypes originating from the depletion of the aforementioned proteins may stem from defects in LAP, not autophagy, especially in the context of pathogen control and disposal of cell corpses (Heckmann et al., 2017). Interestingly, the orthologs of ATG9 and ATG16L1 also mediate autophagy-independent pro-phagocytic effects in Dictyostelium discoideum, a simple eukaryote that transitions from unicellular to multicellular life over the course of its vital cycle (Tung et al., 2010, Xiong et al., 2015). Along with the notion that autophagy is conserved in both unicellular eukaryotes and plants (Üstün et al., 2017), this observation suggests that both the autophagy-dependent and autophagy-independent functions of the autophagic machinery may have appeared early during evolution.
Conventional and Non-conventional Secretion
Conventional protein secretion involves the anterograde transport of ER-derived vesicles to the GA, vesicle trafficking from the cis though the trans GA, and ultimately, fusion of vesicles released from the trans GA with the plasma membrane (Braakman and Bulleid, 2011). Although this process has been intensively investigated, only recently it has become clear that, at least in some cell types, components of the autophagic machinery are required for conventional protein secretion.
Mice with a brain-specific co-deletion of Ulk1 and Ulk2 are born at Mendelian ratios yet die rapidly after birth (40% in the first 24 h) owing to massive degeneration of pyramidal neurons in the CA1 region (Joo et al., 2016). Mice with conditional Ulk1/Ulk2 co-deletion in the brain surviving the first 24 h after birth resemble their Atg5- and Atg7-deficient counterparts as they display abnormal limb-clasping reflexes, but they do not develop cerebellar ataxia, suggesting that the neuronal phenotype caused by the lack of Ulk1 and Ulk2 may not result from an autophagic defect (Joo et al., 2016). Indeed, Ulk1 −/− Ulk2 −/− neurons do not exhibit accumulation of autophagic substrates such as sequestosome 1 (SQSTM1, best known as p62) but manifest signs of the unfolded protein response (Joo et al., 2016, Wang et al., 2018), an intracellular pathway of adaptation to accumulation of unfolded polypeptides in the ER lumen (Galluzzi et al., 2018b). Consistently, both ULK1 and ULK2 can phosphorylate SEC16 homolog A, endoplasmic reticulum export factor (SEC16A) to drive the anterograde transport of ER-derived vesicles to the GA, and this pathway is insensitive to depletion of ATG13 (which is required for the pro-autophagic functions of ULKs), ATG14, and ATG7 (Joo et al., 2016). Of note, neurodegeneration imposed by Ulk1/Ulk2 co-deletion is also accompanied by defective axonal pathfinding affecting multiple areas of the forebrain, and similar alterations cannot be recapitulated by the brain-specific deletion of Atg7 or Rb1cc1 (Wang et al., 2018). However, to what extent these defects in axonal projection reflect secretory alterations in post-synaptic cells as opposed to primary deficiencies in pre-synaptic cells remains to be clarified. In apparent contrast with the observations discussed here above, ULK1 is not required for the secretion of type I interferon (IFN) by plasmacytoid dendritic cells (pDCs) exposed to DNA-immunoglobulin complexes, whereas ATG7 is required (Henault et al., 2012). In this setting, however, ATG7 does not promote anterograde ER-to-GA vesicle trafficking but enables pDCs to take up DNA-immunoglobulin complexes via LAP, followed by Toll-like receptor 9 (TLR9) signaling at acidified phagosomes (Henault et al., 2012). Intriguingly, Atg1, the ortholog of mammalian ULK1 in Drosophila melanogaster, is also involved in JNK-driven secretion of mitogens that underlies apoptosis-induced compensatory proliferation, an effect that does not depend on multiple other components of the autophagy machinery in flies including Atg3, Atg6 (the BECN1 ortholog), Atg8 (the LC3 ortholog), and Vps34 (Li et al., 2016). Moreover, Caenorhabditis elegans lacking the worm homolog of mammalian ULK1 (i.e., unc-51) display axonal defects that resemble those of Ulk1 −/− Ulk2 −/− neurons (Hedgecock et al., 1985). These observations suggest that the autophagy-independent functions of ULKs in conventional secretion evolved early during evolution.
In contrast to Atg5 −/− and Atg7 −/− neurons, colonic goblet cells lacking Atg5, Atg7, or Map1lc3b exhibit a secretory defect causing accumulation of intracellular mucin granules (Patel et al., 2013). In this context, secretory alterations reflect the inability of goblet cells lacking Atg5, Atg7, or Map1lc3b to produce reactive oxygen species (ROS) in sufficient amounts to drive granule exocytosis, secondary to a defect in the formation of NADPH oxidase-competent subcellular compartments at the interface between autophagosomes and endosomes (Patel et al., 2013). Along similar lines, cells unable to form non-conventional ATG3-ATG12 conjugates, which are formed by ATG7 but are not involved in bona fide autophagic responses, accumulate autophagosomes and late endosomes in baseline conditions, coupled to reduced exosomal output (Murrow et al., 2015). A similar phenotype results from the deletion of Atg5 or Atg16l1 (but not Atg7 or Atg14) from mouse embryonic fibroblasts (MEFs) as well as from the deletion of ATG5 from human breast cancer cells, where it limits exosome-driven metastatic dissemination (Guo et al., 2017). However, in the former setting, ATG3-ATG12 appears to drive exosome secretion upon interaction with the exosomal protein programmed cell death 6 interacting protein (PDCD6IP, best known as ALIX) (Murrow et al., 2015), while in the latter scenario, ATG5 appears to disrupt endosomal acidification by disassociating the V1V0-ATPase, resulting in diversion of late endosomes toward secretion (rather than lysosomal degradation) (Guo et al., 2017). In some cases, ATG7 seems to be required for optimal exosome release (potentially linked to its role in ATG3-ATG12 conjugation) (Murrow et al., 2015, Shrivastava et al., 2015), while in other settings, exosome release operates normally in the absence of ATG7 (Guo et al., 2017, Sahu et al., 2011). Irrespective of such unknowns, these observations link multiple components of the autophagy machinery to exosome secretion via autophagy-independent mechanisms.
The hypothesis that autophagy-relevant proteins could be involved in the release of intracellular material into the microenvironment irrespective of lysosomal degradation has first been formulated in the setting of “non-canonical secretion,” a form of secretion of cytosolic entities devoid of leader peptides for ER translocation (Rabouille, 2017). One of the substrates of non-conventional secretion is mature interleukin 1 beta (IL1B, best known as IL-1β), which is produced in the cytosol of cells with pro-inflammatory activity (like macrophages) upon proteolytic maturation of the IL-1β precursor by the NLRP3 inflammasome (Rathinam and Fitzgerald, 2016). Bone-marrow-derived macrophages treated with nigericin (an inflammasome activator) secrete increased amounts of IL-1β in conditions of nutrient deprivation (as compared with nigericin-treated macrophages maintained in control conditions), and IL-1β secretion can be quenched by Atg5 deletion (Dupont et al., 2011). ATG5-dependent IL-1β secretion appears to rely on recognition of two KFERQ-like motifs by heat shock protein 90 alpha family class A member 1 (HSP90AA1), resulting in the relocalization of IL-1β between autophagosomal membranes, potentially explaining why IL-1β does not undergo degradation within autolysosomes in this context (Zhang et al., 2015). Another (hitherto untested) possibility is that ATG5 may prevent IL-1β degradation by subverting autolysosomal acidification, reminiscent of the non-autophagic pathway whereby ATG5 regulates exosome production (Guo et al., 2017). It has been proposed that IL-1β release ultimately depends on the fusion between autophagosomes or autolysosomes and the plasma membrane (Kimura et al., 2017). However, this model cannot be easily reconciled with the fact that gasdermin D (GSDMD), a core component of the molecular mechanism for pyroptosis (Box 2 ), is also required for IL-1β secretion by macrophages (Heilig et al., 2018). How components of the autophagy machinery interface with pyroptosis regulators to control the non-canonical release of IL-1β remains to be elucidated.
Box 2. Principles of Cell Death Regulation in Mammals.
Mammalian cells are provided with a complex molecular machinery that mediates their demise, a process commonly referred as regulated cell death (RCD) (Galluzzi et al., 2018a). RCD occurs in both fully physiological and pathological settings. In the former case, RCD is instrumental to organismal development and homeostasis, as it ensures organ morphogenesis and adult tissue turnover. In the latter case, RCD is a consequence of failing adaptation to stress. That said, stress-driven RCD can also be viewed as a mechanism for the maintenance of organismal homeostasis, as it underlies the removal of damaged, non-functional, and potentially oncogenic cells (Galluzzi et al., 2018b). The molecular mechanisms whereby mammalian cells undergo RCD in physiological and pathological scenarios exhibit considerable overlap. Extrinsic apoptosis is a form of RCD initiated by plasma membrane receptors through activation of CASP8, and precipitated by CASP3 or other executioner caspases, like CASP6 or CASP7. Intrinsic apoptosis also relies on CASP3, CASP6, or CASP7, but it is initiated by perturbations of intracellular homeostasis culminating with activation of pro-apoptotic BH3-only proteins and BAX- or BAK1-dependent mitochondrial outer membrane permeabilization (MOMP). Mitochondrial permeability transition (MPT)-driven regulated necrosis (RN) is initiated by the opening of a mitochondrial supramolecular entity known as permeability transition pore complex (PTPC) in response to oxidative stress or cytosolic Ca2+ overload and is precipitated by peptidylprolyl isomerase F (PPIF, best known as CYPD). Parthanatos is a form of regulated necrosis impinging on PARP1 hyperactivation, which entails a bioenergetic catastrophe coupled to the release of apoptosis-inducing factor mitochondria associated 1 (AIFM1; best known as AIF) from mitochondria and its translocation to the nucleus, where it mediates nucleolytic effects. Necroptosis is regulated by the RIPK3-dependent (and in some instances RIPK1-regulated) phosphorylation of MLKL, resulting in the formation of plasma-membrane-permeabilizing MLKL pores. Ferroptosis is triggered by oxidative perturbations, relies on lipid peroxidation, and is under tonic control by glutathione peroxidase 4 (GPX4). Pyroptosis is a form of RN impinging on the cleavage or gasdermin family members, such as gasdermin D (GSDMD) or GSDME, by inflammatory (i.e., CASP1, CASP4, CASP5, or CASP11) and apoptotic (i.e., CASP3) caspases often occurring in the context of IL-1β and IL-18 secretion. Lysosome-dependent cell death (LDCD) is a form of RCD demarcated by primary lysosomal membrane permeabilization (LMP) and precipitated by cathepsins, such as cathepsin B (CTSB) and CTSD. Finally, autophagy-dependent cell death (ADCD) relies on one of multiple components of the autophagy machinery (Galluzzi et al., 2018a). Of note, the etiological involvement of lysosomal degradation of autophagy substrates in ADCD has not been exhaustively verified, at least in mammalian settings. Thus, several variants of ADCD may actually constitute autophagy-independent pathways under the control of one or more components of the molecular apparatus for autophagy.
| Cell Death Mode | Prototypic Trigger | Main Initiator(s) | Main Executor(s) | Endogenous Inhibitor(s) | Defining Event(s) | Main Morphology |
|---|---|---|---|---|---|---|
| ADCD | Various | Various | Various | Unclear | Dependence on autophagy proteins | Vacuolated |
| Extrinsic apoptosis | Death receptor ligation | CASP8 or CASP10 | CASP3, CASP6, and CASP7 | CFLAR and anti-apoptotic BCL2 proteins | Caspase activation | Apoptotic |
| Ferroptosis | System xc inhibition | Iron | Oxidative damage to macromolecules | GPX4 | Lipid peroxidation | Necrotic |
| Intrinsic apoptosis | Intracellular stress | Pro-apoptotic BCL2 proteins | CASP3, CASP6, and CASP7 | Anti-apoptotic BCL2 proteins | MOMP and caspase activation | Apoptotic |
| LDCD | Lysosomotropic agents | Unclear | CTSB and CTSD | CSTB and CSTC | LMP | Apoptotic or necrotic |
| MPT-driven RN | Oxidative stress | PTPC | CYPD | Unclear | MPT | Necrotic |
| Necroptosis | TNFR1 ligation under caspase inhibition | RIPK1 and RIPK3 | MLKL | Unclear | MLKL oligomerization | Necrotic |
| Parthanatos | DNA damage | PARP1 | AIF and bioenergetic catastrophe driven by NAD+ depletion | Unclear | PARP1 hyperactivation | Necrotic |
| Pyroptosis | Inflammasome activation | CASP1, CASP3, CASP4, or CASP5 | GSDMD or GSDME | Unclear | Gasdermin oligomerization | Necrotic |
CFLAR, CASP8 and FADD-like apoptosis regulator; CST, cystatin; TNFR1 (official name, TNFRSF1A), TNF receptor superfamily member 1A.
A similar release of lysosomal material that depends on several components of the molecular apparatus for autophagy (i.e., ATG5, ATG7, ATG4B, and LC3), but does not involve bona fide degradation of autophagy substrates, has been linked to osteoclastic bone resorption (DeSelm et al., 2011). In both non-conventional protein secretion and exosome secretion, members of the RAB protein family mediate the fusions of autophagosomes or lysosomes with the plasma membrane (DeSelm et al., 2011, Dupont et al., 2011, Jaé et al., 2015). Since RAB proteins are also involved in bona fide autophagic responses (Galluzzi et al., 2017a), however, they cannot be employed to molecularly discriminate between autophagy-dependent and autophagy-independent pathways. Clarifying the precise signals whereby formed autophagosomes are routed to secretion (directly or upon fusion with non-acidifying lysosomes) instead of delivering their content to disposal will be instrumental for the development of pharmacological agents with various clinical applications.
Inflammation and Innate Immunity
Bona fide autophagic responses mediate robust anti-inflammatory functions as they dispose of potential triggers of inflammation, including (but not limited to) cytosolic pathogens, micronuclei, and permeabilized mitochondria (Levine et al., 2011). In addition, various components of the autophagy machinery regulate inflammatory responses and contribute to intracellular pathogen control via autophagy-independent pathways.
While both Atg5 −/− MEFs and MEFs expressing an autophagy-incompetent variant of ATG16L1 (ATG16L1-ΔCCD) exhibit defective autophagic responses to starvation, the former produce increased amounts of cytokine C-X-C motif chemokine ligand 1 (CXCL1) upon infection with cytoplasm-invading bacteria (as compared to wild-type MEFs), whereas the latter display a sub-optimal pro-inflammatory response (Sorbara et al., 2013). Knockdown of ATG16L1-ΔCCD restores the ability of autophagy-deficient MEFs to secrete CXCL1 in response to infection (Sorbara et al., 2013), supporting a non-autophagic role of ATG16L1 in the regulation of inflammatory responses. Consistent with this, transient knockdown of ATG16L1, but not ATG5, exacerbated CXCL1 secretion by mouse intestinal epithelial cells challenged with Shigella flexneri, Listeria monocytogenes, or Salmonella enterica serovar Typhimurium, an effect that could not be recapitulated with lysosomal inhibitors (Sorbara et al., 2013). In this setting, both full-length ATG16L1 and ATG16L1-ΔCCD, but neither ATG5 nor ATG9, bind to nucleotide binding oligomerization domain containing 1 (NOD1) and NOD2, two intracellular sensors of bacterial invasion, to limit their ability to initiate receptor interacting serine/threonine kinase 2 (RIPK2) signaling in the presence of specific NOD1/NOD2 ligands or cytoplasmic bacteria detected by NOD1/NOD2, but not to TLR4 agonists or tumor necrosis factor (TNF) (Sorbara et al., 2013). These findings delineate an anti-inflammatory function of ATG16L1 that occurs irrespective of autophagosome formation and lysosomal degradation.
Apparently at odds with its anti-inflammatory role in the course of bacterial infection, ATG16L1 supports the ability of IFN-γ to control infection in an autophagy-independent manner (Hwang et al., 2012, Selleck et al., 2015). In particular, ATG16L1, ATG7, LC3, p62, and calcium binding and coiled-coil domain 2 (CALCOCO2, another protein involved in cargo selection, best known as NDP52) cooperate to encapsulate cytoplasmic Toxoplasma gondii (a eukaryotic parasite) in multi-membraned vesicles that efficiently blunt parasite replication but do not fuse with endosomes or lysosomes (Selleck et al., 2015). In this context, ATG5 appears to promote the recruitment of an IFN-γ-responsive GTPase to parasite-encapsulating vesicles, which is required for the optimal anti-infectious effects of IFN-γ (Zhao et al., 2008). It remains unclear whether the autophagy-independent functions of ATG16L1 and ATG5 in the control of T. gondii infection require the formation of the ATG12-ATG5-ATG16L1 complex. In partial support of this possibility, both ATG7, which catalyzes the ATG12-ATG5 conjugation, and the ATG12-ATG5-ATG16L1 complex are required for the control of murine norovirus infection by macrophages exposed to IFN-γ (Hwang et al., 2012), via a mechanism that does not rely on ATG4-dependent LC3 processing, autophagosome-lysosome fusion, or lysosomal degradation (Hwang et al., 2012). Conversely, expression of ATG5 (but not of ATG7, ATG12, and ATG16L1 nor of ULK1, ULK2, ATG3, ATG4, ATG14, and p62) in lysozyme 2 (LYZ2)+ cells (encompassing monocytes, macrophages, and neutrophils) is required for mice to control infection by Mycobacterium tuberculosis (Kimmey et al., 2015). In this setting, ATG5 mediates autophagy-independent functions that prevent the development of severe lung inflammation driven by the release of multiple cytokines by polymorphic mononuclear cells (PMNs) (Kimmey et al., 2015). Consistent with this, mice with a LYZ2-restricted deletion of Atg5 (as well as mice where Atg5 is deleted only in PMNs) succumb prematurely to M. tuberculosis infection, and this can be rescued by PMN depletion (Kimmey et al., 2015). The precise molecular mechanisms linking ATG5 to reduced inflammatory responses in PMNs challenged to M. tuberculosis remain obscure. However, because ATG7, ATG12, and ATG16L1 are not involved in this process (Kimmey et al., 2015), the findings outlined above raise the intriguing possibility that bona fide autophagic responses may be detrimental in this setting as they could limit the availability of unbound ATG5. This hypothesis awaits experimental verification.
Components of the autophagy initiation complex (Box 1) have also been shown to mediate antiviral effects in an autophagy-independent manner. In particular, the replication of encephalomyocarditis virus (EMCV) and coxsackievirus B3 (CVB3) is enhanced in both Atg13 −/− and Rb1cc1 −/− MEFs (as compared with their wild-type counterparts), but a similar effect cannot be observed upon deletion of Ulk1, Ulk2, Atg101, or Atg7 (Mauthe et al., 2016). Consistently, transgene-enforced overexpression of ATG13 or FIP200 in HEK293 human kidney cancer cells considerably limits viral replication in the absence of ULK1 activation (Mauthe et al., 2016). Of note, the lack of Atg13 and Rb1cc1 does not influence viral entry but rather boosts intracellular replication from 2- to 5-fold (Mauthe et al., 2016). Whether such an effect depends on the ability of autophagy-related factors to regulate intracellular membranes, which is often hijacked by pathogens (see below), remains to be elucidated.
Cell Death
In mammalian cells, bona fide autophagic responses most often mediate robust cytoprotective effects (Boya et al., 2005, Rybstein et al., 2018). Indeed, while several instances of regulated cell death (RCD) (Box 2) rely on one or multiple autophagy-relevant proteins, hence constituting forms of autophagy-dependent cell death (ADCD) according to recent nomenclature guidelines (Galluzzi et al., 2018a), in none of these cases does ADCD involve lysosomal degradation. Thus, at odds with ADCD in lower eukaryotes, which involves bona fide autophagic degradation (Berry and Baehrecke, 2007, Denton et al., 2009), ADCD in mammals stands out as an autophagy-independent pathway under the control of the autophagy machinery.
Neurons subjected to ischemic conditions and various human cancer cell lines exposed to nutrient deprivation or to a cell-permeant peptide that displaces BECN1 from inhibitory interactions with BCL2, apoptosis regulator (BCL2) (Box 1) undergo a variant of ADCD that relies on the plasma membrane Na+/K+ ATPase, which has been dubbed autosis (Liu et al., 2013). Accordingly, neriifolin (a Na+/K+ ATPase inhibitor of the family of cardiac glycosides) mediates robust neuroprotective effects in rats experiencing cerebral hypoxia-ischemia (Liu et al., 2013). Autosis occurs irrespective of BCL2-associated X, apoptosis regulator (BAX)- or BCL2 antagonist/killer 1 (BAK1)-dependent mitochondrial outer membrane permeabilization (MOMP) (Liu et al., 2013), and hence, it does not constitute a form of apoptotic cell death. Rather, autosis relies on ATG5, ATG7, ATG12, ATG14, and BECN1 as well as the PI3K activity of VPS34 (Liu et al., 2013). However, pharmacological inhibition of lysosomal functions fails to rescue human cervical carcinoma HeLa cells from autosis (Liu et al., 2013), strongly suggesting that autosis occurs independently of bona fide autophagic responses.
ATG5, ATG7, and VPS34-dependent PI3P production are required for the necroptotic death (Box 2) of Map3k7 −/− mouse prostate epithelial cells (MPECs), which lack a signal transducer in the pathway linking death receptor signaling to NF-κB activation, best known as TAK1, caused by exposure to TNF superfamily member 10 (TNFSF10, best known as TRAIL) (Goodall et al., 2016). Such necroptotic response is exacerbated, rather than blunted, by chemical inhibitors of lysosomal degradation (Goodall et al., 2016), pointing to a non-autophagic role for ATG5, ATG7, and VPS34 in this setting. Consistently, multiple components of the RIPK1- and RIPK3-containing complex that drives necroptosis colocalize with ATG5, ATG7, and p62 at autophagosomes in Map3k7 −/− MPECs responding to TRAIL (Goodall et al., 2016). Moreover, depletion of p62 from Map3k7 −/− MPECs switches TRAIL-driven necroptosis to apoptosis, which can be rescued with chemical caspase inhibitors (Goodall et al., 2016). Thus, in the absence of TAK1, autophagosomes favor necroptotic cell death by providing a physical platform for the activation of necroptosis, independent of autophagic substrate degradation.
Autophagy regulators also influence non-necroptotic forms of regulated necrosis. Developing Atg9a −/− embryos fail to manifest foci of cell death at the surface of multiple bones, resulting in impaired bone morphogenesis (Imagawa et al., 2016). Such foci of dying cells fail to exhibit CASP3 activation and mixed lineage kinase domain like pseudokinase (MLKL) phosphorylation (Box 2) and can still be observed in Casp9 −/− and Ripk1 −/− embryos, implying that they do not originate from intrinsic apoptosis or necroptosis (Imagawa et al., 2016). Moreover, Atg5 −/− embryos exhibit normal bone morphogenesis at birth, implying that ATG9 regulates a programmed variant of necrosis irrespective of canonical autophagy (Imagawa et al., 2016). Along similar lines, Ulk1 −/− MEFs are protected from hydrogen-peroxide-driven RCD, but this phenotype cannot be recapitulated by the deletion of Atg7 (Joshi et al., 2016). Rather, hydrogen-peroxide-driven RCD can be prevented by chemical inhibition or depletion of poly(ADP-ribose) polymerase 1 (PARP1) (Joshi et al., 2016), a mediator of parthanatos (Box 2). Moreover, ULK1 physically interacts with PARP1, especially in cells responding to oxidative stress, and such interaction stimulates the enzymatic functions of PARP1 to precipitate the bioenergetic catastrophe that underlies parthanatos (Joshi et al., 2016). Consistent with these observations, ATG13 silencing and consequent ULK1 destabilization limits the death of human osteosarcoma cells responding to the topoisomerase inhibitor camptothecin (Gao et al., 2011). Although the involvement of PARP1 in the ability of ULK1 to support camptothecin toxicity has not been clarified, circumstantial evidence exists in support of such a link (Das et al., 2016).
Various autophagy regulators control apoptotic forms of RCD in an autophagy-independent manner. Immunity-related GTPase M (IRGM), the only human paralog of a large cluster of murine IFN-γ-regulated genes (Bekpen et al., 2005), not only participates in the autophagic control of M. tuberculosis driven by IFN-γ (Singh et al., 2006) but, upon infection, also translocates to mitochondria to initiate MOMP (Singh et al., 2010). Importantly, RCD caused by IRGM occurs irrespective of BECN1 or ATG7 but can be rescued by co-deletion of Bax and Bak1 (Singh et al., 2010). Whether the ability of IRGM to trigger MOMP depends on a direct physical interaction with BAX and/or BAK1 versus the inactivation of anti-apoptotic Bcl-2 family members remains to be elucidated. This latter mechanism accounts for the pro-apoptotic effects of ATG12 (Rubinstein et al., 2011). Indeed, ATG12 contains a bona fide BH3 domain that enables ATG12 to interact with BCL2 and MCL1, BCL2 family apoptosis regulator (MCL1), ultimately resulting in BAX/BAK1-dependent RCD irrespective of ATG3, ATG4, and ATG5 (Haller et al., 2014, Rubinstein et al., 2011). Both BECN1 and AMBRA1 also contain a bona fide BH3 domain (Oberstein et al., 2007, Strappazzon et al., 2016). However, while interaction between BECN1 and BCL2 or BCL2-like 1 (BCL2L1, best known as BCL-XL) mainly regulates autophagic responses (Maiuri et al., 2007, Pattingre et al., 2005), a cleavage product of AMBRA1 generated over the course of apoptosis appears to precipitate RCD upon BCL2 binding (Strappazzon et al., 2016). The reasons underlying such a discrepancy are unclear but may relate to the differential binding affinity of the AMBRA1 and BECN1 BH3 domains for Bcl-2 family members. BECN1 also competes with MCL1 for stabilization by ubiquitin specific peptidase 9 X-linked (USP9X) (Elgendy et al., 2014). In this setting, increased levels of MCL1 limit BECN1 deubiquitination, hence favoring BECN1 proteasomal degradation. Such a simultaneous inhibition of autophagy and apoptosis has been documented in progressing melanomas (Elgendy et al., 2014). Conversely, high levels of BECN1 favor the proteasomal degradation of MCL1, culminating with increased cellular susceptibility to apoptotic RCD (Elgendy et al., 2014).
ATG7 can also regulate apoptotic RCD irrespective of degradative autophagy. Specifically, ATG7 can bind tumor protein p53 (TP53, best known as p53) to regulate p53 transcriptional activity (Lee et al., 2012). Consistently, Atg7 −/− MEFs as well as Atg7 −/− MEFs reconstituted with an autophagy-incompetent variant of ATG7 (but not Atg5 −/− and Becn1 −/− MEFs) display defective p53-dependent cyclin dependent kinase inhibitor 1A (CDKN1A, best known as p21) expression and consequent cell-cycle arrest during starvation (Lee et al., 2012). Interestingly, this phenotype is accompanied by accrued ROS generation and oxidative DNA damage, culminating in premature p53-driven, BAX/BAK1-dependent RCD (Lee et al., 2012). Of note, p53 hyperactivation caused by the absence of ATG7 participates in the perinatal lethality of Atg7 −/− mice (Lee et al., 2012). These observations delineate an autophagy-independent mechanism whereby ATG7 regulates the activity of p53, both positively (as a consequence of physical interactions) and negatively (reflecting the establishment of accrued metabolic stress). The ultimate metabolic catastrophe promoted by the absence of ATG7 irrespective of bona fide autophagic responses may reflect the ability of p53 to support the maintenance of metabolic homeostasis at baseline (Kruiswijk et al., 2015). This hypothesis awaits experimental validation.
Genomic Stability and Cell Proliferation
Uvrag +/− embryonic stem cells spontaneously accumulate DNA double-strand breaks and are more sensitive to DNA-damaging agents than their Uvrag +/+ counterparts (Zhao et al., 2012). Similarly, human melanoma cells depleted of UVRAG display increased sensitivity to UV-driven DNA photolesions (Yang et al., 2016). Moreover, UVRAG-deficient MEFs are prone to centrosome abnormalities and consequent mitotic defects that favor aneuploidy (Zhao et al., 2012). Thus, UVRAG supports genetic and genomic stability. Notably, neither of these phenotypes is the result of autophagic defects associated with UVRAG downregulation (Yang et al., 2016, Zhao et al., 2012), even though bona fide autophagic responses are known to contribute to the maintenance of genomic homeostasis (Karantza-Wadsworth et al., 2007, Mathew et al., 2009). Indeed, Atg5 −/− MEFs are equally sensitive to UVRAG depletion with regards to DNA damage and centrosome dysfunction as their wild-type counterparts (Yang et al., 2016, Zhao et al., 2012). Rather, the genoprotective effects of UVRAG reflect its ability to support non-homologous end-joining (a form of DNA repair specific for DSBs) by binding to the catalytic subunit of the DNA-dependent protein kinase (DNA-PK) complex, to promote the repair of UV-driven DNA photolesions following interaction with damage-specific DNA binding protein 1 (DDB1), and to regulate centrosome functions upon binding to centrosomal protein 63 (CEP63) (Yang et al., 2016, Zhao et al., 2012). All these activities are molecularly and functionally independent of the capacity of UVRAG to bind BECN1 and hence stimulate autophagy (Yang et al., 2016, Zhao et al., 2012). In line with this notion, UVRAG is often affected by monoallelic mutations in colorectal and gastric carcinomas with high degrees of genomic instability (so-called microsatellite instable), but these mutations fail to affect autophagic responses (Knævelsrud et al., 2010). Similarly, a truncated variant of UVRAG expressed by some CRCs has been associated with an autophagy-independent DNA repair defect and consequent increased sensitivity to genotoxic chemotherapy (He et al., 2015). BECN1 appears to share with UVRAG the ability to regulate centrosome functions in an ATG5-independent manner, especially in the context of DNA damage (Park et al., 2014). Whether the physical interaction between UVRAG and BECN1 is required for centrosome regulation by UVRAG remains to be elucidated.
Uvrag −/− T cells also display defects in homeostatic proliferation that cannot be related to the autophagic functions of UVRAG and cannot be explained by the alterations imposed by the absence of Atg5 or Atg7 (encoding another core component, the autophagy machinery) (Afzal et al., 2015). Thus, Uvrag ablation compromises CD8+ T cell responses to lymphocytic choriomeningitis virus (LMCV) in the absence of major alterations in autophagic flux (Afzal et al., 2015). The deletion of Atg5 or Atg7 similarly impairs the survival of effector CD8+ T cells and their ability to establish immunological memory upon challenge (Puleston et al., 2014, Xu et al., 2014). Moreover, the conditional deletion of Atg3, Atg5, or Atg7 from the CD4+ cellular compartment, which encompasses CD4+ T cells as well as multiple populations of innate lymphoid cells (ILCs), or from the NKp46+ compartment, corresponding to ILCs only, imposes a major defect in homeostatic proliferation to multiple ILC populations (O’Sullivan et al., 2016, Pei et al., 2015). Importantly, although the defects imposed on T lymphocytes and ILCs by the deletion of Atg3, Atg5, or Atg7 are generally attributed to compromised autophagic responses (Clarke and Simon, 2019), no strict experimental evidence in support of this interpretation is currently available. Thus, it is tempting to speculate that ATG3, ATG5, and ATG7 resemble UVRAG in its ability to support T cell homeostasis irrespective of bona fide autophagic responses.
Pathogen Invasion
A variety of viruses, including several members of the enterovirus genus, have evolved strategies to hijack the molecular machinery for autophagy, which often mediates antiviral effects by promoting the lysosomal degradation of cytosolic virions or components thereof, (Choi et al., 2018) to their own benefit. Such strategies go beyond the simple inhibition of autophagic flux, which would suppress the autophagic disposal of cytosolic virions but at the same time would render host cells considerably more sensitive to undergo premature apoptotic cell death and potentially compromise viral replication (Galluzzi et al., 2008, Liang et al., 2015, Stewart and Cookson, 2016). Rather, they involve the modulation of various components of the autophagic apparatus resulting in (1) increased availability of autophagosomes or other membranes that support viral replication via LC3 (Alirezaei et al., 2015, Wong et al., 2008); (2) diversion of the autophagic flux from lysosomal degradation, as a consequence of compromised interaction between synaptosome associated protein 29 (SNP29) and pleckstrin homology and RUN domain containing M1 (PLEKHM1), which normally underlies autophagosome-to-lysosome fusion (Mohamud et al., 2018, Tian et al., 2018); (3) early release of virus-loaded exosome-like microvesicles that bear autophagosomal markers (Robinson et al., 2014, Sin et al., 2017); and (4) ultimately, activation of apoptotic cell death for massive viral dissemination (Xin et al., 2014).
The modulation of LC3 or other components of the autophagy machinery as a means to hijack intracellular membranes for viral replication in an autophagy-independent manner is an extraordinarily common mechanism (as an estimate, 36% of the autophagy-related cellular proteome influence the replication of one or multiple viruses irrespective of autophagic degradation) (Mauthe et al., 2016) shared by multiple RNA viruses other than enteroviruses, including the equine arteritis virus (EAV) (Monastyrska et al., 2013), the hepatitis C virus (Shrivastava et al., 2015), and coronaviruses (Reggiori et al., 2010). Similarly, LC3 appears to support the intracellular propagation of the bacterial parasite Chlamydia trachomatis, by a mechanism that can be fully segregated from increased autophagic flux, which is detrimental to the pathogen (Al-Younes et al., 2011). Many DNA viruses, including members of the adenoviridae and herpesviridae families, have also evolved strategies to control various components of the autophagy machinery to their own benefit. In this case, however, modulation seems focused on actual autophagic flux rather than on autophagy-independent signaling pathways (Rodriguez-Rocha et al., 2011, Yin et al., 2017). We surmise that this discrepancy is only apparent, potentially originating from the autophagy-biased approach with which the phenotypes linked to genetic or pharmacological inhibition of single components of the autophagy machinery have been interpreted so far.
The obligate intracellular bacterium Legionella pneumophila encodes RavZ, a protease that, upon secretion to the cytosol of infected cells, specifically hydrolyzes the amide bond between the C-terminal glycine and the adjacent aromatic residue of all mammalian MAP1LC3 family members (Choy et al., 2012). Such a cleavage irreversibly prevents the (re-)lipidation of MAP1LC3 proteins, hence inhibiting autophagy (Choy et al., 2012). However, given that L. pneumophila lives within a replicative organelle derived from the phagosome, it is unclear why inhibition of autophagy, per se, would be beneficial to the pathogen. As an alternative, RavZ may operate to prevent LAP, thus inhibiting the fusion of the replicative organelle with lysosomes and consequent pathogen eradication. As soil amoeba are the primary host of Legionella spp. (Richards et al., 2013), it is tempting to speculate that LAP is a major non-canonical function for autophagy regulators in such hosts.
Conclusions
Most, if not all, components of the molecular apparatus for autophagy mediate non-autophagic functions (Figure 3 ). Although at this point in time some proteins involved in autophagy have not yet been attributed non-autophagic roles (e.g., ATG2), we surmise that this only reflects our limited knowledge of their biology and that all components of the autophagy proteome de facto serve multiple functions. Precisely characterizing the autophagy-independent functions of the autophagy machinery and their pathophysiological relevance calls for the design of refined experimental strategies that fully uncouple autophagic versus non-autophagic activities, such as the generation of mice genetically engineered to express a variant of FIP200 that cannot bind ATG13 (Chen et al., 2016). This approach relies on in-depth structure-to-function relationship data that are not available for all autophagy-relevant proteins and their interactors. Indeed, considerable attention has been given so far to the structural characterization of single proteins and protein complexes linked to degradative autophagy, including ATG3 (Yamada et al., 2007), ATG4B (Sugawara et al., 2005), BECN1 (Li et al., 2012b), VPS34 (Miller et al., 2010, Rostislavleva et al., 2015), the ATG12-ATG5:ATG16L1 complex (Noda et al., 2008), LC3B (Sugawara et al., 2004), and several others. Although these studies provided profound mechanistic insights into bona fide autophagic responses (Li et al., 2012b, Satoo et al., 2009) and fostered the development of targeted inhibitors (Miller et al., 2010, Qiu et al., 2016), experimental design was often, if not always, biased toward autophagy-relevant domains and interactions. That said, it may not always be feasible to molecularly dissociate the autophagic and non-autophagic functions of a specific protein, even in the presence of detailed data on structure and physical interactions.
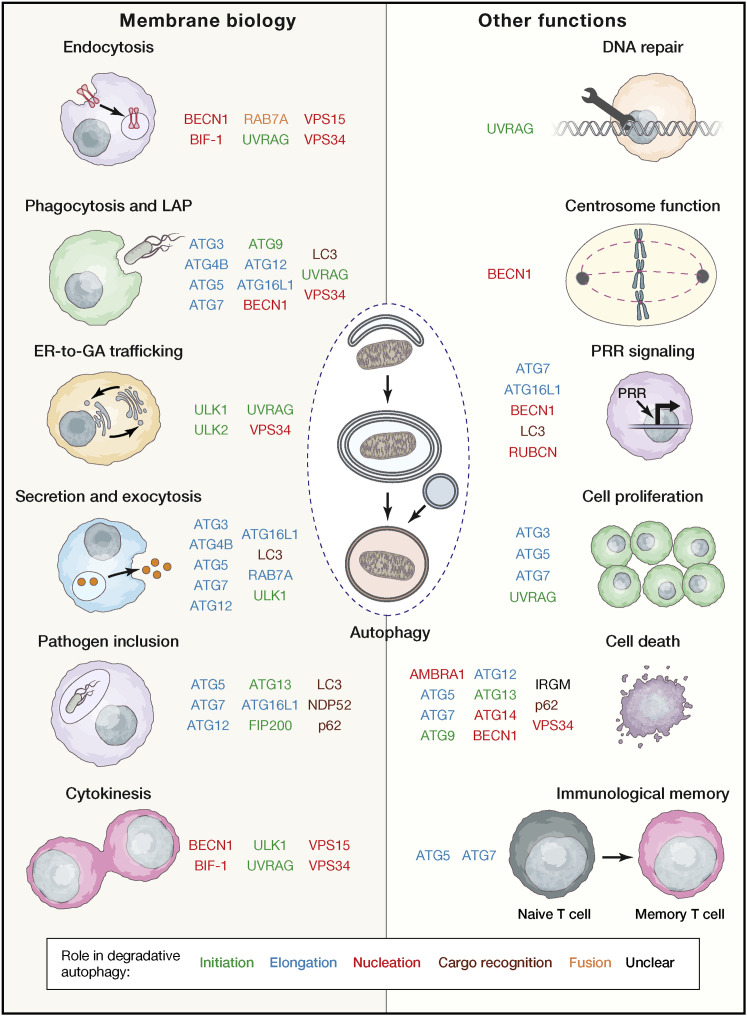
Figure 3.
Non-autophagic Functions of the Autophagy Apparatus
A large number of autophagy-relevant proteins mediate non-autophagic effects related to membrane biology and other cellular functions. Interestingly, many of these proteins operate in early steps of bona fide autophagic responses (e.g., initiation, nucleation, and elongation).
ER, endoplasmic reticulum; GA, Golgi apparatus; LAP, LC3-associated phagocytosis; PRR, pattern recognition receptor.
An additional layer of complexity originates from the fact that several proteins that participate in multiple cellular functions exist in limiting amounts, implying that the inhibition of one pathway may result in the hyperactivation of another one. For instance, displacement of UVRAG from RINT1, which inhibits retrograde GA-to-ER vesicular trafficking, results in the activation of autophagy as a consequence of increased UVRAG availability for binding to BECN1 (He et al., 2013). Moreover, it is likely that the molecular links between degradative autophagy and non-autophagic cellular functions may exhibit considerable degrees of context dependence (i.e., they may vary considerably depending on cell type, differentiation stage, etc.). An intriguing possibility is that cells may actively route disposable cytosolic material to autophagic degradation versus non-canonical secretion as a means to control the emission of damage signals into the microenvironment (Galluzzi et al., 2017c, Yatim et al., 2017). Although preclinical data in support of this notion is emerging, additional work is required to clarify the mutual regulation of autophagy and non-autophagic pathways and its pathophysiological implications.
Finally, autophagy is a redundant process that can occur via a plethora of mechanisms, including several variants of macroautophagy that do not rely on canonical regulators (e.g., they occur irrespective of ATG5 or ATG7) (Honda et al., 2014, Ma et al., 2015, Nishida et al., 2009) as well as microautophagy and CMA (Galluzzi et al., 2017a). Thus, other variants of autophagy can compensate for the lack of macroautophagy in the degradation of some substrates, such as ferritin (Goodwin et al., 2017, Mancias et al., 2014). Importantly, the molecular machinery involved in the degradation of ferritin in this scenario involves regulators of macroautophagy including ATG9, FIP200, and VPS34 (Goodwin et al., 2017, Mancias et al., 2014). Thus, pharmacological inhibitors or genetic interventions specific for any of these proteins cannot be used to identify the precise mechanism underlying the autophagic degradation of ferritin.
Of note, components of the autophagy machinery appear to mediate non-autophagic functions mostly, although not exclusively, in the context of (1) vesicle uptake, trafficking, release, and other processes involving membrane rearrangements (e.g., cytokinesis); and (2) mechanisms for the innate and adaptive control of invading pathogens (including the regulated death of infected cells) (Table 2). Thus, membrane trafficking (in the extended sense of the term) and pathogen control constitute the most ancient evolutionary hubs for autophagy to interact with non-autophagic functions. In line with this notion, autophagy-independent effects for the orthologs of mammalian ULK1, UVRAG, BECN1, ATG9, and ATG16L1 in endocytosis, phagocytosis, and secretion have been documented in lower eukaryotes (Hedgecock et al., 1985, Lee et al., 2011, Li et al., 2016, Shravage et al., 2013, Tung et al., 2010, Xiong et al., 2015). Moreover, the majority of autophagy-related proteins with non-autophagic functions operate at early phases of degradative autophagy (i.e., initiation, nucleation, and elongation) (Table 1). Taken together, these observations may suggest that an ancient molecular machinery regulating basic membrane functions (e.g., phospholipid insertion, curvature regulation) in lower eukaryotes may have diversified to control broad cellular processes (e.g., phagocytosis, vesicular trafficking, autophagy) while retaining shared molecular components.
In summary, autophagy stands out as a key process for the lysosomal degradation of cytosolic entities that is highly interconnected with several other biological functions. Such a connection does not only reflect the key role of bona fide autophagic responses in the maintenance of metabolic homeostasis but also the ability of multiple autophagy-relevant proteins to mediate non-autophagic functions. We surmise that elucidating such molecular crosstalk in detail will provide important insights into the pathophysiology of multiple disorders and potentially foster the identification of actionable therapeutic targets.
Acknowledgments
We thank Camille Daviaud (Weill Cornell Medical College, New York, NY, USA) for help with figure preparation. L.G. is supported by a Breakthrough Level 2 grant from the US Department of Defense (DoD), Breast Cancer Research Program (BRCP) [#BC180476P1]; by a startup grant from the Department of Radiation Oncology at Weill Cornell Medicine (New York, USA); by industrial collaborations with Lytix (Oslo, Norway) and Phosplatin (New York, USA); and by donations from Phosplatin (New York, USA), the Luke Heller TECPR2 Foundation (Boston, USA), and Sotio a.s. (Prague, Czech Republic). D.R.G. is supported by grants from the US National Institutes of Health and by ALSAC.
Author Contributions
L.G. and D.R.G. conceived the paper. L.G. wrote the first version of the manuscript and designed the display items with constant input from D.R.G. L.G. integrated comments from the reviewers. Both authors approved the final version of the article.
Declaration of Interests
L.G. provides remunerated consulting to OmniSEQ (Buffalo, NY, USA), Astra Zeneca (Gaithersburg, MD, USA), VL47 (New York, NY, USA), and the Luke Heller TECPR2 Foundation (Boston, MA, USA), and he is member of the Scientific Advisory Committee of OmniSEQ (Buffalo, NY, USA). D.R.G. has no conflicts of interest to declare.
Contributor Information
Lorenzo Galluzzi, Email: deadoc80@gmail.com.
Douglas R. Green, Email: douglas.green@stjude.org.
References
- Afzal S., Hao Z., Itsumi M., Abouelkheer Y., Brenner D., Gao Y., Wakeham A., Hong C., Li W.Y., Sylvester J. Autophagy-independent functions of UVRAG are essential for peripheral naive T-cell homeostasis. Proc. Natl. Acad. Sci. USA. 2015;112:1119–1124. doi: 10.1073/pnas.1423588112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Younes H.M., Al-Zeer M.A., Khalil H., Gussmann J., Karlas A., Machuy N., Brinkmann V., Braun P.R., Meyer T.F. Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy. 2011;7:814–828. doi: 10.4161/auto.7.8.15597. [DOI] [PubMed] [Google Scholar]
- Alirezaei M., Flynn C.T., Wood M.R., Harkins S., Whitton J.L. Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo. Autophagy. 2015;11:1389–1407. doi: 10.1080/15548627.2015.1063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C., Hunn J.P., Rohde C., Parvanova I., Guethlein L., Dunn D.M., Glowalla E., Leptin M., Howard J.C. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D.L., Baehrecke E.H. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P., González-Polo R.A., Casares N., Perfettini J.L., Dessen P., Larochette N., Métivier D., Meley D., Souquere S., Yoshimori T. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., Bulleid N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- Chen S., Wang C., Yeo S., Liang C.C., Okamoto T., Sun S., Wen J., Guan J.L. Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev. 2016;30:856–869. doi: 10.1101/gad.276428.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Bowman J.W., Jung J.U. Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 2018;16:341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy A., Dancourt J., Mugo B., O’Connor T.J., Isberg R.R., Melia T.J., Roy C.R. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.J., Simon A.K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 2019;19:170–183. doi: 10.1038/s41577-018-0095-2. [DOI] [PubMed] [Google Scholar]
- Cunha L.D., Yang M., Carter R., Guy C., Harris L., Crawford J.C., Quarato G., Boada-Romero E., Kalkavan H., Johnson M.D.L. LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell. 2018;175:429–441.e16. doi: 10.1016/j.cell.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.K., Rehman I., Ghosh A., Sengupta S., Majumdar P., Jana B., Das B.B. Poly(ADP-ribose) polymers regulate DNA topoisomerase I (Top1) nuclear dynamics and camptothecin sensitivity in living cells. Nucleic Acids Res. 2016;44:8363–8375. doi: 10.1093/nar/gkw665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D., Shravage B., Simin R., Mills K., Berry D.L., Baehrecke E.H., Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSelm C.J., Miller B.C., Zou W., Beatty W.L., van Meel E., Takahata Y., Klumperman J., Tooze S.A., Teitelbaum S.L., Virgin H.W. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy M., Ciro M., Abdel-Aziz A.K., Belmonte G., Dal Zuffo R., Mercurio C., Miracco C., Lanfrancone L., Foiani M., Minucci S. Beclin 1 restrains tumorigenesis through Mcl-1 destabilization in an autophagy-independent reciprocal manner. Nat. Commun. 2014;5:5637. doi: 10.1038/ncomms6637. [DOI] [PubMed] [Google Scholar]
- Evans R.J., Sundaramurthy V., Frickel E.M. The Interplay of Host Autophagy and Eukaryotic Pathogens. Front. Cell Dev. Biol. 2018;6:118. doi: 10.3389/fcell.2018.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Button R., Anichtchik O., Luo S. Visualization and Measurement of Multiple Components of the Autophagy Flux. In: Turksen K., editor. Autophagy in Differentiation and Tissue Maintenance. Humana Press; 2018. pp. 1–12. [Google Scholar]
- Fernández A.F., Sebti S., Wei Y., Zou Z., Shi M., McMillan K.L., He C., Ting T., Liu Y., Chiang W.C. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558:136–140. doi: 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K., Ulferts R., Jacquin E., Veith T., Gammoh N., Arasteh J.M., Mayer U., Carding S.R., Wileman T., Beale R., Florey O. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018;37:e97840. doi: 10.15252/embj.201797840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Brenner C., Morselli E., Touat Z., Kroemer G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Bravo-San Pedro J.M., Levine B., Green D.R., Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017;16:487–511. doi: 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Buqué A., Kepp O., Zitvogel L., Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Yamazaki T., Kroemer G. Linking cellular stress responses to systemic homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:731–745. doi: 10.1038/s41580-018-0068-0. [DOI] [PubMed] [Google Scholar]
- Gao W., Shen Z., Shang L., Wang X. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ. 2011;18:1598–1607. doi: 10.1038/cdd.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall M.L., Fitzwalter B.E., Zahedi S., Wu M., Rodriguez D., Mulcahy-Levy J.M., Green D.R., Morgan M., Cramer S.D., Thorburn A. The Autophagy Machinery Controls Cell Death Switching between Apoptosis and Necroptosis. Dev. Cell. 2016;37:337–349. doi: 10.1016/j.devcel.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J.M., Dowdle W.E., DeJesus R., Wang Z., Bergman P., Kobylarz M., Lindeman A., Xavier R.J., McAllister G., Nyfeler B. Autophagy-Independent Lysosomal Targeting Regulated by ULK1/2-FIP200 and ATG9. Cell Rep. 2017;20:2341–2356. doi: 10.1016/j.celrep.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.R., Galluzzi L., Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Chitiprolu M., Roncevic L., Javalet C., Hemming F.J., Trung M.T., Meng L., Latreille E., Tanese de Souza C., McCulloch D. Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell. 2017;43:716–730.e7. doi: 10.1016/j.devcel.2017.11.018. [DOI] [PubMed] [Google Scholar]
- Haller M., Hock A.K., Giampazolias E., Oberst A., Green D.R., Debnath J., Ryan K.M., Vousden K.H., Tait S.W. Ubiquitination and proteasomal degradation of ATG12 regulates its proapoptotic activity. Autophagy. 2014;10:2269–2278. doi: 10.4161/15548627.2014.981914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Bassik M.C., Moresi V., Sun K., Wei Y., Zou Z., An Z., Loh J., Fisher J., Sun Q. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Ni D., Ma B., Lee J.H., Zhang T., Ghozalli I., Pirooz S.D., Zhao Z., Bharatham N., Li B. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat. Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Zhao Z., Yang Y., O’Connell D., Zhang X., Oh S., Ma B., Lee J.H., Zhang T., Varghese B. Truncating mutation in the autophagy gene UVRAG confers oncogenic properties and chemosensitivity in colorectal cancers. Nat. Commun. 2015;6:7839. doi: 10.1038/ncomms8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann B.L., Boada-Romero E., Cunha L.D., Magne J., Green D.R. LC3-Associated Phagocytosis and Inflammation. J. Mol. Biol. 2017;429:3561–3576. doi: 10.1016/j.jmb.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E.M., Culotti J.G., Thomson J.N., Perkins L.A. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Heilig R., Dick M.S., Sborgi L., Meunier E., Hiller S., Broz P. The Gasdermin-D pore acts as a conduit for IL-1β secretion in mice. Eur. J. Immunol. 2018;48:584–592. doi: 10.1002/eji.201747404. [DOI] [PubMed] [Google Scholar]
- Henault J., Martinez J., Riggs J.M., Tian J., Mehta P., Clarke L., Sasai M., Latz E., Brinkmann M.M., Iwasaki A. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Arakawa S., Nishida Y., Yamaguchi H., Ishii E., Shimizu S. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat. Commun. 2014;5:4004. doi: 10.1038/ncomms5004. [DOI] [PubMed] [Google Scholar]
- Hwang S., Maloney N.S., Bruinsma M.W., Goel G., Duan E., Zhang L., Shrestha B., Diamond M.S., Dani A., Sosnovtsev S.V. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa Y., Saitoh T., Tsujimoto Y. Vital staining for cell death identifies Atg9a-dependent necrosis in developmental bone formation in mouse. Nat. Commun. 2016;7:13391. doi: 10.1038/ncomms13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaé N., McEwan D.G., Manavski Y., Boon R.A., Dimmeler S. Rab7a and Rab27b control secretion of endothelial microRNA through extracellular vesicles. FEBS Lett. 2015;589(20 Pt B):3182–3188. doi: 10.1016/j.febslet.2015.08.040. [DOI] [PubMed] [Google Scholar]
- Joo J.H., Wang B., Frankel E., Ge L., Xu L., Iyengar R., Li-Harms X., Wright C., Shaw T.I., Lindsten T. The Noncanonical Role of ULK/ATG1 in ER-to-Golgi Trafficking Is Essential for Cellular Homeostasis. Mol. Cell. 2016;62:491–506. doi: 10.1016/j.molcel.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Iyengar R., Joo J.H., Li-Harms X.J., Wright C., Marino R., Winborn B.J., Phillips A., Temirov J., Sciarretta S. Nuclear ULK1 promotes cell death in response to oxidative stress through PARP1. Cell Death Differ. 2016;23:216–230. doi: 10.1038/cdd.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Zhao H., Martinez J., Doggett T.A., Kolesnikov A.V., Tang P.H., Ablonczy Z., Chan C.C., Zhou Z., Green D.R., Ferguson T.A. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154:365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmey J.M., Huynh J.P., Weiss L.A., Park S., Kambal A., Debnath J., Virgin H.W., Stallings C.L. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528:565–569. doi: 10.1038/nature16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Jia J., Kumar S., Choi S.W., Gu Y., Mudd M., Dupont N., Jiang S., Peters R., Farzam F. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 2017;36:42–60. doi: 10.15252/embj.201695081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knævelsrud H., Ahlquist T., Merok M.A., Nesbakken A., Stenmark H., Lothe R.A., Simonsen A. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–870. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- Lee G., Liang C., Park G., Jang C., Jung J.U., Chung J. UVRAG is required for organ rotation by regulating Notch endocytosis in Drosophila. Dev. Biol. 2011;356:588–597. doi: 10.1016/j.ydbio.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.H., Kawai Y., Fergusson M.M., Rovira I.I., Bishop A.J., Motoyama N., Cao L., Finkel T. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–228. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H.W. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.W., Li J., Bao J.K. Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., He L., Che K.H., Funderburk S.F., Pan L., Pan N., Zhang M., Yue Z., Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Lindblad J.L., Perez E., Bergmann A., Fan Y. Autophagy-independent function of Atg1 for apoptosis-induced compensatory proliferation. BMC Biol. 2016;14:70. doi: 10.1186/s12915-016-0293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Oh B.H., Jung J.U. Novel functions of viral anti-apoptotic factors. Nat. Rev. Microbiol. 2015;13:7–12. doi: 10.1038/nrmicro3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shoji-Kawata S., Sumpter R.M., Jr., Wei Y., Ginet V., Zhang L., Posner B., Tran K.A., Green D.R., Xavier R.J. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl. Acad. Sci. USA. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Li J., Xu Y., Yu C., Xu T., Wang H., Liu K., Cao N., Nie B.M., Zhu S.Y. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat. Cell Biol. 2015;17:1379–1387. doi: 10.1038/ncb3256. [DOI] [PubMed] [Google Scholar]
- Maiuri M.C., Le Toumelin G., Criollo A., Rain J.C., Gautier F., Juin P., Tasdemir E., Pierron G., Troulinaki K., Tavernarakis N. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Malireddi R.K., Lu Q., Cunha L.D., Pelletier S., Gingras S., Orchard R., Guan J.L., Tan H., Peng J. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinez J., Cunha L.D., Park S., Yang M., Lu Q., Orchard R., Li Q.Z., Yan M., Janke L., Guy C. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mathew R., Kongara S., Beaudoin B., Karp C.M., Bray K., Degenhardt K., Chen G., Jin S., White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y., Bray K., Reddy A., Bhanot G., Gelinas C. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe M., Langereis M., Jung J., Zhou X., Jones A., Omta W., Tooze S.A., Stork B., Paludan S.R., Ahola T. An siRNA screen for ATG protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. J. Cell Biol. 2016;214:619–635. doi: 10.1083/jcb.201602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight N.C., Zhong Y., Wold M.S., Gong S., Phillips G.R., Dou Z., Zhao Y., Heintz N., Zong W.X., Yue Z. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet. 2014;10:e1004626. doi: 10.1371/journal.pgen.1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Tavshanjian B., Oleksy A., Perisic O., Houseman B.T., Shokat K.M., Williams R.L. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamud Y., Shi J., Qu J., Poon T., Xue Y.C., Deng H., Zhang J., Luo H. Enteroviral Infection Inhibits Autophagic Flux via Disruption of the SNARE Complex to Enhance Viral Replication. Cell Rep. 2018;22:3292–3303. doi: 10.1016/j.celrep.2018.02.090. [DOI] [PubMed] [Google Scholar]
- Monastyrska I., Ulasli M., Rottier P.J., Guan J.L., Reggiori F., de Haan C.A. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013;9:164–174. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz-Feliciano L., Doggett T.A., Zhou Z., Ferguson T.A. RUBCN/rubicon and EGFR regulate lysosomal degradative processes in the retinal pigment epithelium (RPE) of the eye. Autophagy. 2017;13:2072–2085. doi: 10.1080/15548627.2017.1380124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L., Malhotra R., Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 2015;17:300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y.I., Kuranaga E. Caspase-dependent non-apoptotic processes in development. Cell Death Differ. 2017;24:1422–1430. doi: 10.1038/cdd.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Noda N.N., Fujioka Y., Ohsumi Y., Inagaki F. Crystallization of the Atg12-Atg5 conjugate bound to Atg16 by the free-interface diffusion method. J. Synchrotron Radiat. 2008;15:266–268. doi: 10.1107/S0909049507054799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan T.E., Geary C.D., Weizman O.E., Geiger T.L., Rapp M., Dorn G.W., 2nd, Overholtzer M., Sun J.C. Atg5 Is Essential for the Development and Survival of Innate Lymphocytes. Cell Rep. 2016;15:1910–1919. doi: 10.1016/j.celrep.2016.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein A., Jeffrey P.D., Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- Park J.M., Tougeron D., Huang S., Okamoto K., Sinicrope F.A. Beclin 1 and UVRAG confer protection from radiation-induced DNA damage and maintain centrosome stability in colorectal cancer cells. PLoS ONE. 2014;9:e100819. doi: 10.1371/journal.pone.0100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.K., Miyoshi H., Beatty W.L., Head R.D., Malvin N.P., Cadwell K., Guan J.L., Saitoh T., Akira S., Seglen P.O. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32:3130–3144. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Pei B., Zhao M., Miller B.C., Vela J.L., Bruinsma M.W., Virgin H.W., Kronenberg M. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. J Immunol. 2015;194:5872–5884. doi: 10.4049/jimmunol.1402154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooz S.D., He S., Zhang T., Zhang X., Zhao Z., Oh S., O’Connell D., Khalilzadeh P., Amini-Bavil-Olyaee S., Farzan M., Liang C. UVRAG is required for virus entry through combinatorial interaction with the class C-Vps complex and SNAREs. Proc. Natl. Acad. Sci. USA. 2014;111:2716–2721. doi: 10.1073/pnas.1320629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston D.J., Zhang H., Powell T.J., Lipina E., Sims S., Panse I., Watson A.S., Cerundolo V., Townsend A.R., Klenerman P., Simon A.K. Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife. 2014;3 doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Kuhn B., Aebi J., Lin X., Ding H., Zhou Z., Xu Z., Xu D., Han L., Liu C. Discovery of Fluoromethylketone-Based Peptidomimetics as Covalent ATG4B (Autophagin-1) Inhibitors. ACS Med. Chem. Lett. 2016;7:802–806. doi: 10.1021/acsmedchemlett.6b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017;27:230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Rai S., Arasteh M., Jefferson M., Pearson T., Wang Y., Zhang W., Bicsak B., Divekar D., Powell P.P., Nauman R. The ATG5-binding and coiled coil domains of ATG16L1 maintain autophagy and tissue homeostasis in mice independently of the WD domain required for LC3-associated phagocytosis. Autophagy. 2018;15:599–612. doi: 10.1080/15548627.2018.1534507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Fitzgerald K.A. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell. 2016;165:792–800. doi: 10.1016/j.cell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A., Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A.M., Von Dwingelo J.E., Price C.T., Abu Kwaik Y. Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence. 2013;4:307–314. doi: 10.4161/viru.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.M., Tsueng G., Sin J., Mangale V., Rahawi S., McIntyre L.L., Williams W., Kha N., Cruz C., Hancock B.M. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014;10:e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rocha H., Gomez-Gutierrez J.G., Garcia-Garcia A., Rao X.M., Chen L., McMasters K.M., Zhou H.S. Adenoviruses induce autophagy to promote virus replication and oncolysis. Virology. 2011;416:9–15. doi: 10.1016/j.virol.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R.A., Janusis J., Leonard D., Bellvé K.D., Fogarty K.E., Baehrecke E.H., Corvera S., Shaw L.M. Beclin 1 regulates growth factor receptor signaling in breast cancer. Oncogene. 2015;34:5352–5362. doi: 10.1038/onc.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostislavleva K., Soler N., Ohashi Y., Zhang L., Pardon E., Burke J.E., Masson G.R., Johnson C., Steyaert J., Ktistakis N.T., Williams R.L. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein A.D., Eisenstein M., Ber Y., Bialik S., Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Rybstein M.D., Bravo-San Pedro J.M., Kroemer G., Galluzzi L. The autophagic network and cancer. Nat. Cell Biol. 2018;20:243–251. doi: 10.1038/s41556-018-0042-2. [DOI] [PubMed] [Google Scholar]
- Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., Green D.R. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Satoo K., Noda N.N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck E.M., Orchard R.C., Lassen K.G., Beatty W.L., Xavier R.J., Levine B., Virgin H.W., Sibley L.D. A Noncanonical Autophagy Pathway Restricts Toxoplasma gondii Growth in a Strain-Specific Manner in IFN-γ-Activated Human Cells. MBio. 2015;6 doi: 10.1128/mBio.01157-15. e01157–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shravage B.V., Hill J.H., Powers C.M., Wu L., Baehrecke E.H. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013;140:1321–1329. doi: 10.1242/dev.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S., Devhare P., Sujijantarat N., Steele R., Kwon Y.C., Ray R., Ray R.B. Knockdown of Autophagy Inhibits Infectious Hepatitis C Virus Release by the Exosomal Pathway. J. Virol. 2015;90:1387–1396. doi: 10.1128/JVI.02383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin J., McIntyre L., Stotland A., Feuer R., Gottlieb R.A. Coxsackievirus B Escapes the Infected Cell in Ejected Mitophagosomes. J. Virol. 2017;91 doi: 10.1128/JVI.01347-17. e01347–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.B., Davis A.S., Taylor G.A., Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Singh S.B., Ornatowski W., Vergne I., Naylor J., Delgado M., Roberts E., Ponpuak M., Master S., Pilli M., White E. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat. Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Letai A., Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019;20:175–193. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbara M.T., Ellison L.K., Ramjeet M., Travassos L.H., Jones N.L., Girardin S.E., Philpott D.J. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity. 2013;39:858–873. doi: 10.1016/j.immuni.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Sousa C.M., Biancur D.E., Wang X., Halbrook C.J., Sherman M.H., Zhang L., Kremer D., Hwang R.F., Witkiewicz A.K., Ying H. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M.K., Cookson B.T. Evasion and interference: intracellular pathogens modulate caspase-dependent inflammatory responses. Nat. Rev. Microbiol. 2016;14:346–359. doi: 10.1038/nrmicro.2016.50. [DOI] [PubMed] [Google Scholar]
- Storz J.F. Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet. 2016;17:239–250. doi: 10.1038/nrg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strappazzon F., Di Rita A., Cianfanelli V., D’Orazio M., Nazio F., Fimia G.M., Cecconi F. Prosurvival AMBRA1 turns into a proapoptotic BH3-like protein during mitochondrial apoptosis. Autophagy. 2016;12:963–975. doi: 10.1080/15548627.2016.1164359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 2005;280:40058–40065. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- Tang H., Sebti S., Titone R., Zhou Y., Isidoro C., Ross T.S., Hibshoosh H., Xiao G., Packer M., Xie Y., Levine B. Decreased BECN1 mRNA Expression in Human Breast Cancer is Associated with Estrogen Receptor-Negative Subtypes and Poor Prognosis. EBioMedicine. 2015;2:255–263. doi: 10.1016/j.ebiom.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen S.B., Pedersen N.M., Liestøl K., Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 2010;316:3368–3378. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Tian L., Yang Y., Li C., Chen J., Li Z., Li X., Li S., Wu F., Hu Z., Yang Z. The cytotoxicity of coxsackievirus B3 is associated with a blockage of autophagic flux mediated by reduced syntaxin 17 expression. Cell Death Dis. 2018;9:242. doi: 10.1038/s41419-018-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- Tung S.M., Unal C., Ley A., Peña C., Tunggal B., Noegel A.A., Krut O., Steinert M., Eichinger L. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell. Microbiol. 2010;12:765–780. doi: 10.1111/j.1462-5822.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- Üstün S., Hafrén A., Hofius D. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 2017;40:122–130. doi: 10.1016/j.pbi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Wang R.C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., White M., Reichelt J., Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Iyengar R., Li-Harms X., Joo J.H., Wright C., Lavado A., Horner L., Yang M., Guan J.L., Frase S. The autophagy-inducing kinases, ULK1 and ULK2, regulate axon guidance in the developing mouse forebrain via a noncanonical pathway. Autophagy. 2018;14:796–811. doi: 10.1080/15548627.2017.1386820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Zou Z., Becker N., Anderson M., Sumpter R., Xiao G., Kinch L., Koduru P., Christudass C.S., Veltri R.W. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Zhang J., Si X., Gao G., Mao I., McManus B.M., Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Xiao Z., Ma X., He F., Yao H., Liu Z. Coxsackievirus B3 induces crosstalk between autophagy and apoptosis to benefit its release after replicating in autophagosomes through a mechanism involving caspase cleavage of autophagy-related proteins. Infect Genet Evol. 2014;26:95–102. doi: 10.1016/j.meegid.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Xiong Q., Ünal C., Matthias J., Steinert M., Eichinger L. The phenotypes of ATG9, ATG16 and ATG9/16 knock-out mutants imply autophagy-dependent and -independent functions. Open Biol. 2015;5:150008. doi: 10.1098/rsob.150008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Araki K., Li S., Han J.H., Ye L., Tan W.G., Konieczny B.T., Bruinsma M.W., Martinez J., Pearce E.L. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Suzuki N.N., Hanada T., Ichimura Y., Kumeta H., Fujioka Y., Ohsumi Y., Inagaki F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- Yang Y., He S., Wang Q., Li F., Kwak M.J., Chen S., O’Connell D., Zhang T., Pirooz S.D., Jeon Y. Autophagic UVRAG Promotes UV-Induced Photolesion Repair by Activation of the CRL4(DDB2) E3 Ligase. Mol. Cell. 2016;62:507–519. doi: 10.1016/j.molcel.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Jang G.B., Yang X., Wang Q., He S., Li S., Quach C., Zhao S., Li F., Yuan Z. Central role of autophagic UVRAG in melanogenesis and the suntan response. Proc. Natl. Acad. Sci. USA. 2018;115:E7728–E7737. doi: 10.1073/pnas.1803303115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatim N., Cullen S., Albert M.L. Dying cells actively regulate adaptive immune responses. Nat. Rev. Immunol. 2017;17:262–275. doi: 10.1038/nri.2017.9. [DOI] [PubMed] [Google Scholar]
- Yin H.C., Zhao L.L., Li S.Q., Niu Y.J., Jiang X.J., Xu L.J., Lu T.F., Han L.X., Liu S.W., Chen H.Y. Autophagy activated by duck enteritis virus infection positively affects its replication. J. Gen. Virol. 2017;98:486–495. doi: 10.1099/jgv.0.000696. [DOI] [PubMed] [Google Scholar]
- You S.Y., Park Y.S., Jeon H.J., Cho D.H., Jeon H.B., Kim S.H., Chang J.W., Kim J.S., Oh J.S. Beclin-1 knockdown shows abscission failure but not autophagy defect during oocyte meiotic maturation. Cell Cycle. 2016;15:1611–1619. doi: 10.1080/15384101.2016.1181235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Kenny S.J., Ge L., Xu K., Schekman R. Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion. eLife. 2015;4:e11205. doi: 10.7554/eLife.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Fux B., Goodwin M., Dunay I.R., Strong D., Miller B.C., Cadwell K., Delgado M.A., Ponpuak M., Green K.G. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Oh S., Li D., Ni D., Pirooz S.D., Lee J.H., Yang S., Lee J.Y., Ghozalli I., Costanzo V. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Dev. Cell. 2012;22:1001–1016. doi: 10.1016/j.devcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]