Abstract
During the immune response, striking the right balance between positive and negative regulation is critical to effectively mount an anti-microbial defense while preventing detrimental effects from exacerbated immune activation. Intra-cellular immune signaling is tightly regulated by various post-translational modifications, which allow for this dynamic response. One of the post-translational modifiers critical for immune control is ubiquitin, which can be covalently conjugated to lysines in target molecules, thereby altering their functional properties. This is achieved in a process involving E3 ligases which determine ubiquitination target specificity.
One of the most prominent E3 ligase families is that of the tripartite motif (TRIM) proteins, which counts over 70 members in humans. Over the last years, various studies have contributed to the notion that many members of this protein family are important immune regulators. Recent studies into the mechanisms by which some of the TRIMs regulate the innate immune system have uncovered important immune regulatory roles of both covalently attached, as well as unanchored poly-ubiquitin chains. This review highlights TRIM evolution, recent findings in TRIM-mediated immune regulation, and provides an outlook to current research hurdles and future directions.
Abbreviations: 5′ppp-RNA, 5′-triphosphate RNA; AIM2, Absent in melanoma 2; ASC, apoptosis-associated speck-like protein containing a CARD; CARD, caspase activation and recruitment domain; CLR, C-type lectin receptor; DC, dendritic cell; DDX41, DEAD-box protein 41; dsRNA, double-stranded RNA; GART, gene encoding phosphoribosylglycinamide formyltransferase; IFITM, interferon-induced transmembrane protein; IFN, interferon; IFNαR, IFNα receptor; IKK, IκB kinase; IL, interleukin; iNOS, inducible nitric oxide synthetase; IRF, IFN regulatory factor; ISG, interferon-stimulated gene; ISG15, IFN-stimulated gene 15; JAK, Janus kinase; MAVS, mitochondrial antiviral signaling; MDA5, melanoma differentiation-associated protein 5; MEF, mouse embryonic fibroblast; MHC, major histocompatibility complex; MΦ, macrophage; MYD88, myeloid differentiation primary response gene 88; NEDD8, neural precursor cell expressed developmentally down-regulated 8; NEMO, NFκB essential modulator; NFκB, nuclear factor κB; NK, natural killer cell; NLR, NOD-like receptor; NS1, influenza virus non structural 1; OAS, 3–5 oligo adenylate synthetase; PAMP, pathogen-associated molecular pattern; PBMC, peripheral blood mononuclear cell; PHD, plant homeo domain; PI3KC2β, phosphatidylinositol 3 kinase C2β; PIN1, prolyl isomerase 1; PRR, pattern recognition receptor (PRR); RBCC, RING-BBox-coiled coil (= tripartite motif); RIG-I, retinoic acid inducible I; RING, really interesting new gene; RLR, RIG-I-like receptor; ROS, reactive oxygen species; SeV, Sendai virus; STAT, stimulator and activator of transcription; STING, stimulator of interferon genes; SUMO, small ubiquitin-like modifier; TAB, TGFβ-activated kinase binding protein; TAK, TGFβ-activated kinase; TBK1, TANK-binding kinase; TCR, T cell receptor; TLR, toll-like receptor; TNF, tumor necrosis factor; TRAF, TNF receptor associated factor; TRIF, TIR-domain-containing adapter-inducing IFN; TRIM, tripartite motif protein; TUB, TRIM-ubiquitin body; Ub, ubiquitin
Keywords: E3 ligase, TRIMmunity, Ubiquitin, TRIM protein, Interferon
1. Regulation of immune balance
The immune system is at its rudimentary level divided in an innate and adaptive part. Innate immune responses can be mounted by virtually all nucleated cells in the body and are critical for anti-microbial defense [1], [2]. Besides being crucial in various non-immune cells, the innate responses are particularly developed in several key innate immune cells from myeloid origin (such as monocytes, macrophages (MΦ), dendritic cells (DC), mast cells, neutrophils, basophils, eosinophils), and natural killer cells from lymphoid origin [1], [2]. The innate response is mounted very rapidly (within hours) and lasts for about four days until the adaptive response by T and B cells is engaged. This innate response is critical for initial survival and controlling infection until the slower but specific adaptive response has been mounted. Moreover, innate immune cells create a cytokine milieu which facilitates the activation of the adaptive response [3]. The innate immune compartment includes various biologically important sensors and effector components to detect and limit microbial infection, respectively [4]. The importance of these systems for microbial control is underpinned by the fact that almost all currently circulating pathogens have developed multiple strategies to circumvent these cellular recognition and innate immunity branches [5].
Correct spatio-temporal regulation of immune signaling is critical to adequately respond to pathogen infection, while preventing auto-immunity and hyper-inflammatory disease [6], [7]. At the inter-cellular level this balance is for an important part determined by how different cell types communicate through cytokines and cell–cell interactions. At the intra-cellular level in all these various cell types lay complex, interconnected cell signaling networks which in turn are controlled in a dynamic fashion by positively and negatively regulating molecular complexes.
As for all cellular signaling, the formation, activity, localization and stability of these intra-cellular communication hubs are controlled by post-translational modifications. In that regard, phosphorylation plays an abundant, critical role in virtually all stages of immune cell signaling. Additionally, in recent years it has become clear that also conjugation with the post-translational modifier ubiquitin is wide-spread and essential for positive and negative immune signaling and hence maintaining immune balance [8].
2. Ubiquitin-mediated immune regulation
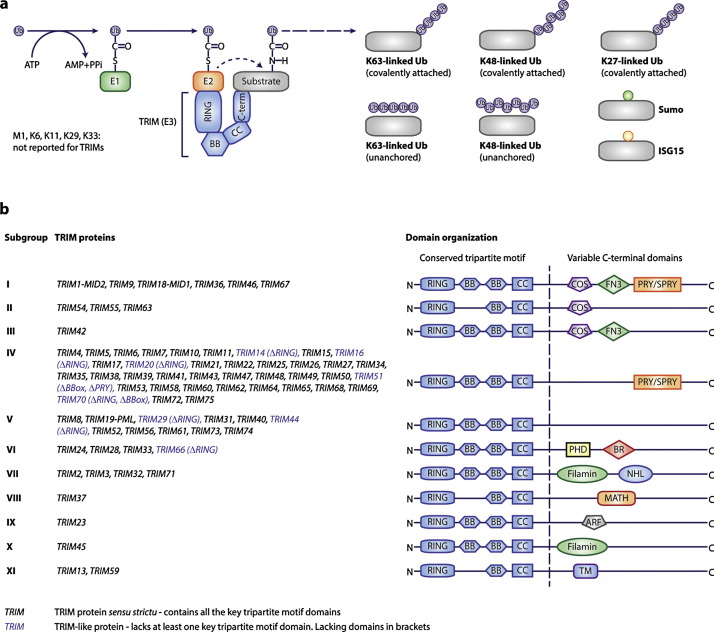
Ubiquitin is a 8.5 kDa protein, which can be covalently attached through its highly conserved C-terminal di-glycine motif onto lysines in target proteins by an isopeptide bond [9]. This ubiquitination takes place in an orchestrated fashion (Fig. 1a): (i) ubiquitin is conjugated to a cysteine residue of an E1 activating enzyme through a thioester bond, (ii) is subsequently transferred to an E2 conjugating enzyme by thioester bond, to (iii) ultimately be transferred to the targeted substrate lysine through the action of an E3 ligating enzyme.
Fig. 1.
(a) Overview of ubiquitin conjugation by RING E3 ligases such as TRIM proteins. After ubiquitin activation by the E1 enzyme, ubiquitin is covalently attached to an E2 conjugase through a thio-ester bond. Subsequently, the TRIM E3 ligases bind this E2 complex through their RING domain, while determining specificity by associating with the ubiquitination target often through one of its C-terminal domains. This brings the ubiquitin-loaded E2 in close proximity to the intended target, after which the ubiquitin moiety is directly transferred to a lysine residue in the target protein by an isopeptide bond. Subsequently, additional ubiquitins can be conjugated onto internal lysines in ubiquitin or its N-terminal methionine (M1). The same ubiquitin chains can also be synthesized without being covalently coupled to their targets (unanchored ubiquitin chains). (b) Overview of the domain organization of TRIM family members.
The most numerous class of E3 ligases – RING E3s – facilitates the transfer of ubiquitin directly from the E2 conjugase onto the target protein [10]. The number of enzymes involved in ubiquitin transfer follows a pyramid-like shape: ∼1–3 E1 activating enzymes, ∼30–35 E2 conjugating enzymes, and >600 E3 ligases [11]. This distribution fits the notion that E3 ligases are the main determinants of target specificity [9].
What makes ubiquitin-mediated regulation stand out from other post-translational modifications is that ubiquitin itself can be ubiquitinated and form poly-ubiquitin chains. Ubiquitin has itself seven internal lysines (K6, K11, K23, K27, K33, K48, K63) on which different chain types can be formed, in addition to head-to-tail linear (M1) ubiquitin chains. In the case of RING E3s, the chain type is predominantly determined by the associated E2 enzyme [9].
Taken together, ubiquitin has nine different forms by which it can modify targets proteins: as a single ubiquitin moiety or eight different chain types. Each of these different ubiquitin chain constellations can alter various different characteristics of their target proteins, ranging from stability, to sub-cellular localization and activation status. This provides a highly dynamic and versatile platform to regulate cell signaling as the conjugated “Ub-code” can be read-out by different cellular protein complexes containing Ub-binding molecules and effector molecules such as e.g. kinases or proteases [9]. Hence, the same Ub chain type on distinct proteins can differentially affect the characteristics of these molecules.
Functionally, K48 and K63-linked poly-ubiquitin chains are the best characterized; proteins covalently modified with K48-linked poly-ubiquitin are targeted for proteasomal degradation, whereas proteins covalently modified with K63-linked poly-ubiquitin, in general, become functionally activated. It is important to point out that the study of most other types of poly-ubiquitin chains has been complicated by the lack of poly-ubiquitin chain-type-specific reagents. The importance of the ubiquitination process in immune signaling is also highlighted by the fact that many viruses have acquired methods to counteract the immune response by inhibiting the ubiquitin system [12].
In addition to the above described covalently attached poly-ubiquitin chains, in recent years increasing evidence has accumulated indicating that also unanchored poly-ubiquitin chains can have important activating functions [13], [14], [15]. The variety of modifications and effects of the ubiquitination system is further increased by the presence of several ubiquitin-like (Ubl) molecules. Although their primary structures are often only distantly homologous to ubiquitin, structural studies have shown that all Ubls adopt a ubiquitin-like fold highly reminiscent of ubiquitin itself. Like ubiquitin, Ubls are covalently conjugated to lysine residues on target proteins through a conserved Ubl C-terminal motif, thereby altering their target's properties in various ways [9].
Similar to the ubiquitin conjugation machinery, other Ubls are conjugated through an E1–E2–E3 system. In most reported cases, the E1 and E2 enzymes are specific for one Ubl-type, whereas certain E3s functionally conjugate more than one Ubl. It is not uncommon that this switch in Ubl conjugation is part of a regulatory mechanism to alter its target's characteristics [16].
The major Ubls which have been implemented in affecting immune regulation pathways are ISG15, SUMO, and NEDD8 [17], [18], [19], [20], [21], [22]. Similar to ubiquitin, SUMO and NEDD8 have the reported ability to forms polymeric chains, although their relevance in vivo and how well-spread their involvement in various biological processes is remains to be determined in more detail [23], [24], [25].
3. The tripartite motif (TRIM) E3 ligase family
In recent years, an increasing number of studies have implicated the E3 ligase family of tripartite motif (TRIM) proteins in regulation of immune signaling and conferring direct anti-viral activity [26], [27], [28]. These proteins derive their name from their common N-terminal tripartite RBCC motif consisting of a RING domain, one or two BBox domains and a coiled-coil domain. Based on their variable C-terminal domains, they have been classified into eleven different sub-groups (Fig. 1b). In line with important functions in immune regulation, many TRIMs have been reported to be interferon (IFN)-inducible (Fig. 2 ) [26], [29]. Moreover, a substantial number of TRIM genes are organized in gene clusters, of which especially the short arm of chromosome 6 stands out to harbor an array of recognized antiviral and immune-regulatory TRIMs (Fig. 2).
Fig. 2.
Chromosomal location of human TRIM genes. TRIM proteins induced by influenza virus infection [26] or IFN stimulation [29] are indicated with orange circles and yellow stars, respectively.
Several TRIMs have been recognized as critical intrinsic antiviral restriction factors by directly acting as antiviral effector molecules. The best studied antiviral TRIM, is the alpha isoform of TRIM5, which has been under heavy positive selection pressure [30]. Functional studies have shown that TRIM5α can directly target components of retro- and lentiviral life cycles to confer antiviral effects by ubiquitin-dependent and -independent means [31], [32], [33], [34], and may confer species-specific restriction. For example, while human TRIM5α restricts murine leukemia virus infection, it does not prevent HIV-1 infection [35]. In contrast, Old World monkey TRIM5α restricts infection with HIV-1, while being ineffective against infection with its simian counterpart SIV [36], [37]. In addition, to restriction of retroviruses, several other TRIMs have been reported to confer antiviral activity against members of most major virus families [38]. However, as recently discussed the biological relevance of some of these overexpression-based studies will need to be established in vivo [39].
TRIM proteins are defined by the presence of an N-terminal RBCC tripartite motif (Fig. 1b), yet members of the family exist which lack one or more of these RBCC domains. Nevertheless, these “TRIM-like” proteins generally share their C-terminal domain layout with other members of their respective TRIM sub-group. Importantly, analysis of alternatively spliced TRIM mRNAs has indicated that for over a third of all TRIMs isoforms may be expressed lacking at least one of the RBCC domains. These data suggest that the pool of functional TRIM protein forms may be 2–3 times larger than the mere number of TRIM genes. Moreover, it indicates that the TRIM family is seemingly more defined by having at least a partial RBCC in combination with one of the eleven C-terminal domain constellations. Also functionally, various “TRIM-like” members have reported immune regulatory functions similar to their full-RBCC relatives (discussed below). For that reason, we use the TRIM designation for all family members.
The functional core of the RING domain is a Cys3-His-Cys4 zinc finger, which binds two zinc cations. The RING domain is a well-known docking site for E2 conjugases in E3 ligase enzymes. Hence, a substantial number of the TRIM family members have been demonstrated to confer E3 ligase activity for ubiquitin or UbLs, such as SUMO, ISG15 and NEDD8. However, the E3 potential of many TRIMs and their relevance for cell biology remains to be experimentally demonstrated.
All TRIM proteins contain a BBox2 domain, while some also contain a BBox1 domain which differs in its spacing of zinc-binding residues. Both BBox types are zinc-finger structures highly similar to the RING domain, and the PHD domain found in the C-terminus of group VI TRIMs [40], [41]. Although mutations in TRIM BBoxes have been associated with auto-immune diseases in humans, very little is known about their structural and functional roles in cellular regulation. As a result of the high resemblance of BBoxes to RING domains, it has been suggested that BBoxes could in some TRIMs provide an E2 binding site similar to RINGs and thereby confer E3 ligase activity to RINGless TRIMs. In support of this notion, recent work demonstrated that RINGless TRIM16 confers ubiquitin E3 ligase activity in vitro [42]. In addition to this ubiquitin-dependent function, ubiquitin-independent functions have been described for TRIM BBoxes, such as for TRIM20 to keep the ASC inflammasome adaptor in an auto-inhibited state [43], and for TRIM14 where it may fulfill an scaffolding role in MAVS-mediated signaling [44].
Several TRIM proteins have been reported to form homo-dimers, which in most cases is required for their biological function and the formation of sub-cellular bodies [45]. The coiled-coil (CC) domain is necessary and sufficient for oligomerization of TRIM proteins [45], [46], [47]. Although predominantly the formation of TRIM homo-oligomers has been reported, systematic studies have revealed that TRIM hetero-oligomers can be formed at least experimentally, potentially adding to the diversification of their biological functions [42], [48].
The C-terminal domains found in TRIM proteins are diverse. The most prominent C-terminal TRIM domain is the PRY-SPRY domain (also known as B30.2), which is present in members of the most numerous TRIM sub-group IV (Fig. 1b). The number of TRIMs bearing this domain expanded and evolved rapidly in recent evolution. The reported functions of the PRY-SPRY domain are diverse, yet predominantly this domain mediates protein-interactions [38]. In most instances this protein-interaction entails binding of ubiquitination targets and determining E3 ligase specificity as discussed for individual TRIMs in detail below. Moreover, the PRY-SPRY domain is critical for the direct antiviral restriction activity of certain TRIMs such as TRIM5α [49].
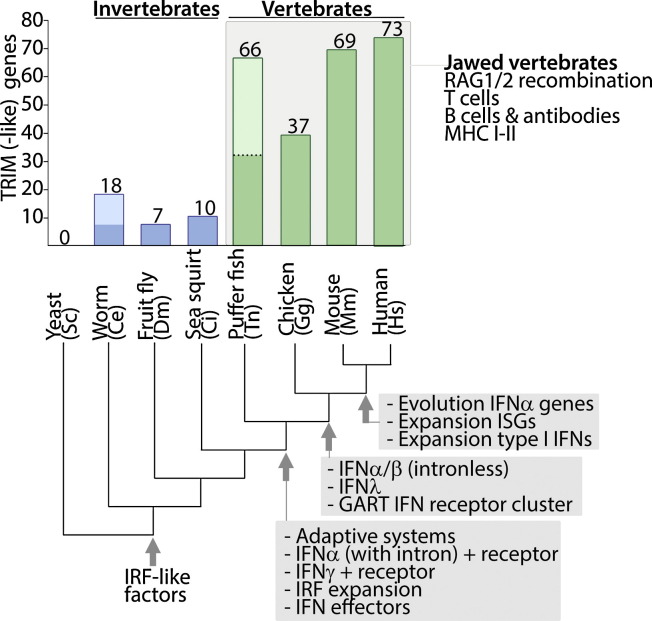
In addition to direct antiviral effectors, various TRIM proteins have been recognized as important immune signaling mediators. Although all non-plant, multi-cellular eukaryotes have TRIM genes, their number dramatically expanded relatively recently in evolution with the split-off of jawed vertebrates (Fig. 3 ). Since this TRIM expansion shows striking parallels with the development of innate and adaptive immune traits it has been speculated that the TRIM family may have expanded to provide regulatory mechanisms for the increasingly complex immune system.
Fig. 3.
Number of TRIM genes plotted for organisms representative of various evolutionary stages. TRIM genes likely to have arisen in a species specific manner after split-off from a common ancestor are indicated in lighter color. Hallmarks in immune system evolution are indicated at the tree branch point where they were introduced in a common ancestor.
4. TRIM family evolution
Lower invertebrates ranging from flies to organisms such as sea squirts (invertebrate) and lampreys (jawless vertebrate), have in general a relative low number of TRIM genes (5–10; Fig. 3). However, certain exceptions of species with higher TRIM gene content have been identified, which may have resulted from genetic duplications in those species, independently from the gene sets in common ancestors. Phylogenetic analyses have indicated that the relative high TRIM gene number in e.g. the nematode C. elegans with 18 TRIM genes (vs. 6–10 in the fruit fly and sea squirt) may have arisen from a species-specific multiplication [50].
Jawless fish such as lampreys have still relatively low numbers of TRIM genes, yet all jawed fish and mammals have expanded families of >60 TRIM genes. TRIMs have been under heavy selection pressure in various fish species, similar to TRIM5 in humans and monkeys, indicating that they may play important, fish-specific roles in anti-microbial defense [51], [52]. This is underpinned by the notion that puffer fish have 66 TRIM genes (similar to mammals), whereas zebra fish have over 200 TRIM genes [51], [53], the antimicrobial function of which remains to be determined.
Phylogenetic analysis of the puffer fish and zebra fish genomes has indicated that despite their high number of TRIMs, only ∼30 of them have counterparts in mammals [51]. This suggests that although the main TRIM classes were already present in the common ancestor of fish and tetrapods, various fish-specific TRIMs have evolved from them independently of mammalian TRIM evolution [51].
Interestingly, the chicken genome only holds 37 TRIM genes [50], even though they are evolutionary closer to mammals than fish. Analyses on other avian species are sparse, and it is thus difficult to say whether chicken have an aberrantly low number of TRIM genes compared to mammals or whether other avian species have a TRIM gene count of 30–40 similar to what may have been in the common ancestor of fish and tetrapods, suggesting that two TRIM expansions may have occurred in evolution: one at the rise of jawed vertebrates from ∼10 to ∼30 TRIMs, and a second one independently for fish and mammals: from ∼30 TRIMs in the common ancestor to ∼60–70 in puffer fish and mammals, and >200 in fish with larger genomes such as zebra fish.
The evolution and expansion of the TRIM family has striking parallels with the evolution of the adaptive immune system, as well as the type I IFN (IFN-I) system [30], [54]. Since many members of the TRIM family have been implicated in regulation of the IFN-I system, it is not inconceivable that these protein families may have in part co-evolved. In fact, plotting of the TRIM gene numbers in species representing various stages of eukaryotic evolution suggests that the increases in TRIM genes shows several similarities with the rapid evolution of the IFN system (Fig. 3). One example of these similarities is the expansion of the ancestors of the IFN regulatory factors (IRF) -which in vertebrates drive the transcription of IFNs. Although ancestral IRFs were already present in early invertebrates, their number rapidly expanded and evolved under heavy positive selection from a common ancestor to the jawed vertebrates similar to TRIMs (Fig. 3) [55].
In the same evolutionary time frame as this IRF expansion, also the ancestral intron-containing IFNα genes with corresponding IFNαR, ancestral type II IFN and receptor, and ISGs were introduced [56]. Although the common ancestor of fish and tetrapods introduced a comprehensive IFN system in both evolutionary lines, the IFN system continued to evolve rapidly in both fish and tetrapods after their split-off [56], [57]. While fish have maintained ancestral IFN genes containing introns, amniote (e.g. avian and mammalian) IFNs lost their introns and in addition developed a type III pathogen-inducible, antimicrobial IFNλ system [56]. Despite the knowledge of the chicken ISG repertoire being limited, published work on the OAS and IFITM families of ISGs have indicated these ISGs groups are more developed and abundant in mammals than in avian species [58], [59].
Similarly, cytokine receptors have evolved rapidly and divergently in fish and amniotes [60]. One of the hallmarks has been the formation of an IFNαR, IFNγR/IL10R gene cluster downstream of the GART gene on chromosome 21, already present in chicken, but further developed to form a synteny group in mammals. Additionally, the IFNα/β genes have been under heavy selection pressure, while also additional type I IFNs (such as IFNδ, IFNɛ, IFNτ and IFNω) with both immune and non-immune functions arose in mammals [56], [61]. This, together with the increasing experimental evidence linking TRIM function to the IFN-I system has led to the hypothesis that TRIMs may have evolved as a component of the innate IFN response.
Importantly, all jawed vertebrates have a similar set of adaptive immune traits, which also diversified, expanded and evolved after the split-off from jawless vertebrates [54], [56]. Thus the introduction of key adaptive components, such as T and B cell receptors, immunoglobulins, MHC and Rag genes also evolved in a similar evolutionary time frame as the TRIM family [54]. The fact that these systems show paralleled evolution may indicate that the TRIM family expanded to regulate the increasingly complex immune system, perhaps both indirectly at the level of innate cytokine expression involved in coordinating the adaptive responses, as well as directly in T and B cells.
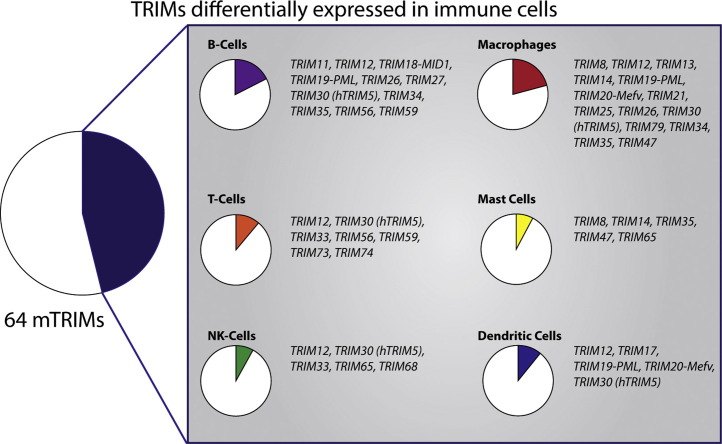
So far, most evidence indicates roles for TRIM proteins in the innate compartment. However, since uncontrolled innate immune activation also affects adaptive immune activation, the possibility remains that the TRIM family expanded to control the innate immune compartment in the face of the arising adaptive system. Additionally, the co-expansion of the TRIM family and adaptive immune mechanism indicates that certain TRIMs may have direct roles in adaptive immunity. So far there is little experimental evidence linking TRIM function to the adaptive immune system, although fifteen TRIMs are highly expressed in T and/or B cells (Fig. 4 ). In fact, a subset of TRIMs has been previously shown to be highly expressed in CD4 T cells with immune-modulatory roles such as Th2 and IL10-Treg cells as compared to MΦ and DCs [26]. Thus far, only a specific function has been described for TRIM27 in negative regulation of Ca2+ influx and cytokine production during TCR engagement by targeting phosphatidylinositol 3 kinase C2β (PI3KC2β) for K48-mediated proteasomal degradation [62], and TRIM28 which in CD4 T cells by yet unknown mechanisms regulates IL2 production and cell proliferation upon TCR engagement [63]. Future studies on TRIMs in adaptive immune cells are needed to further elucidate the roles of other TRIMs in lymphoid cells.
Fig. 4.
Cell-type restricted TRIM mRNA expression. The mouse MOE430 dataset (GRCMA normalized) was analyzed using BioGPS.org. TRIMs expressed >3 times the mean in the selected immune cell types (compared to all other cell types tested) are plotted.
All amniotes have roughly the same immune system in terms of cell types and immune-relevant organs, which would not explain a secondary expansion of the TRIM family from ∼30 to 40 TRIM genes in chicken to >60 in mammals. However, despite a similar overall structure, still many differences exist between avian and mammalian immune system. A detailed comparison is hindered by relatively limited functional knowledge on the avian side, however, it is clear that several important differences between the avian and mammalian immune system exist. These differences may have functional significance; for example, studies have shown that avian strains of influenza viruses encode for non-structural-1 (NS1) proteins that are more efficient in binding and inhibiting TRIM25-mediated IFN induction in chickens than human influenza virus strains, suggesting species-specific adaptation [64].
Taken together, these observations contribute to the notion that the number of TRIM genes developed rapidly in recent evolution with compelling parallels to innate and adaptive immune expansion and development. Importantly, recent analysis identified several novel TRIM genes exclusively found in humans, chimpanzees, gorillas and bonobos [65]. This indicates that even differences exist in the TRIM repertoire of these primates and other higher mammals with highly similar immune systems, suggesting that these TRIMs are under specific selection pressures such as from specific pathogens, rather than being only immune regulators. Nevertheless, in recent years many TRIM proteins have been reported to have immune regulatory activities as discussed below, supporting the notion that the TRIM family as a whole may have expanded in evolution to provide a means to fine-tune cell signaling of the immune response.
5. TRIM spatial regulation and cell-type specific expression
Different TRIMs have been reported to localize to various sub-cellular compartments, some of which are relocalized upon infection, suggesting that their activation or function may be spatially controlled [28], [45]. Upon exogenous expression, many TRIMs form cytoplasmic bodies, which do not co-localize with sub-cellular markers for most-common organelles [45]. Recent work on TRIM6, which also facilitates the formation of these cytoplasmic structures, showed that unanchored poly-ubiquitin synthesized by TRIM6 is crucial for the formation of these TRIM-ubiquitin bodies (TUB) and their role in immune signaling [14]. It is tempting to speculate that these TUBs may be structures providing a sub-cellular scaffolding platform to recruit and confine TRIMs and their targets, although more studies are needed to investigate the general nature of this finding.
It is important to note that the large TRIM6 TUBs found by exogenous expression are smaller and less well-defined in the endogenous setting [14], indicating that the levels of expression may affect the size and shape of these structures. Future work is needed to determine the existence and relevance of these cytoplasmic ubiquitin-containing bodies formed by other TRIMs. In addition to ubiquitin as a component of TRIM-cytoplasmic bodies, quantitative proteomics analysis of TRIM32 interactors, identified 14-3-3 proteins as potential regulators of the TRIM32 oligomeric state and its sub-cellular localization [66]. Hence, TRIM-body formation and disassembly may be regulated in a 14-3-3-dependent manner, although again the general nature of this requires further investigation.
Expression profiling has indicated that virtually all cell types express mRNA for multiple TRIMs, yet the TRIM repertoire can vary considerably per cell type. Analysis of mouse TRIM expression indicated that nearly half of all TRIMs is expressed in a relatively cell-type or tissue-specific manner (MOE430 dataset, GRCMA normalized; analyzed using BioGPS.org), with above three times mean expression of TRIM mRNA in various key myeloid and lymphoid immune cell types (Fig. 4). In addition, different subset of TRIMs was also shown to be differentially expressed in plasmacytoid DCs (pDC) and in CD4 T cells whereas a different subset was induced in MΦs and DCs by TLR stimulation and virus infection in a IFN-I dependent manner [22]. These preferential expression patterns add to the notion that TRIMs may have evolved as immune regulators and play important regulatory roles in these immune cells. However, it also indicates that the cell-regulatory roles of individual TRIMs may be highly cell-type specific and thus future investigation using specific primary cell types and cell-type specific knock-out approaches.
6. The IFN system
Arguably the most critical innate control axes for anti-viral control are the type I (IFN-I; IFNα/β) and type III (IFNλ) interferon cytokine systems. IFN-I is produced by virtually all nucleated cell types [4]. In the face of viral infection, IFN-I production is critical to limit viral spread. Mice lacking key components of these systems succumb to infection with otherwise sub-lethal virus doses as they are unable to limited viral replication until the adaptive immune system has been mounted, which is often essential for viral clearance [67].
It should be noted that for bacterial infections the effects of IFN-I can vary from beneficial to detrimental to the host, depending on bacterial species and strain [68], [69]. The IFN-I system is also critical for shaping other parts of the innate and adaptive immune branches [2], whereas pro-longed or aberrant IFN-I production contributes to auto-immune reactions [6].
Similar to IFN-I, type III IFN plays a major role in antiviral control. This cytokine is also expressed by most cell types upon stimulation, yet expression of its receptor (IFN-λR1/IL10R2) is restricted to epithelial tissues [70]. Despite having a different receptor from IFN-I, IFN-III JAK-STAT signaling down-stream of its receptor is virtually the same as for IFN-I [70]. In contrast to IFN-I and IFN-III during viral infections, type II IFN (IFNγ) is in general considered more critical for the control of bacterial infections. This cytokine is predominantly secreted by T helper 1 (Th1) and NK cells during infection, and activates mechanisms in macrophages critical for anti-bacterial immunity against phagocytosed bacteria. The induction of the phagosome NADPH-oxidase complex which synthesizes reactive oxygen species (ROS) and inducible nitric oxide synthetase (iNOS) are main effector functions by which IFNγ confers anti-bacterial infection[71], which is underpinned by the fact that individuals with mutations in these systems are hyper-susceptible to bacterial infections [72].
7. TRIM-mediated regulation of cytokine induction and signaling
Signaling associated with IFN-I induction and response follows a common strategy similar to other major innate immune cytokine induction and signaling pathways [4]. Pathogen sensing occurs when pathogen-associated molecular patterns (PAMPs) contained in microbial products are recognized by pattern recognition receptors (PRR), leading to the recruitment of adaptor proteins, such as TIR-domain-containing adapter-inducing IFN (TRIF), Myeloid differentiation primary response gene 88 (MyD88) and TNF receptor associated factors (TRAF), or MAVS in the mitochondria. Subsequently, multiple kinases are activated resulting in phosphorylation of transcription factors which in turn translocate to the nucleus and drive pro-inflammatory cytokine and/or IFN expression [4]. TRIMs have been shown to be involved in any one of these signaling steps in several different cytokine pathways.
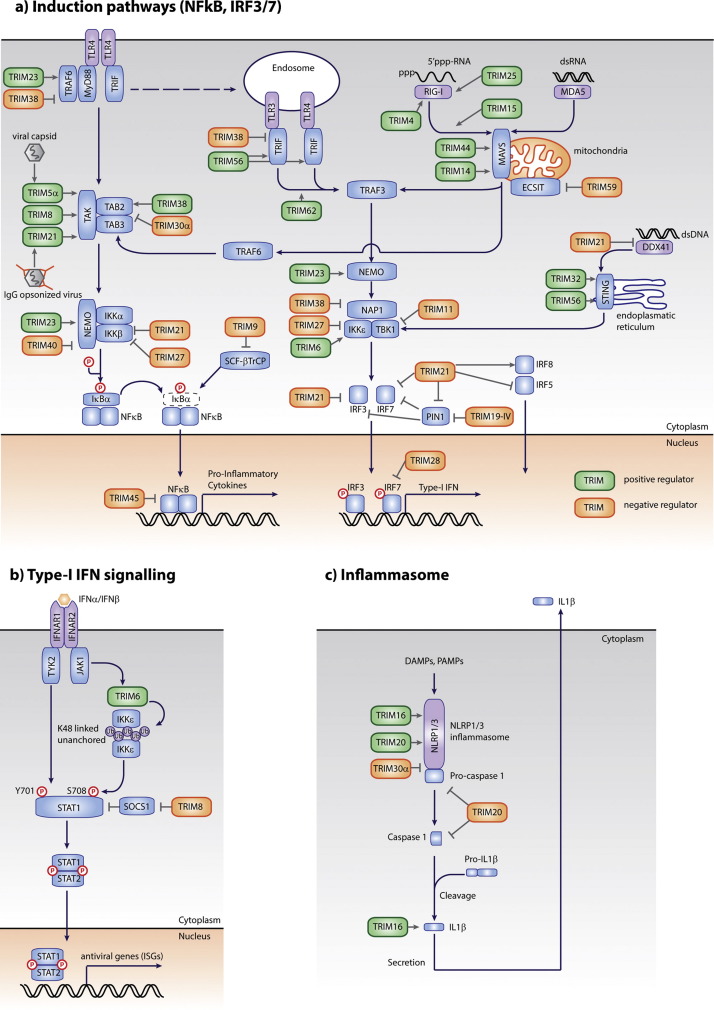
Since by far most TRIMs currently reported to facilitate immune regulation have been associated with IFN-I and inflammasome signaling, these two axes will be further discussed below. We will discuss here a subset of these TRIMs to highlight commonalities, unique features, or experimental evidence. For completion, a summary of all reported immune-regulating TRIMs and their proposed targets has been included (Table S1).
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.cytogfr.2014.08.001.
7.1. TRIMs as receptors in immune signaling
Cells can sense invading microbes by different PRRs, including Toll-like receptors (TLR), C-type lectin receptors (CLR), RIG-I-like receptors (RLR) or, NOD-like receptors (NLR). All of these PRRs recognize molecular patterns either not present in healthy cells, or separated by sub-cellular compartmentalization. So far, most immune regulatory TRIM proteins have been associated with regulating immune signaling. However, in recent years two TRIMs have been proposed to facilitate pathogen recognition either directly or in an immune receptor capacity.
Firstly, recent work showed that TRIM5 induces NFκB activation by synthesis of K63 Ub chains which non-covalently associate with the TAK1-containing kinase complex (Fig. 5a) [13]. Infection experiments using retroviruses with differential TRIM5 binding affinities, and in vitro assays with oxidized cylinders of viral capsid, indicated that TRIM5 recognizes the oligomeric retroviral capsid lattice through its PRY-SPRY domain. This in turn triggers the synthesis of unanchored K63 ubiquitin chains which activate the NFκB pathway and pro-inflammatory cytokine response through the TAB/TAK kinase complex [13]. By these means TRIM5 has been proposed to directly act as a pattern recognition receptor.
Fig. 5.
Overview of all TRIM proteins reported to date to modulate (a) IFN-I induction, (b) IFN-I signaling, and (c) inflammasome activity. TRIMs enhancing the activity/function of the indicated proteins are indicated in green, whereas negative regulators are orange. Pathogen-recognition receptors are indicated in purple; signaling molecules are in blue.
Additionally, TRIM21 has been recognized to have the properties of a cytoplasmic Fc receptor [73], by interacting through its SPRY domain with the Fc region of antibody-opsonized non-enveloped viruses and bacteria [74], [75]. Although the exact molecular mechanism remains elusive, adenovirus infection in Trim21 −/− mice and MEFs demonstrated that TRIM21 is critical for the anti-microbial response against opsonized pathogens and subsequent pro-inflammatory and IFN-I cytokine induction. Together, these hallmark studies provided the first experimental data to the long-standing speculation that some TRIMs could directly act as receptors, bridging pathogen recognition to the induction of cell signaling. An important question remains whether this feature is exclusive to TRIM5 and TRIM21, or whether other PRY-SPRY domain containing TRIMs can also function as cytoplasmic receptors.
7.2. TRIMs regulating PRR activation
Even though TRIM5 and TRIM21 have been reported to directly act as receptors, a much larger number of TRIMs has been shown to regulate the activity of PRRs, especially in the context of viral infection. In actively infected cells, viral replication products are sensed by cytoplasmic sensors. RIG-I and the closely related MDA-5 are activated by 5′-triphosphate RNA (5′ppp-RNA) and longer double-strand RNA (dsRNA) molecules, respectively [76], [77], [78], [79], [80]. Both of these PAMPs are common products synthesized by viruses during infection.
RIG-I and MDA-5 contain each two CARD domains, which need to become exposed and homo-oligomerize for activation and assembly into a signaling complex with the CARD domains of the mitochondria-anchored adaptor protein MAVS [81], [82]. RNA-binding is insufficient for CARD oligomerization and activation of RIG-I and MDA-5 [83], [84]; RIG-I requires K63-linked poly-ubiquitin chains for oligomerization, activation, subsequent MAVS association and signal transduction [83]. Oligomerization of the CARD domains of MDA-5 was reported to be exclusively through filament formation on long dsRNA in structural studies [84], whereas in contrast biochemical approaches identified a ubiquitin-dependent oligomerization of MDA-5 similar to RIG-I [85].
TRIM25 was one of the first proteins to be recognized as a key regulator of RIG-I and has been the subject of intense investigation in recent years (Fig. 5a). Studies using MEFs from Trim25 −/− mice and reconstituted in vitro assays have shown that TRIM25 mediates K63-linked poly-ubiquitination required for RIG-I activation [15], [76]. However, controversy has remained whether these K63 ubiquitin chains are covalently anchored to RIG-I or not.
A recent crystal structure of RIG-I in complex with K63 poly-ubiquitin indicated that both chain types are feasible for activation [83], yet further study will be required to determine which is the most prominent activating oligomer in cells and mice in vivo. It should be noted that although the relevance of TRIM25 in RIG-I activation has been well established in cells, experiments in Trim25 −/− mice to address its relevance for immune signaling have not been performed to our knowledge. Hence, the main cell types in which TRIM25 plays a biologically relevant role during infections in vivo remains to be determined. This is particularly important since the main E3 ligase for RIG-I activation could be redundant or differ per cell type, the notion of which is underpinned by the fact that other E3 ligases – such as Riplet and TRIM4 – have been found to associate with RIG-I, synthesize K63 poly-ubiquitin and under experimental conditions mediate its activation [86], [87].
In addition to RNA recognition, dsDNA during virus infections can be sensed by cytoplasmic sensors such as AIM2 and DDX41 [88], [89]. DDX41 activation triggers IFNβ induction, whereas the dsDNA sensor AIM2 is dispensable for IFNβ induction and predominantly activates the inflammasome resulting in pro-inflammatory IL1β production [88]. In addition, DDX41 senses cyclic di-nucleotides during intra-cellular bacterial infections [90].
A recent study identified TRIM21 as a negative regulator of DDX41 activity (Fig. 5a) [91]. Using Trim21 −/− mice and their bone marrow-derived DCs, the authors demonstrated that TRIM21 ablation resulted in ∼1 log lower DNA virus titers during infection as the result of increased IFN-I and pro-inflammatory cytokine expression. Mechanistically, TRIM21 binds DDX41 through its PRY-SPRY domain thereby mediating K48 poly-ubiquitination of DDX41 and its subsequent degradation under resting conditions. K48 ubiquitination of DDX41 was almost completely abolished in Trim21 −/− bone marrow-derived DCs, indicating that it is the predominant E3 ligase for DDX41 degradation in these cells. Similar to DDX41, the cytoplasmic sensor for the PAMP muramyl dipeptide, NOD2, has also been reported to be negatively regulated by K48 ubiquitination and proteasomal degradation. In that case, TRIM27 is the responsible E3 enzyme which also mediates target recognition by its PRY-SPRY domain [92].
Together these studies show that TRIMs are important factors in both the positive and negative regulation of PRR activity. It remains to be seen if other TRIMs play redundant roles in this type of regulation. In addition, future studies will also need to address the mechanisms by which these TRIMs are themselves activated during PAMP recognition by PRRs.
7.3. TRIMs regulating immune adaptor molecules
After activation of cytoplasmic PRRs such as RIG-I and MDA5, various proteins are recruited into signaling complexes on the mitochondria-associated MAVS protein, which transduces the signal by mediating the activation of TBK1 and/or IKKɛ kinases. Subsequently, these kinases phosphorylate the transcription factors IRF3 and IRF7, which mediates their nuclear translocation and subsequent IFNβ transcription. A number of TRIMs has been reported to act at the level of these immune adaptor molecules.
TRIM14 is a sub-group IV, PRY-SPRY containing protein, which lacks a RING domain. This TRIM member has been reported as a positive regulator of the MAVS-induced IRF3- and NFκB-regulated pathways [44]. Co-localization studies indicated that TRIM14 increasingly co-localized with MAVS on mitochondria upon IFNβ induction by SeV infection. Expression studies using TRIM14 BBox point mutants identified three closely located residues in this domain essential for mitochondrial localization and IRF3-dependent reporter activation [44]. Additionally, the PRY-SPRY domain was found to be essential for MAVS interaction. While this RINGless TRIM was not reported to mediate E3 ligase activity, its own K63 poly-ubiquitination has been reported to be essential for NEMO recruitment and signal transduction.
Similar to TRIM14, sub-group V TRIM44 has been shown to interact with MAVS through its BBox, and enhance SeV-induced reporter activation in the HEK-293T cell line in over-expression and knockdown experiments [93]. Although the mechanism of positive regulation will need further investigation, the data indicated that TRIM44 may reduce K48 poly-ubiquitination of MAVS, thereby enhancing its stability.
In two TRIM-family-wide studies TRIM15 was identified as a positive regulator of the IRF3 and AP1-mediated pathways, but not NFκB [27], [28]. Although the molecular mechanism of signal transduction regulation remains to be determined, knockdown in primary mouse PBMC MΦ and stimulation in cell lines with various expression constructs, supported a positively regulatory role for TRIM15 at the level of the MAVS adapter requiring its PRY-SPRY domain [27]. Interestingly, TRIM15 knockdown specifically affected IRF3-mediated, but not NFκB-mediated transcription while MAVS signaling controls both [27], indicating that additional factors may be at play to skew cell signaling to specific pathways as has been suggested for other TRIMs [28].
The dsDNA sensor DDX41 also activates TBK1 and subsequent IFNβ induction through the ER-associated adaptor molecule STING[89]. Both TRIM32 and TRIM56 have been reported to positively regulate STING-mediated cell signaling [94], [95]. Despite these TRIMs belonging to two quite different sub-groups hallmarked by NHL domains in their C-terminus (TRIM32) or no recognized conserved C-terminal domains (TRIM56), overexpression and knockdown studies in cell lines indicated that both TRIMs enhance STING activation by K63 ubiquitination, albeit at different lysine residues [94], [95], indicating that different TRIMs may indeed play redundant regulatory roles by different molecular mechanisms.
TLRs rely on members of the TRAF family of E3 ubiquitin ligases to bridge receptor activation with down-stream cell signaling [96]. In the case of TRAF6, which is recruited by MyD88 to TLR4, K63 poly-ubiquitin synthesized by TRAF6 activates its down-stream kinase complex containing TAK1/TAB2/TAB3 [97]. TRIM38 was shown to interact with TRAF6 through its PRY-SPRY domain, thereby negatively regulating TRAF6 function by facilitating TRAF6 K48 poly-ubiquitination and degradation [98]. Consistent with this notion, knockdown in primary peritoneal MΦs increased pro-inflammatory cytokine expression, whereas this was reduced by over-expression. This indicates that TRIMs can also regulate the activity of other non-TRIM ubiquitin ligases in these signaling pathways. Importantly, the fact that TRIMs may form cellular complexes with other ubiquitin ligases could complicate the interpretation of some experiments, as it would be difficult to differentiate which E3-ligase is responsible for modifying the target protein.
7.4. TRIMs regulating the kinase layers
The kinase complex downstream of TRAF6 containing TAK1/TAB2/TAB3 in turn activates a hetero-trimeric kinase complex consisting of IKKα, IKKβ and NEMO (aka IKKγ), resulting in phosphorylation and subsequent ubiquitin-dependent degradation of IκBα, the inhibitor of NFκB. This releases NFκB, which translocates to the nucleus and enhances IFNβ expression, as well as pro-inflammatory cytokine production. Various steps of the NFκB and IRF activation pathways have been demonstrated to be heavily controlled by ubiquitin at the kinase level [8], [99].
As described above, in human cells TRIM5 has been identified as a major regulator of the TAB/TAK kinase complex by mediating K63 poly-ubiquitin chain synthesis [13]. However, the relevance of TRIM5-mediated signaling has never been addressed in vivo, with the main reason being that mice lack a TRIM5 gene. Mice do however express the highly homologous mouse-specific TRIM30α. Interestingly, studies using in vivo siRNA-mediated knockdown and transgenic mice overexpressing TRIM30 have demonstrated an opposite function for TRIM30 as a negative regulator of the TAB2/TAB3 complex compared to human TRIM5 [13], [100]. TRIM30 is predominantly expressed in mouse lymphoid tissues in e.g. MΦ and pDCs [26]. In bone-marrow DCs it is only detectable after stimulation of TLR-signaling [100]. Expression studies demonstrated that TRIM30 interacts with TAB2 and TAB3 and mediated their degradation in a ubiquitin-dependent manner by targeting the kinases for lysosomal degradation [100].
In addition to a role for TRIM30 in DCs, NFκB activation was attenuated in CD4 T cells of Trim30 −/− mice upon T cell receptor engagement, resulting in reduced expression of the pro-survival cytokine IL2 [101]. In contrast to what would be expected from reduced IL2 production, CD4 T cell proliferation was elevated in the knockout mice, suggesting that TRIM30 may regulate both cytokine expression and proliferation independently.
NAP1 is a TBK1-adaptor protein required for TBK1/IKKɛ kinase complex assembly and their ultimate phosphorylation of IRF3/7 [102]. TRIM38 has been shown to negatively regulate NAP1 levels in primary mouse peritoneal MΦs by mediating NAP1 K48 poly-ubiquitination and its subsequent proteasomal degradation [103]. In agreement with this, knockdown of TRIM38 in mice and peritoneal MΦs attenuated LPS-induced IFNβ expression and control of VSV infection [103]. A ubiquitin-dependent negative regulating function for TRIM38 was also described by another group, although the molecular target and means of inhibition differed from the one described above in peritoneal MΦs [104]. In contrast, over-expression and knockdown of TRIM38 in non-immune cell lines favored a model in which TRIM38 recruits TAB2 to lysosomes through its PRY-SPRY domain, and ubiquitinates it, thereby targeting it for lysosomal degradation [104]. Future studies using conditional knockout mice will need to be performed to address the cell-type specific relevance of TRIM38 in cytokine regulation and the molecular mechanism employed.
Our recent work indicates that TRIM6 is also a critical component of IFN production but not NFκB-dependent pro-inflammatory cytokines in primary human DCs upon TLR4 stimulation, as well as in the human epithelial cell line A549 upon influenza, EMCV and Sendai virus infection [14]. These data together with overexpression studies suggested that TRIM6 might act at the level of the IKKɛ/TBK1 kinases or the transcription factors IRF3/IRF7. Interestingly, knockdown of TRIM6 in vivo in mice and subsequent infection with influenza virus did not show differences in IFN production suggesting that TRIM6 may have different functions in human and mouse species or there might be tissue/cell type-specific differences in TRIM6 function [14].
Lastly, TRIM23 has been reported to ubiquitinate and thereby positively regulate NEMO [105]. In agreement with this, TRIM23 knockdown MEFs were about 10-fold more susceptible to VSV infection. What makes this particular TRIM worth mentioning is that it is the only group IX TRIM in the family and is characterized by a C-terminal ARF domain. It is this domain which was found to interact with the N-terminal coiled-coil 1 and C-terminal leucine-zipper in NEMO. A second interesting aspect is that the authors found that NEMO is ubiquitinated by TRIM23 on five lysine residues, exclusively by the non-canonical K27-linked poly-Ub chains [105], which is the only instance of TRIM-mediated K27 ubiquitination reported thus far.
Similar to the IFNβ induction pathway, IFN signaling follows a cell signaling structure with a central kinase activation level (Fig. 5b). Upon secretion from activated cells, IFNβ and IFNα bind and signal through the same IFNαR, resulting in the upregulation of several hundreds of IFN-stimulated genes (ISG). Upon binding of the IFN-I to the IFNαR, the receptor-associated kinases JAK1 and TYK2 phosphorylate the key transcription factors STAT1 at residue Y701 and STAT2 at Y690. Subsequently, STAT1 hetero-trimerizes with STAT2 and IRF9 into the ISGF3 complex. This complex translocates to the nucleus and drives ISG expression [106]. In parallel, the kinase IKKɛ is activated and phosphorylates a secondary residue, S708, on STAT1 [107].
In contrast to IFNβ, IFNγ engages its own receptor, which results in the activation of the IFNγR-associated JAK1 and JAK2 kinases, subsequent phosphorylation of STAT1 at Y701, STAT1 homo-dimerization and nuclear translocation. STAT1 homo-dimers associate with gamma-associated sequences (GAS) and drive the transcription of IFNγ-stimulated genes. IKKɛ-mediated STAT1 S708 phosphorylation is thought to prevent STAT1 homo-dimerization, but not STAT1-STAT2 hetero-dimerization, thereby skewing cell signaling toward ISRE-driven transcription by IFN-I [107], [108]. Moreover, S708 phosphorylation favors the transcription of a specific subset of antiviral ISGs, possibly by altering ISGF3 conformation and increasing the affinity for certain IFN-stimulated response elements (ISRE).
Recently, our labs demonstrated TRIM6 as a key regulator in IKKɛ activation and STAT1-mediated ISG induction using in vivo knock-down in mice, primary human DCs and in vitro assays [14]. Using these approaches combined with the deubiquitinating enzyme IsoT, which specifically degrades unanchored poly-ubiquitin chains by recognizing their exposed C-terminal motif, our work identified TRIM6 to activate IKKɛ by mediating the synthesis of unanchored K48-linked poly-Ub chains (Fig. 5b). Although covalently attached K48 poly-Ub had been exclusively reported as mediators for proteasomal degradation, these data indicated that their unanchored variety could have the opposite function and mediate kinase multimerization and activation. In addition, this study also provided evidence that unanchored poly-ubiquitin chains are formed in vivo and play an important physiological activating role in antiviral immunity. The question whether these findings are unique to TRIM6 and IKKɛ or are in fact relevant in other cell biological systems is of great interest and will need further future study.
7.5. TRIMs regulating the transcriptional layer
A general feature of cytokine induction pathways is the phosphorylation and subsequent translocation of transcription factors into the nucleus, which drive cytokine production. In the case of IFN-I induction, the TBK1/IKKɛ kinases phosphorylate the transcription factor IRF3 which translocates to the nucleus to drive IFNβ mRNA transcription. Moreover, the IKKα/β-NEMO kinase complex mediates the phosphorylation and subsequent degradation of IκBα, thereby releasing its inhibition of NFκB and allowing it to translocate to the nucleus and enhance IFNβ transcription.
In addition to the proposed role of TRIM21 as an Ig-receptor, several studies have implicated TRIM21 in cytokine induction. However, several groups have reported conflicting data, which has made it difficult to interpret what the relevance of TRIM21 for cytokine responses is in tissue culture and in vivo. Cell culture-based studies have identified TRIM21 as both a positive regulator of IRF3 and thus IFNβ induction by binding to PIN1 through its PRY-SPRY domain and thereby preventing subsequent IRF3 K48 poly-ubiquitination and degradation [109]. In contrast, other groups reported negative regulatory functions for TRIM21 by IRF3 and IRF7 K48 poly-ubiquitination and degradation in cell lines and mouse bone-marrow derived MΦs [110], [111].
To clarify this discrepancy and shed further light on possible differential regulation by TRIM21 during different stimuli, TRIM21 knockout mice were generated by two different groups, which yielded contradictory results. On the one hand, Trim21 −/− mice were reported to develop systemic auto-immune symptoms upon ear puncture with a highly inflammatory Th17 cytokine signature, which was reversed by crossing of these mice to an IL23/p19 deficient background [112]. On the other hand, other groups found that TRIM21 −/− mice did not have any signs of auto-inflammatory disease, even upon ear puncture [113], [114]. While Trim21 −/− MEFs derived from these mice did have an elevated pro-inflammatory cytokine signature, these responses were indistinguishable in wt and Trim21 −/− bone marrow-derived DCs and MΦs [114]. In line with this finding that perhaps TRIM21 only affects cytokine induction in certain cell types like fibroblasts, cytokine induction in the brains of adenovirus infected Trim21 −/− mice was similar to wt counterparts [75]. Although differences in animal housing and pathogen exposure could account for the phenotypic differences found in these studies, it has been suggested that the strategies used to make the knockout mice in these studies differed and some of the Th17-axis inflammatory phenotype could be due to the synthesis of a truncated TRIM21 form lacking its C-terminal half [112], [113].
7.6. TRIMs regulating inflammasomes
Innate myeloid cells also express components of an oligomeric complex known as the inflammasome [115]. While TLR engagement and subsequent NFκB activation mediates the synthesis of an unprocessed pro-IL1β cytokine, a secondary step controlled by the inflammasome is required for cleavage into active, pro-inflammatory IL1β. The composition of the inflammasome oligomer varies depending on the stimuli activating the cell, yet ultimately results in Caspase1 activation, and subsequent cleavage and activation of pro-IL1β. Mature IL1β in turn has pro-inflammatory effects on other cells including induction of other cytokines and promotes a form of anti-microbial response-associated programmed cell death known as pyroptosis [116].
The three best characterized inflammasome complexes include NLRP1 (recognizes bacterial muramyl dipeptide), NLRP3 (recognizes e.g. asbestos, alum, uric acid), and AIM2 with its adaptor ASC (recognizes dsDNA) components. So far, three TRIMs have been found to be involved in inflammasome regulation: TRIM16, TRIM20 and mouse-specific TRIM30α (Figure 5c). Their biological relevance is underpinned by the fact that mutations in the human TRIMs have been associated with auto-immune pathologies [7].
Elevated expression of TRIM30 in transgenic mice limited IL1β production and neutrophil influx upon mono-sodium-urate stimulation in a NLRP3 peritonitis model [117]. Similarly, overexpression of TRIM30 in bone-marrow MΦs, attenuated inflammasome activation. Mechanistically, this was dependent on reactive oxygen species (ROS) production, which in TRIM30 knockdown MΦs was linked to an increase in Caspase 1 activity and production of mature IL1β [117].
Human TRIM20 is a group IV TRIM without RING domain and is predominantly expressed by myeloid cells. Mutations in TRIM20 have been linked to familial Mediterranean fever (FMF), although it has for many years not been fully understood whether a TRIM20 loss-of-function or gain-of-function was responsible for this auto-immune disease. A comparative study by Chae and co-workers has directly addressed the question as to whether FMF in mice is caused by loss- or gain of TRIM20 function [118]. In their experiments, Trim20 −/− mice did not have any auto-inflammatory phenotype and were phenotypically normal, whereas knock-in mice carrying TRIM20 PRY-SPRY mutations found in human FMF patients developed spontaneous inflammatory disease, which cross-ins with Il1br −/− or Asc −/− mice revealed was completely ASC and IL1β dependent [118]. Hence, these findings suggest that FMF may be driven by a gain-of-TRIM20 function rather than a loss-of-TRIM20 function.
In contrast, another group who generated an independent true Trim20 null mouse, reported increased IL1β production in MΦs from these animals [119]. Although the discrepancy between these findings will need further investigation, the differences could be related to the different genotypes of the exon1-2 knockout [118] versus complete null [119], and the potential expression of truncated peptides. However, it is also important to note that in contrast to human TRIM20, the mouse TRIM20 protein lacks an intact PRY-SPRY domain in which mutations have been found in human FMF patients [120]. This could partially explain functional differences between human and mouse TRIM20, since the SPRY domain is important for the interaction of human TRIM20 with Caspase 1 [121].
Studies into the roles of the PYRIN and PRY-SPRY domains in TRIM20 indicated that both may contribute to the observed phenotypes in mice. The PRY-SPRY domain has been reported to negatively regulate the inflammasome and IL1β expression, whereas this is lost in PRY-SPRY point mutants found in FMF patients [121]. In contrast the PYRIN domain has been reported to be cleaved off by Caspase 1 and mediate NFκB activation [121]. In line with this notion, MΦs from mice only expressing an intact PYRIN domains of TRIM20 has increased LPS-induced Caspase1 activation and IL1β production, yet reduced apoptosis [122]. Together, these studies highlight the complexity of TRIM function, which varies depending of the species, the cell types and the transcript variants/isoforms expressed.
8. Concluding remarks
Taken together, the studies discussed in this review add to the growing body of work supporting the notion that many TRIM family members are critical immune regulatory proteins [27], [28]. However, what these studies also emphasize is that the manner in which different TRIMs regulate the immune response is complex. The same TRIMs may have different, multiple or even opposite functions in different signaling pathways and some functions may be species-specific and cell type-specific. Although some studies have focused on the biological relevance of individual TRIM proteins in these primary cell types, the majority of studies have been in cell lines and/or non-immune cells. Hence, an important future effort will lay in addressing the relevance and molecular mechanism of these TRIMs in the regulation of immune signaling in primary cells and in vivo. The identification and study of biological relevance of individual TRIMs in the correct cell type is likely to be especially important considering the restricted expression of many (Fig. 4). Even in mouse studies using different Trim21 knockout alleles, discrepancies in inflammatory phenotypes have been reported, which stresses the importance of generating mice with null mutations or carefully analyzing the expression of the protein expressed from the targeted gene using antibodies which could recognize truncated peptides expressed from exons not targeted in the mouse.
For about a third of all TRIMs mRNA splice variants have been identified which encode proteins lacking at least one of the conserved domains [28]. This complicating factor in addressing the biological roles of individual TRIMs has – with the exception of TRIM19 – remained practically unstudied. At least seven TRIM19 isoforms have been recognized, which all have reported different functions [123]. Investigation of the individual isoforms revealed that only isoforms 4 of TRIM19 (TRIM19-IV) can enhance IRF3 stability and signaling by targeting its negative regulator PIN1 for proteasomal degradation [124].
Another complicating factor in the future generation of TRIM knockout animals may be the fact that many TRIMs have been reported to have functions in cell differentiation, apoptosis and many TRIMs has been linked to development and progression of many cancers [125]. Although many of the cancer-associated TRIMs have been shown to modulate specific cell cycle regulators, it is interesting to note that NFκB-mediated cell regulation and the ubiquitin-proteasome system are two of the three central molecular systems involved in the formation and progression of cancer [126], [127]. Since TRIMs as immune regulatory E3 ligases have been heavily associated with both of these systems, it could be possible that for some cancer-associated TRIMs there is a link to their immune regulatory functions. Furthermore, it emphasizes that their physiological role may only be addressed by generating conditional knockout mice or selective gene targeting approaches to study their roles in vivo.
To address this issue TRIM knockout mice or transgenic mice harboring point mutations in TRIMs could be generated by specifically abolishing the interaction with identified immune regulatory molecules, while not affecting the interaction with other molecules. This will require mapping in specific cells of all major TRIM interaction partners, combined with the identification of key residues essential for TRIM interaction with its binding partners, and the generation of mice where specifically the interaction with the immune molecule of interest is disrupted. Especially in combination with conditional/cell type-specific targeting, such an approach could be very powerful to delineate the biological role of TRIMs for the immune response in vivo with much more resolution and less interference from potentially affecting more than one role of a multi-functional protein.
Furthermore, cell biological dynamics of the recruitment of TRIMs, their targets, ubiquitin, and deubiquitinating enzymes remains poorly understood. The recent introduction of CRISPR/CAS9-based genome engineering has made the generation of endogenous TRIM genes with fluorescent tags or photoactivatable tags much more feasible. In combination with advanced live imaging techniques these approaches will enable studying the regulation in space and time of TRIM proteins under endogenous setting without the risk of artifacts arising from overexpression or the use of artificial cell types.
Taken together, the last decade has brought a wealth of data on immune interaction partners of TRIMs and how these interactors may be controlled by TRIM proteins in ubiquitin-dependent and independent ways. However, unraveling the dynamics of these interactions during immune activation, their roles in different cell types and protein complexes will be one of the challenges for the years to come, which will require bringing together experimental efforts from in vitro, cell culture and in vivo systems.
Biographies

Gijs A. Versteeg is currently a group leader at the Max F. Perutz Laboratories in Vienna, Austria. Gijs has a long-standing interest in understanding how innate immune cell signaling is regulated by the post-translational modifier ubiquitin, and how viruses antagonize these immune responses. He received his Ph.D. from Leiden University studying how coronaviruses regulate interferon induction and performed his post-doctoral training in the laboratory of Adolfo García-Sastre at Icahn School of Medicine at Mount Sinai in New York. Here Dr. Versteeg investigated how tri-partite motif (TRIM) proteins regulate the innate immune system.

Stefan Benke received his MSc from the University of Graz (Austria) where he studied the role of Ca2+ signaling in Parkinson's disease. His scientific interest is focused on disease-related cell signaling. Currently, Stefan is a graduate student at the Max F. Perutz Laboratories of the University of Vienna (Austria) under the supervision of Gijs Versteeg. His present research aims to understand how the Tripartite Motif (TRIM) family of E3 ubiquitin ligases regulates inflammation and the innate immune response.

Adolfo García-Sastre is a professor in the Department of Microbiology and Department of Medicine, and Director of the Global Health & Emerging Pathogens Institute at Icahn School of Medicine at Mount Sinai in New York. He is also Principal Investigator for the Center for Research on Influenza Pathogenesis (CRIP), one of five NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS). For the past 25 years, Adolfo García-Sastre's research interest has been focused on the molecular biology negative strand RNA viruses and the means by which these viruses antagonize the type I interferon system.

Ricardo Rajsbaum is an assistant professor in the Department of Microbiology and Immunology at the University of Texas Medical Branch, Galveston, Texas. Dr. Rajsbaum performed his Ph.D. studies in the laboratory of Anne O’Garra at the MRC National Institute for Medical Research, London, UK, and completed his postdoctoral training at Mount Sinai School of Medicine, New York in the laboratory of Adolfo Garcia-Sastre. Dr. Rajsbaum's interests include regulation of cytokine expression in immune cells, TLR and RIG-I-like receptor signaling, regulation and function of type-I IFNs, and the study of virus–host interactions, with a specific focus on the role of ubiquitin and TRIM proteins in innate antiviral function.
References
- 1.Latz E., Xiao T.S., Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.González-Navajas J.M., Lee J., David M., Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Versteeg G.A., Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganguly D., Haak S., Sisirak V., Reizis B. The role of dendritic cells in autoimmunity. Nat Rev Immunol. 2013;13:566–577. doi: 10.1038/nri3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jefferies C., Wynne C., Higgs R. Antiviral TRIMs: friend or foe in autoimmune and autoinflammatory disease. Nat Rev Immunol. 2011;11:617–625. doi: 10.1038/nri3043. [DOI] [PubMed] [Google Scholar]
- 8.Liu S., Chen Z.J. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husnjak K., Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Ann Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 10.Metzger M.B., Hristova V.A., Weissman A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicke L., Schubert H.L., Hill C.P. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 12.Rajsbaum R., Garcia-Sastre A. Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol. 2013;21:421–429. doi: 10.1016/j.tim.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertel T., Hausmann S., Morger D., Zuger S., Guerra J., Lascano J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajsbaum R., Versteeg G.A., Schmid S., Maestre A.M., Belicha-Villanueva A., Martinez-Romero C. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKK epsilon kinase-mediated antiviral response. Immunity. 2014;40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou W., Zhang D.E. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 17.Amir R.E., Iwai K., Ciechanover A. The NEDD8 pathway is essential for SCFβ-TrCP-mediated ubiquitination and processing of the NF-κB precursor p105. J Biol Chem. 2002;277:23253–23259. doi: 10.1074/jbc.M200967200. [DOI] [PubMed] [Google Scholar]
- 18.Arimoto K., Konishi H., Shimotohno K. UbcH8 regulates ubiquitin and ISG15 conjugation to RIG-I. Mol Immunol. 2008;45:1078–1084. doi: 10.1016/j.molimm.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Begitt A., Droescher M., Knobeloch K.P., Vinkemeier U. SUMO conjugation of STAT1 protects cells from hyperresponsiveness to IFNgamma. Blood. 2011;118:1002–1007. doi: 10.1182/blood-2011-04-347930. [DOI] [PubMed] [Google Scholar]
- 20.Kim M.J., Hwang S.Y., Imaizumi T., Yoo J.Y. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regad T., Chelbi-Alix M.K. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 22.Vatsyayan J., Qing G., Xiao G., Hu J. SUMO1 modification of NF-kappaB2/p100 is essential for stimuli-induced p100 phosphorylation and processing. EMBO Rep. 2008;9:885–890. doi: 10.1038/embor.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones J., Wu K., Yang Y., Guerrero C., Nillegoda N., Pan Z.Q. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srikumar T., Lewicki M.C., Costanzo M., Tkach J.M., van Bakel H., Tsui K. Global analysis of SUMO chain function reveals multiple roles in chromatin regulation. J Cell Biol. 2013;201:145–163. doi: 10.1083/jcb.201210019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xirodimas D.P., Sundqvist A., Nakamura A., Shen L., Botting C., Hay R.T. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajsbaum R., Stoye J.P., O’Garra A. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol. 2008;38:619–630. doi: 10.1002/eji.200737916. [DOI] [PubMed] [Google Scholar]
- 27.Uchil P.D., Hinz A., Siegel S., Coenen-Stass A., Pertel T., Luban J. TRIM protein mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol. 2013;87:257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Versteeg G.A., Rajsbaum R., Sanchez-Aparicio M.T., Maestre A.M., Valdiviezo J., Shi M. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carthagena L., Bergamaschi A., Luna J.M., David A., Uchil P.D., Margottin-Goguet F. Human TRIM gene expression in response to interferons. PLoS ONE. 2009;4:e4894. doi: 10.1371/journal.pone.0004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demuth J.P., De Bie T., Stajich J.E., Cristianini N., Hahn M.W. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukic Z., Hausmann S., Sebastian S., Rucci J., Sastri J., Robia S.L. TRIM5alpha associates with proteasomal subunits in cells while in complex with HIV-1 virions. Retrovirology. 2011;8:93. doi: 10.1186/1742-4690-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roa A., Hayashi F., Yang Y., Lienlaf M., Zhou J., Shi J. RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J Virol. 2012;86:1717–1727. doi: 10.1128/JVI.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kutluay S.B., Perez-Caballero D., Bieniasz P.D. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLoS Pathog. 2013;9:e1003214. doi: 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yap M.W., Nisole S., Lynch C., Stoye J.P. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci U S A. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayah D.M., Sokolskaja E., Berthoux L., Luban J., Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 37.Wilson S.J., Webb B.L., Ylinen L.M., Verschoor E., Heeney J.L., Towers G.J. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci U S A. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nisole S., Stoye J.P., Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 39.Rajsbaum R., Garcia-Sastre A., Versteeg G.A. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massiah M.A., Matts J.A., Short K.M., Simmons B.N., Singireddy S., Yi Z. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J Mol Biol. 2007;369:1–10. doi: 10.1016/j.jmb.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Short K.M., Cox T.C. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 2006;281:8970–8980. doi: 10.1074/jbc.M512755200. [DOI] [PubMed] [Google Scholar]
- 42.Bell J.L., Malyukova A., Holien J.K., Koach J., Parker M.W., Kavallaris M. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS ONE. 2012;7:e37470. doi: 10.1371/journal.pone.0037470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J.W., Fernandes-Alnemri T., Datta P., Wu J., Juliana C., Solorzano L. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol Cell. 2007;28:214–227. doi: 10.1016/j.molcel.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Z., Jia X., Xue Q., Dou Z., Ma Y., Zhao Z. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I–like receptor-mediated innate immune response. Proc Natl Acad Sci U S A. 2014;111:E245–E254. doi: 10.1073/pnas.1316941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cainarca S., Messali S., Ballabio A., Meroni G. Functional characterization of the Opitz syndrome gene product (midin): evidence for homodimerization and association with microtubules throughout the cell cycle. Hum Mol Genet. 1999;8:1387–1396. doi: 10.1093/hmg/8.8.1387. [DOI] [PubMed] [Google Scholar]
- 47.Cao T., Borden K.L., Freemont P.S., Etkin L.D. Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J Cell Sci. 1997;110(Pt 14):1563–1571. doi: 10.1242/jcs.110.14.1563. [DOI] [PubMed] [Google Scholar]
- 48.Napolitano L.M., Meroni G. TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- 49.Yap M.W., Nisole S., Stoye J.P. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 50.Meroni G. Genomics and evolution of the TRIM gene family. Adv Exp Med Biol. 2012;770:1–9. doi: 10.1007/978-1-4614-5398-7_1. [DOI] [PubMed] [Google Scholar]
- 51.Boudinot P., van der Aa L.M., Jouneau L., Du Pasquier L., Pontarotti P., Briolat V. Origin and evolution of TRIM proteins: new insights from the complete TRIM repertoire of zebrafish and pufferfish. PLoS ONE. 2011;6:e22022. doi: 10.1371/journal.pone.0022022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Aa L.M., Levraud J.P., Yahmi M., Lauret E., Briolat V., Herbomel P. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009;7:7. doi: 10.1186/1741-7007-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sardiello M., Cairo S., Fontanella B., Ballabio A., Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flajnik M.F., Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line. Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Nehyba J., Hrdlickova R., Bose H.R. Dynamic evolution of immune system regulators: the history of the interferon regulatory factor family. Mol Biol Evol. 2009;26:2539–2550. doi: 10.1093/molbev/msp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu L., Yang L., Liu W. Distinct evolution process among type I interferon in mammals. Protein Cell. 2013;4:383–392. doi: 10.1007/s13238-013-3021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langevin C., Aleksejeva E., Passoni G., Palha N., Levraud J-P., Boudinot P. The antiviral innate immune response in fish: evolution and conservation of the IFN system. J Mol Biol. 2013;425:4904–4920. doi: 10.1016/j.jmb.2013.09.033. [DOI] [PubMed] [Google Scholar]
- 58.Hickford D., Frankenberg S., Shaw G., Renfree M.B. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genom. 2012;13:155. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar S., Mitnik C., Valente G., Floyd-Smith G. Expansion and molecular evolution of the interferon-induced 2′–5′ oligoadenylate synthetase gene family. Mol Biol Evol. 2000;17:738–750. doi: 10.1093/oxfordjournals.molbev.a026352. [DOI] [PubMed] [Google Scholar]
- 60.Reboul J., Gardiner K., Monneron D., Uze G., Lutfalla G. Comparative genomic analysis of the interferon/interleukin-10 receptor gene cluster. Genome Res. 1999;9:242–250. [PMC free article] [PubMed] [Google Scholar]
- 61.Manry J., Laval G., Patin E., Fornarino S., Itan Y., Fumagalli M. Evolutionary genetic dissection of human interferons. J Exp Med. 2011;208:2747–2759. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cai X., Srivastava S., Sun Y., Li Z., Wu H., Zuvela-Jelaska L. Tripartite motif containing protein 27 negatively regulates CD4 T cells by ubiquitinating and inhibiting the class II PI3K-C2beta. Proc Natl Acad Sci U S A. 2011;108:20072–20077. doi: 10.1073/pnas.1111233109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chikuma S., Suita N., Okazaki I.M., Shibayama S., Honjo T. TRIM28 prevents autoinflammatory T cell development in vivo. Nat Immunol. 2012;13:596–603. doi: 10.1038/ni.2293. [DOI] [PubMed] [Google Scholar]
- 64.Rajsbaum R., Albrecht R.A., Wang M.K., Maharaj N.P., Versteeg G.A., Nistal-Villan E. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han K., Lou D.I., Sawyer S.L. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011;7:e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ichimura T., Taoka M., Shoji I., Kato H., Sato T., Hatakeyama S. 14-3-3 proteins sequester a pool of soluble TRIM32 ubiquitin ligase to repress autoubiquitylation and cytoplasmic body formation. J Cell Sci. 2013;126:2014–2026. doi: 10.1242/jcs.122069. [DOI] [PubMed] [Google Scholar]
- 67.Fensterl V., Wetzel J.L., Ramachandran S., Ogino T., Stohlman S.A., Bergmann C.C. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mancuso G., Midiri A., Biondo C., Beninati C., Zummo S., Galbo R. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- 69.Robinson N., McComb S., Mulligan R., Dudani R., Krishnan L., Sad S. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol. 2012;13:954–962. doi: 10.1038/ni.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li M., Liu X., Zhou Y., Su S.B. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol. 2009;86:23–32. doi: 10.1189/jlb.1208761. [DOI] [PubMed] [Google Scholar]
- 71.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 72.Kuhns D.B., Alvord W.G., Heller T., Feld J.J., Pike K.M., Marciano B.E. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med. 2010;363:2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keeble A.H., Khan Z., Forster A., James L.C. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci U S A. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McEwan W.A., Tam J.C., Watkinson R.E., Bidgood S.R., Mallery D.L., James L.C. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14:327–336. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaysburd M., Watkinson R.E., Cooper H., Reed M., O’Connell K., Smith J. Intracellular antibody receptor TRIM21 prevents fatal viral infection. Proc Natl Acad Sci U S A. 2013;110:12397–12401. doi: 10.1073/pnas.1301918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 77.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 78.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 80.Runge S., Sparrer K.M.J., Lässig C., Hembach K., Baum A., García-Sastre A. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., Chen Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 83.Peisley A., Wu B., Xu H., Chen Z.J., Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 85.Jiang X., Kinch L.N., Brautigam C.A., Chen X., Du F., Grishin N.V. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oshiumi H., Miyashita M., Inoue N., Okabe M., Matsumoto M., Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8:496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 87.Yan J., Li Q., Mao A.P., Hu M.M., Shu H.B. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J Mol Cell Biol. 2014;6:154–163. doi: 10.1093/jmcb/mju005. [DOI] [PubMed] [Google Scholar]
- 88.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y-J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parvatiyar K., Zhang Z., Teles R.M., Ouyang S., Jiang Y., Iyer S.S. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Z., Bao M., Lu N., Weng L., Yuan B., Liu Y.J. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14:172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zurek B., Schoultz I., Neerincx A., Napolitano L.M., Birkner K., Bennek E. TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PLoS ONE. 2012;7:e41255. doi: 10.1371/journal.pone.0041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang B., Wang J., Wang Y., Zhou H., Wu X., Tian Z. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol. 2013;190:3613–3619. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 94.Zhang J., Hu M.M., Wang Y.Y., Shu H.B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 96.Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8:7. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kanayama A., Seth R.B., Sun L., Ea C-K., Hong M., Shaito A. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 98.Zhao W., Wang L., Zhang M., Yuan C., Gao C. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J Immunol. 2012;188:2567–2574. doi: 10.4049/jimmunol.1103255. [DOI] [PubMed] [Google Scholar]
- 99.Liu S., Chen J., Cai X., Wu J., Chen X., Wu Y.T. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shi M., Deng W., Bi E., Mao K., Ji Y., Lin G. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 101.Choi U.Y., Hur J.Y., Lee M.S., Zhang Q., Choi W.Y., Kim L.K. Tripartite motif-containing protein 30 modulates TCR-activated proliferation and effector functions in CD4+ T cells. PLoS ONE. 2014;9:e95805. doi: 10.1371/journal.pone.0095805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chau T.-L., Gioia R., Gatot J.-S., Patrascu F., Carpentier I., Chapelle J.-P. Are the IKKs and IKK-related kinases TBK1 and IKK-ɛ similarly activated? Trends Biochem Sci. 2008;33:171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 103.Zhao W., Wang L., Zhang M., Wang P., Yuan C., Qi J. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-beta production and antiviral response by targeting NAP1. J Immunol. 2012;188:5311–5318. doi: 10.4049/jimmunol.1103506. [DOI] [PubMed] [Google Scholar]
- 104.Hu M.M., Yang Q., Zhang J., Liu S.M., Zhang Y., Lin H. TRIM38 inhibits TNFalpha- and IL-1beta-triggered NF-kappaB activation by mediating lysosome-dependent degradation of TAB2/3. Proc Natl Acad Sci U S A. 2014;111:1509–1514. doi: 10.1073/pnas.1318227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arimoto K., Funami K., Saeki Y., Tanaka K., Okawa K., Takeuchi O. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci U S A. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 107.Tenoever B.R., Ng S.L., Chua M.A., McWhirter S.M., Garcia-Sastre A., Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 108.Ng S.L., Friedman B.A., Schmid S., Gertz J., Myers R.M., Tenoever B.R. IkappaB kinase epsilon (IKK(epsilon)) regulates the balance between type I and type II interferon responses. Proc Natl Acad Sci U S A. 2011;108:21170–21175. doi: 10.1073/pnas.1119137109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang K., Shi H.X., Liu X.Y., Shan Y.F., Wei B., Chen S. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol. 2009;182:3782–3792. doi: 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- 110.Higgs R., Gabhann J.N., Larbi N.B., Breen E.P., Fitzgerald K.A., Jefferies C.A. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Higgs R., Lazzari E., Wynne C., Ni Gabhann J., Espinosa A., Wahren-Herlenius M. Self protection from anti-viral responses – Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral Toll-Like receptors. PLoS ONE. 2010;5:e11776. doi: 10.1371/journal.pone.0011776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F.J. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23–Th17 pathway. J Exp Med. 2009;206:1661–1671. doi: 10.1084/jem.20090585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ozato K., Yoshimi R., Chang T-H., Wang H., Atsumi T., Morse H.C. Comment on gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-κB-dependent cytokine expression in fibroblasts. J Immunol. 2009;183:7619. doi: 10.4049/jimmunol.0990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yoshimi R., Chang T.H., Wang H., Atsumi T., Morse H.C., 3rd, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182:7527–7538. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lamkanfi M., Dixit V.M. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 116.Latz E. The inflammasomes: mechanisms of activation and function. Curr Opin Immunol. 2010;22:28–33. doi: 10.1016/j.coi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hu Y., Mao K., Zeng Y., Chen S., Tao Z., Yang C. Tripartite-motif protein 30 negatively regulates NLRP3 inflammasome activation by modulating reactive oxygen species production. J Immunol. 2010;185:7699–7705. doi: 10.4049/jimmunol.1001099. [DOI] [PubMed] [Google Scholar]
- 118.Chae J.J., Cho Y.H., Lee G.S., Cheng J., Liu P.P., Feigenbaum L. Gain-of-function pyrin mutations induce NLRP3 protein-independent interleukin-1beta activation and severe autoinflammation in mice. Immunity. 2011;34:755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hesker P.R., Nguyen M., Kovarova M., Ting J.P.Y., Koller B.H. Genetic loss of murine pyrin, the familial Mediterranean fever protein, increases interleukin-1β levels. PLoS ONE. 2012;7:e51105. doi: 10.1371/journal.pone.0051105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chae J.J., Centola M., Aksentijevich I., Dutra A., Tran M., Wood G. Isolation, genomic organization, and expression analysis of the mouse and rat homologs of MEFV, the gene for familial Mediterranean fever. Mamm Genome. 2000;11:428–435. doi: 10.1007/s003350010082. [DOI] [PubMed] [Google Scholar]
- 121.Chae J.J., Wood G., Masters S.L., Richard K., Park G., Smith B.J. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc Natl Acad Sci U S A. 2006;103:9982–9987. doi: 10.1073/pnas.0602081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chae J.J., Komarow H.D., Cheng J., Wood G., Raben N., Liu P.P. Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 123.Maarifi G., Chelbi-Alix M.K., Nisole S. PML control of cytokine signaling. Cytokine Growth Factor Rev. 2014;25:551–561. doi: 10.1016/j.cytogfr.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 124.El Asmi F., Maroui M.A., Dutrieux J., Blondel D., Nisole S., Chelbi-Alix M.K. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 2014;10:e1003975. doi: 10.1371/journal.ppat.1003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 126.Ben-Neriah Y., Karin M. Inflammation meets cancer, with NF-[kappa]B as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 127.Kumar M.S., Hancock D.C., Molina-Arcas M., Steckel M., East P., Diefenbacher M. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.