Abstract
The combination of cryo-electron microscopy to study large biological assemblies at low resolution with crystallography to determine near atomic structures of assembly fragments is quickly expanding the horizon of structural biology. This technique can be used to advantage in the study of large structures that cannot be crystallized, to follow dynamic processes, and to “purify” samples by visual selection of particles. Factors affecting the quality of cryo-electron microscopy maps and limits of accuracy in fitting known structural fragments are discussed.
Introduction
Three-dimensional structure determinations based on cryo-electron microscopy (cryo-EM) have become a standard tool of structural biology in recent years. Just as in the practice of crystallography (Rodgers, 2001), the technique of freezing samples in vitrious ice for EM analysis (Baker et al., 1999, Dubochet et al., 1988) has made it possible to obtain two-dimensional projected images with minimal distortion or artifacts. Provided there is a plentiful supply of near identical frozen particles in random orientations, these projections can be combined to form three-dimensional images. The resolution of these images has been improving rapidly, largely because of improvements in reconstruction techniques (Tao and Zhang, 2000, van Heel et al., 2000). In particular, efforts have centered on the accurate determination of the contrast function that corrects the two-dimensional images for the experimental out-of-focus distance, the accurate determination of the relative orientation of the projected images, and the use of a far greater number of particles. As a result, it is now quite usual for cryo-EM image reconstructions to have an estimated resolution of 10 Å, and sometimes even as good as 7 Å, with expectations of reconstructions going to 4 Å (Henderson, 2004, van Heel et al., 2000) soon. These improvements have made it possible to accurately fit atomic resolution crystal structures of molecular fragments into the lower resolution EM density to produce “pseudo” atomic structures of the complex (Figure 1 ). Although the fitting procedure is frequently done “by hand” using visual graphics programs, such as O (Kleywegt et al., 2001), there are considerable advantages in performing these operations by computer (Jiang et al., 2001, Roseman, 2000, Rossmann et al., 2001, Volkmann and Hanein, 2003, Wriggers and Chacón, 2001, Wriggers et al., 1999) in order to establish the uniqueness and quality of the fit (especially at resolutions lower than about 15 Å) and to determine whether the crystal structure can be treated as a rigid model or should be modified by permitting some bending or hinge motions.
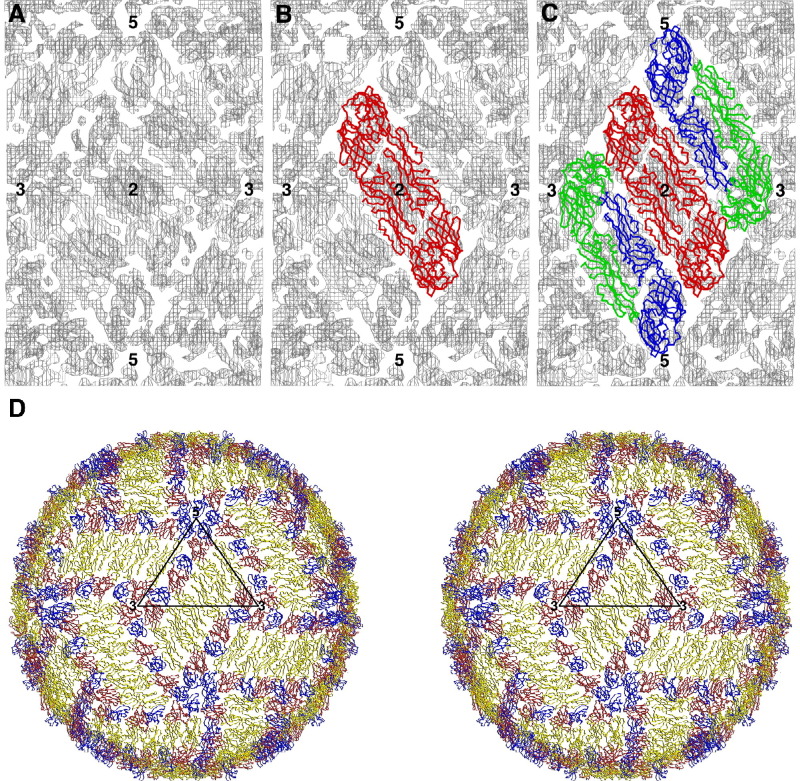
Figure 1.
Fitting the Icosahedrally Averaged, Cryo-EM Density of Dengue Virus at 9 Å Resolution with the Crystal Structure of the Envelope (E) Ectodomain Protein Dimer Cα Backbone
(A) The cryo-EM density (Kuhn et al., 2002; Zhang et al., 2003) and the position of icosahedral 5-fold, 3-fold, and 2-fold symmetry axes surrounding one asymmetric unit. (B) One E dimer (Modis et al., 2004; Rey et al., 1995; Zhang et al., 2004) fitted with its 2-fold axis coincident with an icosahedral 2-fold axis. (C) A second E dimer fitted into the remaining density. (D) The icosahedral symmetry has been used to generate the whole of the top surface of the virus shown as a stereo diagram. Each E monomer is colored red (domain I), yellow (domain II), and blue (domain III).
In discussing the problems and advantages of combining structural techniques, we will take examples primarily from our own experience, although other examples now abound. Furthermore, we will concentrate especially, but not exclusively, on our studies of dengue virus (Kuhn et al., 2002, Zhang et al., 2003) and the tailed bacteriophages ϕ29 (Tao et al., 1998), and T4 (Kostyuchenko et al., 2003, Leiman et al., 2004).
When Is Cryo-EM Essential?
Crystals of macromolecules are limited to those cases where significantly strong contacts can be made between molecules that create the infinite lattice. The larger and structurally more complicated the molecule or molecular assembly, the less likely it is that the molecule can be packed into a crystal, such that the surface area of contact between molecules is large compared with the total area of the molecular assembly itself. An obvious example is bacteriophage T4 (Figure 2 ). This phage has a dsDNA genome consisting of about 168 kbp, representing about 300 open reading frames of which about 40 code for structural proteins, many of which occur in multiple copies in the virion. Crystallization can also be difficult or impossible when the structure of interest has components that are flexible, implying that each structure has specific regions on its surface that vary from molecule to molecule. This might be caused by a hinge between domains (e.g., the fibers of T4; Figure 2) or a surface carbohydrate moiety (Figure 3 ).
Figure 2.
Diagrammatic Representation of Bacteriophage T4
The dsDNA genome is protected by the head capsid. The head is attached to the tail, a highly specialized and extremely efficient phage component required for infecting the E. coli host. The hexagonally shaped baseplate is situated at the distal end of the tail. The baseplate coordinates the movement of the six long tail fibers that initially sense the presence of the host, the short tail fibers that unfold from underneath the baseplate to firmly anchor on the E. coli surface, and the tail sheath surrounding the tail tube that contracts, thereby ejecting DNA into the host. The numbers identify the gene products of the various proteins that are in the assembled virion. It would be difficult to place this complex virus into a well-packed crystal lattice both because of its shape and because of the variably angled fibers (Eiserling and Black, 1994; Leiman et al., 2003).
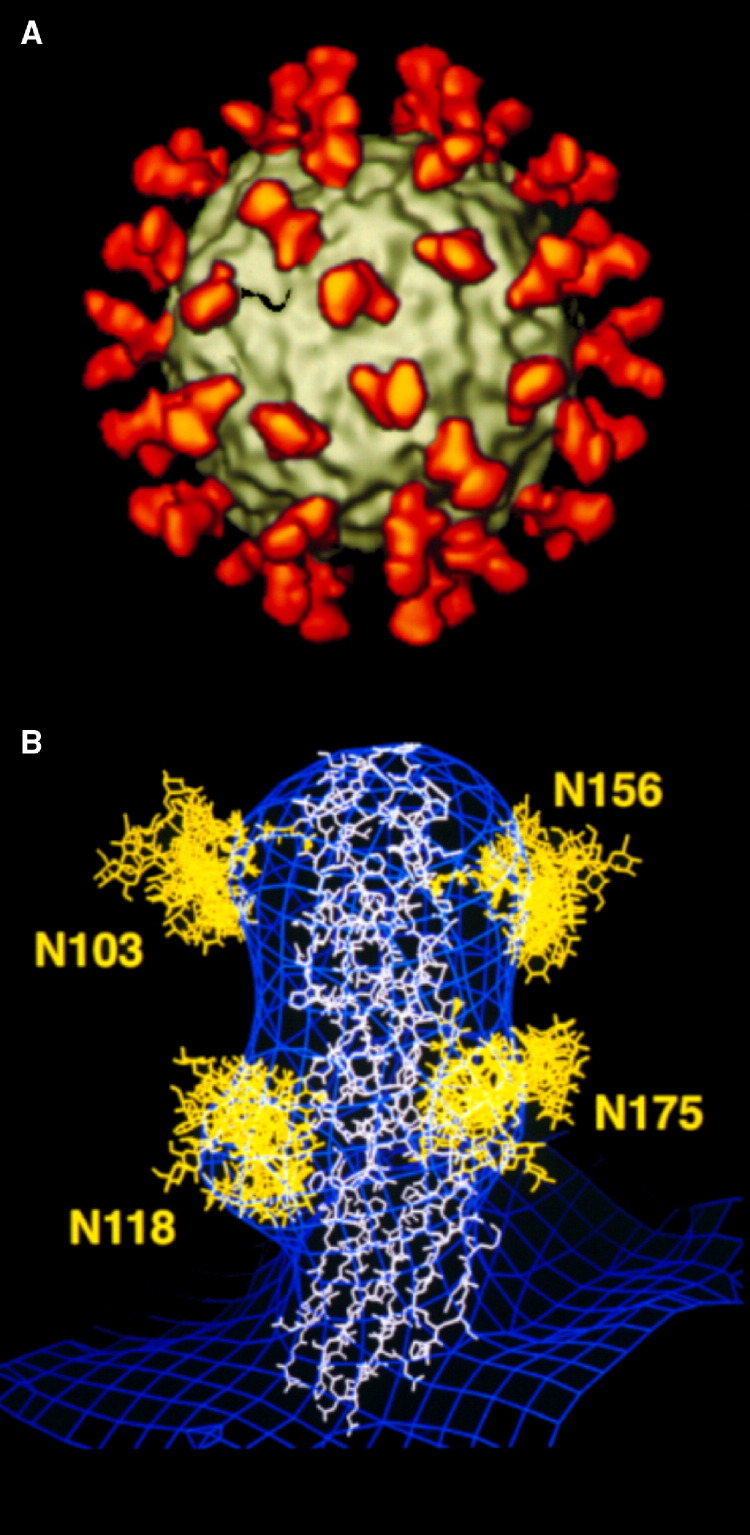
Figure 3.
The Structure of Human Rhinovirus 14 Complexed with Its Cellular Receptor Molecule, Intercellular Adhesion Molecule 1, at 26 Å Resolution
(A) The icosahedral virus (gray) complexed with the two amino-terminal domains of intercellular adhesion molecule 1 (ICAM1) (red) at all icosahedrally equivalent positions. (B) The cryo-EM density of the complex (blue), fitted with the X-ray crystal structure of the two amino-terminal domains decorated with multiple conformations of the carbohydrates at the four potential N-glycosylation sites. The Asn residues at positions 103, 118, and 156, but not 175, had to be mutated in order to form viable crystals. However, note the bulges of the blue density indicating the presence of the carbohydrate moieties in the wild-type ICAM1 used to form the complex. (Modified and reprinted by permission from Kolatkar et al., 1999, Macmillan Publishers Ltd.)
Another form of flexibility that can inhibit crystallization is the presence of lipid membranes, as occurs in many viruses. However, cryo-EM has made it possible to visualize not only the virus structure as a whole, but also the membrane proteins in situ (Figure 4 ), as opposed to the usually artificial hydrophobic environments used for crystallizing membrane proteins. Amino acid sequence information and model building have produced a reasonable structure showing how the transmembrane helices interact with their surrounding lipid (Zhang et al., 2002).
Figure 4.
The Membrane Structure of Dengue Virus
(A) A central cross-section through the cryo-EM density at 9.5 Å resolution showing the E glycoprotein ectodomain, the lipid bilayer, and the internal nucleocapsid. (B) Radial density section at a radius of 185 Å, showing higher density blacker than lower density, with the superimposed envelopes of the fitted E ectodomain. Note the four blacker regions associated similarly with each monomer corresponding to four transmembrane helices per monomer. (C) Diagrammatic side view of the E protein (domains I, II, and III). Domain III connects with the EH1 and EH2 helices of the stem region in the outer lipid leaflet, and ET1 and ET2 antiparallel transmembrane helices. Also shown are the two antiparallel transmembrane helices MH1 and MH2 of the membrane protein. (Reprinted with permission from Zhang et al., 2003, Nature Publishing Group.)
Crystallization also fails in the study of labile complexes that would degrade or come apart in long crystallization processes. An example is the complex of a virus with its receptor and accessory receptors. A virus must not only recognize its cellular receptor, but the recognition event has to initiate virus entry and possibly virus uncoating. For instance, the complex of human rhinovirus 14 with ICAM1 (Figure 3; Kolatkar et al., 1999) was formed by incubating the virus with the two-domain receptor for a few hours and then freezing within a few minutes before the virus degraded, but long enough to permit complex formation. The degradation could be slowed by cooling to 4°C, but certainly not for long enough to allow crystallization.
Crystallization requires significant amount of sample to search for conditions that produce well-diffracting crystals. For cryo-EM, it is only necessary to have enough sample to collect sufficient data to produce a reconstruction which might amount to 105 particles or so to attain resolutions of better than 10 Å, perhaps. In comparison, a minimally sized crystal with dimensions of about 200 μm in each direction would contain about 1010 particles of 1,000 Å diameter. A further advantage of cryo-EM is that sample purity is not as critical as is required for crystallization, as images of the molecular assemblies being studied can be selected from the micrographs even when mixed with other molecules. A number of examples come to mind. With present techniques, purified flaviviruses, the less stable types such as dengue virus (Figure 5 ) and especially yellow fever virus, are often mixtures of good and broken particles that would be impossible to crystallize. Or, in studying virus-receptor or virus-antibody complexes, it is often necessary to have excess ligand present to assure saturation of all sites on the virus. Another frequent occurrence is that there are two or more different modifications of the sample under study that are difficult to separate.
Figure 5.
Cryo-EM Micrograph of Mature Dengue Virus
Note the many broken particles, indicated by arrows, that can be neglected for an image reconstruction, but that are likely to inhibit crystallization attempts.
A further example of the power of being able to select specific images on a micrograph is in the study of dynamic processes such as stages in virus assembly including DNA packaging of proheads. This process was used in the analysis of dsDNA packaging into the proheads of the small tailed ϕ29 phage (Figure 6 ; Morais et al., 2001, Simpson et al., 2000). The packaging process was stopped by freezing about 2 minutes after initiation. The original micrographs showed roughly two types of particles, those that appeared to be empty and those that appeared to be partially filled. Separate image reconstructions showed not only the partial presence of DNA in the fuller particles, but also a significantly different and larger structure around the unique vertex containing the portal for DNA entry. It was concluded that the additional density is due to the ATPase (gene product 16) known to be essential for DNA packaging. The portal vertex density could be fitted with the crystal structure of the dodecameric “connector” (Simpson et al., 2000), the central component of the DNA-packaging machine, and the difference density of the structural prohead RNA (pRNA; Figure 7 ). The resultant model has provided a hypothesis on how the packaging motor works (Simpson et al., 2000).
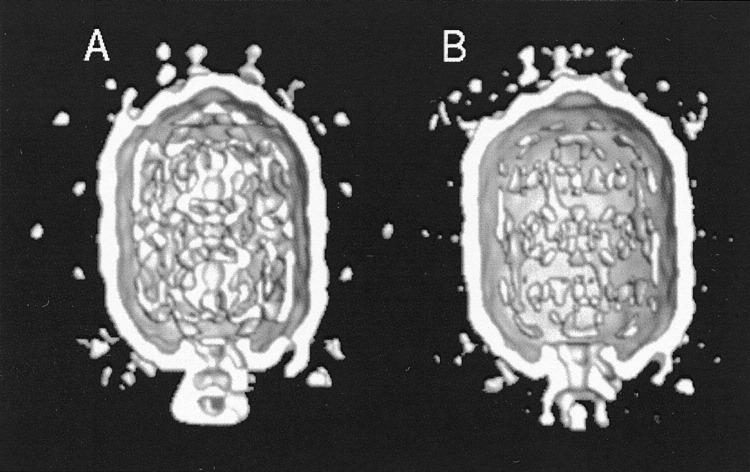
Figure 6.
Cryo-EM Reconstruction of ϕ29 Prohead
The packaging reaction was stopped by freezing 2 minutes after initiation. Particles partially packaged with genomic DNA (A) and empty (B) were selected by eye for each reconstruction from the same micrographs. Note the additional density, around the special pentagonal entry vertex, representing the ATPase (gp16) required to hydrolyze ATP for DNA packaging (Morais et al., 2001; Tao et al., 1998).
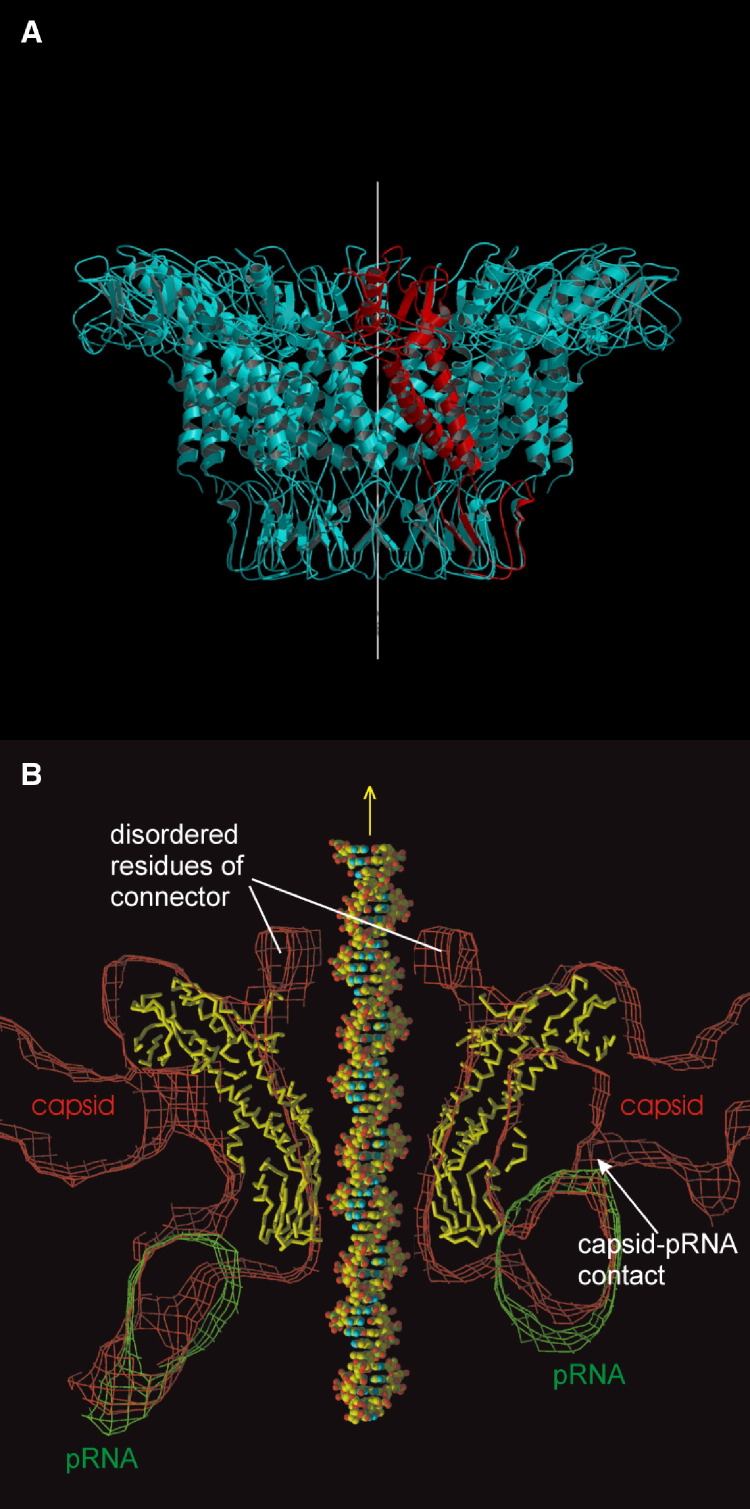
Figure 7.
The Structure of the ϕ29 DNA-Packaging Machine
(A) The crystal structure of the dodecameric connector (blue with one monomer picked out in red). (B) Fit of the Cα trace of the connector (yellow) into the cryo-EM density of the prohead. Shown also is the fit of the pRNA derived from a difference map between prohead and RNAase-treated connector (Morais et al., 2001; Tao et al., 1998). (Reprinted with permission from Simpson et al., 2000, Nature Publishing Group.)
Crystallization requires the presence of a large number of essentially identical particles. Although this is also required for single-particle reconstructions, the tomographic technique does permit the reconstruction of three-dimensional images to low resolution (Baumeister and Steven, 2000, Grunewald et al., 2003). In this technique, the EM grid is exposed to various tilt angles, allowing for the collection of a series of images projected in different directions for the particles on the grid. The limitations are, however, that the exposures have to be few and low dose to avoid excessive radiation damage, resulting in low-resolution reconstructions. Nevertheless, there is promise of three-dimensional analyses of whole cells and pleomorphic, membrane-enveloped viruses, such as influenza or coronaviruses.
Factors that Control Resolution of a Cryo-EM Reconstruction
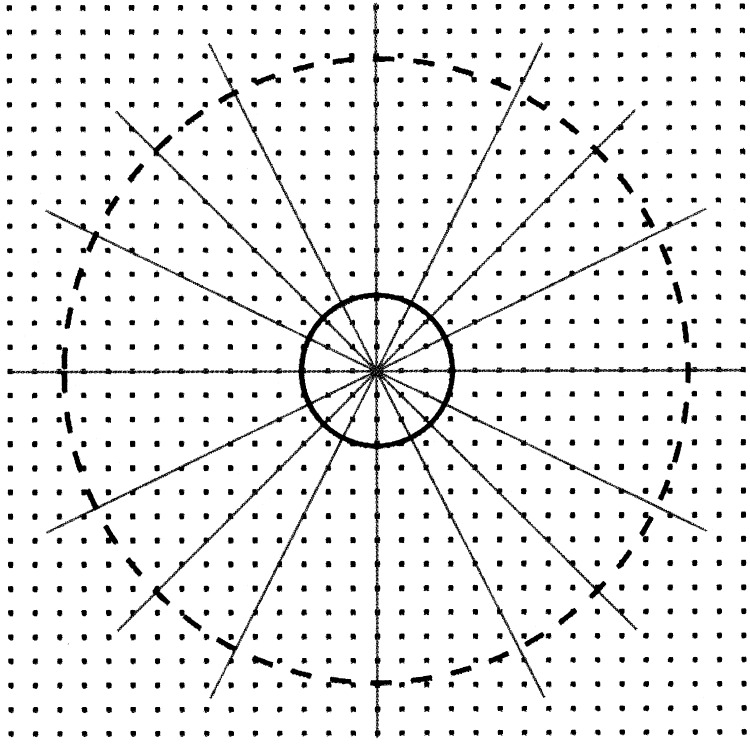
The limit of resolution for which actual data are available on a particular micrograph or for a specific particle can be assessed by looking at the averaged Fourier transformed distribution (van Heel et al., 2000). However, final resolution of a particular reconstruction depends on many factors, including the completeness with which the Fourier transform (reciprocal space) of the reconstructed image is sampled. Each two-dimensional particle image is equivalent to a central section of reciprocal space skewed perpendicular to the direction of projection. Thus, the first few images in random orientation rapidly sample the central, low-resolution volume of reciprocal space. As the number of images is increased, the probability of sampling an extensive volume at higher resolution increases (Figure 8 ). Although higher symmetry implies that fewer images would be required to attain a desired resolution, the larger the particle (corresponding to a smaller reciprocal cell), the greater will be the need for more particles (Rossmann et al., 2001). Another factor that impacts the quality of the reconstructed image is the out-of-focus distance used in recording the micrograph. This distance determines the resolution at which the contrast transfer function has amplitude close to zero. Thus, if all images were taken at the same out-of-focus distance, there would be shells of resolution where there would be few effective data. Hence, it is necessary to combine images taken at various out-of-focus distances. In addition, the higher resolution data can be emphasized by the application of an “inverse temperature factor” correction. Not surprisingly, as resolution limits are being pushed outward, the need for techniques that automatically select particles on micrographs become essential (Glaeser, 2004, Nicolson and Glaeser, 2001, Zhu et al., 2004).
Figure 8.
A Representation of Reciprocal Space with Lines Showing the Random Position of the Projection Planes of Randomly Selected Particle Projections
When relatively few particle images are included in a reconstruction, only a low-resolution region of reciprocal space has moderately complete sampling (inside solid circle). Many more particle orientations are required for a fuller sampling at higher resolution (inside dashed circle).
Given a good sample and the most perfect instrumentation conditions, such as lack of astigmatism, mechanical or magnetic vibrations, thermal motions of the specimen, and more, there are also other factors that determine the quality of the reconstruction. These include the accuracy with which the contrast function is determined, the accuracy with which the relative orientation and position of the particles are determined, the evaluation of the background that underlies every particle image, rejection of poorly formed particle images, and the degree of similarity (homogeneity) of the particles themselves. Where particles have symmetry, it is critical to impose the correct point group, for otherwise the result will be blurred. The reconstruction of the T4 head capsid (Fokine et al., 2004) required 5-fold averaging about the long axis causing the 6-fold symmetric tail to be blurred (Figure 9 ). Similarly, the reconstruction of the ϕ29 prohead, using 5-fold symmetry, showed a good and strong image of the pRNA (Morais et al., 2001, Tao et al., 1998), in contrast to earlier conclusions (Guo et al., 1998).
Figure 9.

Reconstruction of the T4 Head Capsid Using 5-Fold Symmetry
Different proteins are identified by different colors: the major capsid protein, gp23, is blue, the vertex protein is magenta, the highly antigenic outer capsid proteins (hoc) are colored yellow, and the small outer capsid proteins (soc) are colored white. The scale bar represents 100 Å. Note the blurring of the tail, which has 6-fold symmetry (adapted from Fokine et al., 2004).
The Accuracy with which Atomic Structures Can Be Positioned in Cryo-EM Density
Various types of models can be used for establishing the structure associated with a cryo-EM reconstruction. At lower resolution (worse than about 12 Å), it is necessary to interpret the density in terms of the structures of whole proteins or fairly large components of the molecular assembly. As the resolution improves, it is increasingly possible to search for domain repositionings (Zhang et al., 2004) and secondary structural features, such as helices (Jiang et al., 2001) or β sheets (Kong and Ma, 2003), thereby accounting for conformational changes that might occur when component structures are assembled into a complex (Tama et al., 2002).
A variety of criteria can be used when fitting rigid molecular structures into cryo-EM density as implemented in the EMfit program (Rossmann et al., 2001). These need to be suitably weighted to produce a combined overall criterion of best fit. In order to place the different criteria onto equivalent scales, it is convenient to express each measure as a dimensionless quantity representing the number of standard deviations a specific fit is above average. The standard deviations themselves can be determined by analyzing a series of random fits (Rossmann et al., 2001). Examples of different criteria are the mean height of density at their fitted positions, the number of steric clashes between symmetry-related molecules or between different molecules, the number of atoms that are outside the boundary of the available density, and the chemical sense of the interaction between fitted fragments. Other types of information can also be considered, such as the distance between known positions in the density map (e.g., carbohydrate sites found in difference maps; Zhang et al., 2002) and the corresponding residue position in the fitted structure, or the distance between the carboxy-terminal Cα atom of one domain and the amino-terminal Cα atom of the following, independently fitted, domain. The greater the number of consistent restraints, the greater is the probability of an accurate fit. However, as the resolution of the map improves, more reliance can be placed on the shape of the density as implemented in the Situs program (Wriggers et al., 1999).
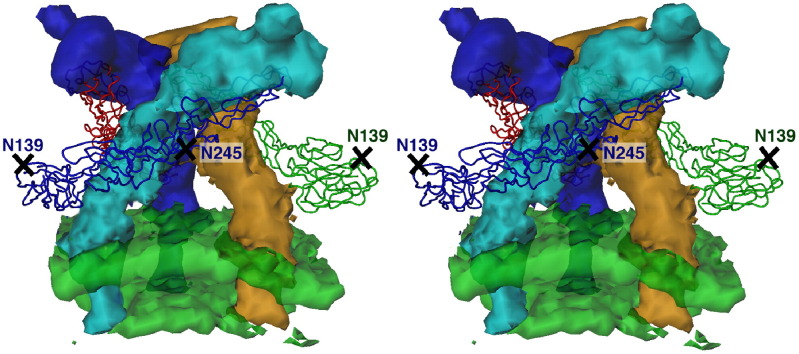
Many larger biological assemblies are composed of a variety of different proteins. The crystal structure of only some of these proteins might be available for fitting into the cryo-EM density of the complex. The boundary between subunits is possibly not well defined in some places at the available resolution. Thus, fitting of the available crystal structures in the absence of others can readily lead to some density at the borderline between the subunits being interpreted by the atoms of the known structure, whereas in reality the density might belong to the neighboring subunit belonging to a protein of a yet undetermined structure. This situation can sometimes be helped by using other indicators or markers, as was the case for separating the density belonging to the known crystal structure of the E1 glycoprotein of Sindbis virus and the density of the unknown crystal structure of the E2 glycoprotein (Figure 10 ; Zhang et al., 2002).
Figure 10.
The Trimeric (E1E2)3 Spike of Sindbis Virus
The known crystal structure of E1 was fitted into the cryo-EM density, assuming 3-fold symmetry, aided by the glycosylation sites at residues Asn139 and Asn245. The carbohydrate positions (crosses) had been determined from difference maps between wild-type and deglycosylated virus. After the E1 molecules had been fitted, the density at all grid points in the map that were within 4 Å of each atom in E1 was set to zero, leaving the density of E2. Hence, the three E2 molecules (cyan, brown, and blue) were shown to be long and thin (Zhang et al., 2002).
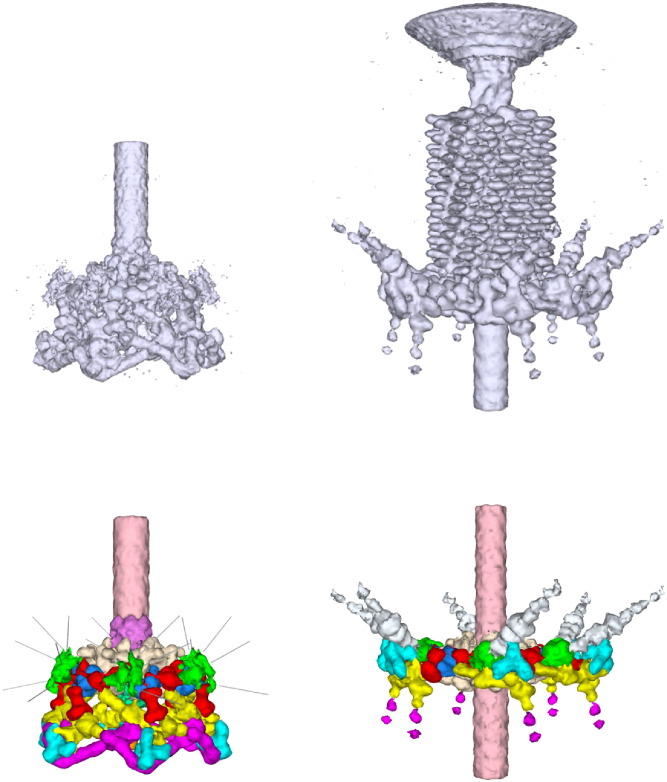
A similar technique was applied to the study of the bacteriophage T4 baseplate (Figure 11 ). This hexagonal structure has a diameter of about 500 Å and a height of 270 Å. The baseplate has a “hexagonal shape” in the infectious virus, but is able to undergo major conformational changes during the process of infecting an E. coli cell, ending up as a “star-shaped” structure. It is composed of about 15 different proteins, each with multiple copies. The crystal structure of six of these proteins is known. The known structures could be readily fitted into the cryo-EM maps and the position and shape of some of the other proteins could be deduced from biochemical and other data (Kostyuchenko et al., 2003, Leiman et al., 2004). It was found that the known crystal structures could be fitted about equally well to both the hexagonal and star-shaped structures, implying that the conformational change is primarily produced by the individual proteins slipping and sliding over each other, without themselves undergoing any major conformational alteration. Although the fitting was fairly straightforward, nevertheless the hybrid technique of combining crystal and cryo-EM data is unlikely to be able to detect small conformational changes in the main chain and especially in side chain structures. Thus, the interaction between the proteins in the baseplate, clearly essential for understanding the baseplate function, remains for now only vaguely known.
Figure 11.
The Cryo-EM Densities (Top) of the Hexagonal (Left) and Star-Shaped (Right) Bacteriophage T4 Baseplate
These densities were interpreted (bottom) by fitting the crystal structures of gene products (gp) 12 (magenta), 11 (blue), 8 (dark blue), 9 (green), 5 (mostly obscured), and 27 (obscured). The position and shape of other proteins (gp 6, 7, 10, 25, 48, 53, and 54) could then be interpreted from biochemical information (Kostyuchenko et al., 2003; Leiman et al., 2004).
Conclusion
The power of combining crystallography with electron microscopy is starting to extend structural knowledge to larger structural assemblies and the dynamic processes that underlie biological functions. Higher resolution cryo-EM results will yield better information at pseudoatomic resolution, while lower resolution cryo-EM results of larger complexes have become an essential tool of cell biology.
Acknowledgments
The structural examples given here are derived from a large number of collaborators, postdoctoral researchers, graduate students, and technical assistants who are recognized by virtue of the quoted references. Similarly, the sources of financial support are numerous, but successive National Institutes of Health grants have supported work on the eukaryotic viruses, while successive National Science Foundation grants and a Human Frontier Science Program grant have supported the bacteriophage studies. We thank Cheryl Towell for her help in preparing this manuscript.
Published: March 8, 2005
References
- Baker T.S., Olson N.H., Fuller S.D. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Steven A.C. Macromolecular electron microscopy in the era of structural genomics. Trends Biochem. Sci. 2000;25:624–631. doi: 10.1016/s0968-0004(00)01720-5. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Adrian M., Chang J.J., Homo J.C., Lepault J., McDowall A.W., Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Eiserling F.A., Black L.W. In: Molecular Biology of Bacteriophage T4. Karam J.D., editor. American Society for Microbiology; Washington, DC: 1994. Pathways in T4 morphogenesis; pp. 209–212. [Google Scholar]
- Fokine A., Chipman P.R., Leiman P.G., Mesyanzhinov V.V., Rao V.B., Rossmann M.G. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser R.M. Historical background: why is it important to improve automated particle selection methods? J. Struct. Biol. 2004;145:15–18. doi: 10.1016/j.jsb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Grunewald K., Medalia O., Gross A., Steven A.C., Baumeister W. Prospects of electron cryotomography to visualize macromolecular complexes inside cellular compartments: implications of crowding. Biophys. Chem. 2003;100:577–591. doi: 10.1016/s0301-4622(02)00307-1. [DOI] [PubMed] [Google Scholar]
- Guo P., Zhang C., Chen C., Garver K., Trottier M. Inter-RNA interaction of phage ϕ29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Henderson R. Realizing the potential of electron cryo-microscopy. Q. Rev. Biophys. 2004;37:3–13. doi: 10.1017/s0033583504003920. [DOI] [PubMed] [Google Scholar]
- Jiang W., Baker M.L., Ludtke S.J., Chiu W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J., Zou J.Y., Kjeldgaard M., Jones T.A. In: International Tables for Crystallography, Volume F, Crystallography of Biological Macromolecules. Rossmann M.G., Arnold E., editors. Kluwer Academic Publishers; Dordrecht/Boston/London: 2001. Around O; pp. 353–356. [Google Scholar]
- Kolatkar P.R., Bella J., Olson N.H., Bator C.M., Baker T.S., Rossmann M.G. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Ma J. A structural-informatics approach for mining β-sheets: locating sheets in intermediate-resolution density maps. J. Mol. Biol. 2003;332:399–413. doi: 10.1016/s0022-2836(03)00859-3. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko V.A., Leiman P.G., Chipman P.R., Kanamaru S., van Raaij M.J., Arisaka F., Mesyanzhinov V.V., Rossmann M.G. Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol. 2003;10:688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G., et al. Structure of the dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman P.G., Kanamaru S., Mesyanzhinov V.V., Arisaka F., Rossmann M.G. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman P.G., Chipman P.R., Kostyuchenko V.A., Mesyanzhinov V.V., Rossmann M.G. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Modis Y., Ogata S., Clements D., Harrison S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 2004;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais M.C., Tao Y., Olson N.H., Grimes S., Jardine P.J., Anderson D.L., Baker T.S., Rossmann M.G. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage ϕ29. J. Struct. Biol. 2001;136:190–200. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson W.V., Glaeser R.M. Review: automatic particle detection in electron microscopy. J. Struct. Biol. 2001;133:90–101. doi: 10.1006/jsbi.2001.4348. [DOI] [PubMed] [Google Scholar]
- Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rodgers D.W. In: International Tables for Crystallography, Volume F, Crystallography of Biological Macromolecules. Rossmann M.G., Arnold E., editors. Kluwer Academic Publishers; Dordrecht/Boston/London: 2001. Cryocrystallography techniques and devices; pp. 202–208. [Google Scholar]
- Roseman A.M. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1332–1340. doi: 10.1107/s0907444900010908. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Bernal R., Pletnev S.V. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- Simpson A.A., Tao Y., Leiman P.G., Badasso M.O., He Y., Jardine P.J., Olson N.H., Morais M.C., Grimes S., Anderson D.L., et al. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F., Wriggers W., Brooks C.L., III. Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- Tao Y., Zhang W. Recent developments in cryo-electron microscopy reconstruction of single particles. Curr. Opin. Struct. Biol. 2000;10:127–136. doi: 10.1016/s0959-440x(00)00139-1. [DOI] [PubMed] [Google Scholar]
- Tao Y., Olson N.H., Xu W., Anderson D.L., Rossmann M.G., Baker T.S. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M., Gowen B., Matadeen R., Orlova E.V., Finn R., Pape T., Cohen D., Stark H., Schmidt R., Schatz M. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Volkmann N., Hanein D. Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol. 2003;374:204–225. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
- Wriggers W., Chacón P. Modeling tricks and fitting techniques for multiresolution structures. Structure. 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- Wriggers W., Milligan R.A., McCammon J.A. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–189. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Zhang W., Mukhopadhyay S., Pletnev S.V., Baker T.S., Kuhn R.J., Rossmann M.G. Placement of the structural proteins in Sindbis virus. J. Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chipman P.R., Corver J., Johnson P.R., Zhang Y., Mukhopadhyay S., Baker T.S., Strauss J.H., Rossmann M.G., Kuhn R.J. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang W., Ogata S., Clements D., Strauss J.H., Baker T.S., Kuhn R.J., Rossmann M.G. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Carragher B., Glaeser R.M., Fellmann D., Bajaj C., Bern M., Mouche F., de Haas F., Hall R.J., Kriegman D.J., et al. Automatic particle selection: results of a comparative study. J. Struct. Biol. 2004;145:3–14. doi: 10.1016/j.jsb.2003.09.033. [DOI] [PubMed] [Google Scholar]