Abstract
A new assay, composed of the NGEN RVA (Nanogen, Inc., San Diego, CA; Prodesse, Inc., Waukesha, WI), which is a pair of analyte-specific reagents that allow the multiplex reverse transcriptase polymerase chain reaction (RT-PCR) and electronic microarray detection of influenza virus A and B, respiratory syncytial virus A and B, and human parainfluenza virus types 1, 2, and 3, was evaluated in comparison with the Hexaplex (Prodesse), a multiplex RT-PCR–enzyme hybridization assay. Comparisons included the detection of respiratory viruses from whole-virus stocks (ATCC) and from frozen pediatric respiratory specimens collected at Children's Hospital of Wisconsin between 1991 and October 1998. After the retesting of six indeterminants and 20 discrepants, overall agreement improved to 96% on the positives and 100% on negatives, with only eight specimens still discrepant. The RVA reagents allow a rapid, sensitive, and specific assay for detecting seven of the most common respiratory viruses in children.
Introduction
Many microbial agents cause upper and lower respiratory infections (LRI) (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18), including a growing list of viruses. However, the majority of identified causes of LRI in children and viral LRI in adults are caused by 8 to 10 viruses. The seven most common viruses are: influenza virus A and B, respiratory syncytial virus (RSV) A and B, and human parainfluenza virus (HPIV) types 1, 2, and 3 (8, 9, 15, 17). Human metapneumovirus (A and B), adenoviruses (51 serotypes, subspecies A to F), human coronaviruses (229E, OC43, NL63, and others), and rhinoviruses (>100 serotypes) also cause LRI in children and adults, but their exact proportion of illness is still being determined (9, 10, 11, 12, 13, 15, 16, 17, 18, 19).
Molecular detection of common community-acquired respiratory viruses has been widely accepted as the “gold standard” in terms of sensitivity and specificity compared to cell culture and rapid antigen methods (4, 8, 13, 15, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31). One of the barriers to acceptance of molecular tests has been their high costs, in terms of both reagents and technician time. In addition, LRI caused by these different respiratory viruses cannot be differentiated reliably by clinical examination (1, 2, 3, 4, 8, 9, 15, 32, 33). Multiplex RT-PCR-enzyme hybridization assay (EHA) detection of the seven most common respiratory viruses (Hexaplex; Prodesse, Inc., Waukesha, WI) has been used widely for accurate, rapid, and cost-effective diagnosis of hospitalized children, adults, and immunocompromised patients (8, 15, 20, 21, 22, 23, 24, 25, 26, 27).
Analyte-specific reagents (ASRs) are reagents that laboratories can use to develop and validate their own assays. We used the NGEN RVA ASRs (Nanogen, Inc., San Diego, CA) and an electronic microarray (NanoChip 400) as an assay method in our laboratory to detect and differentiate the amplified products from the same primer sequences in the field-tested multiplex RT-PCR primer mixture used in the Hexaplex assay. This microarray detection platform may offer advantages to mid- and high-throughput laboratories by decreasing technician time and overall assay time. We compared the RVA chip (and NanoChip 400 machine) to EHA as detection methods for multiplex RT-PCR in the detection of influenza virus A and B, RSV A and B, and HPIV-1, -2, and -3.
Materials and methods
Specimens
Respiratory specimens were collected from patients admitted to Children's Hospital of Wisconsin from 1991 through October 1998 and have been described previously (8, 20, 33, 34). Cell culture, enzyme immunosorbent assay, direct-immunofluorescence assay, and Hexaplex testing for respiratory viruses were previously carried out and have been partially reported (8, 20, 33).
RT-PCR
Two-step RT-PCR was carried out following the manufacturer's directions (Prodesse). The cDNA product of this reaction was split and used in two separate multiplex PCRs. One reaction used the Hexaplex “supermix” for the assay, while the other used a similar RVA primer mix with the 3′ primers lacking biotinylation.
EHA detection
Following amplification, 50 μl of material (amplified with Hexaplex supermix) underwent post-PCR purification using the QIAquick DNA purification kit (Qiagen, Chatsworth, CA) (23). Following purification, 5 μl of amplified material was used in an EHA using 96-well neutravidin-coated microtiter plates.
Electronic microarray detection (NanoChip 400 system)
Following amplification, 9 μl of the amplified material was diluted in 63 μl of CAPdown sample buffer A and placed on the NanoChip 400 instrument; no post-PCR processing (desalting or denaturing) was required. The automated detection of the respiratory viruses was carried out on the NanoChip electronic microarray in three distinct steps, which are depicted in Fig. 1 .
Figure 1.
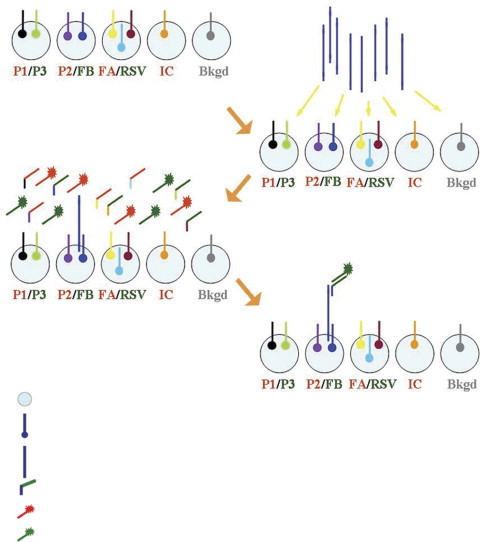
Schematic layout for the detection of seven respiratory viruses on the NanoChip electronic microarray.
Results
Analytical studies
Serial dilutions of ATCC strains of the seven respiratory viruses were tested in both the Hexaplex and RVA assays to determine the limits of detection (LOD) using whole virus (Table 1 ). The Hexaplex demonstrated better overall analytical sensitivity for virus detection than the RVA. The two assays were within 1 log unit of each other for five of the seven viruses. The assays appeared equal at 10−2 50% tissue culture infective dose (TCID50)/ml for HPIV-2, while the RVA assay was more sensitive for influenza virus B at 10−1 TCID50/ml compared to 100 for the Hexaplex. The biggest difference in analytical sensitivity was observed with HPIV-3, where the Hexaplex was 3 log units more sensitive.
Table 1.
Limits of detection of multiplex RT-PCR–EHA (Hex) and multiplex RT-PCR–electronic miroarray (RVA) in serial 10-fold dilutions of ATCC whole virus (TCID50)a
| HPIV-1 |
HPIV-2 |
HPIV-3 |
Influenza virus A |
Influenza virus B |
RSVA |
RSVB |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hexb |
RVAc |
Hex |
RVA |
Hex |
RVA |
Hex |
RVA |
Hex |
RVA |
Hex |
RVA |
Hex |
RVA |
|
| TCID50/ml | ODd | SNe | OD | SN | OD | SN | OD | SN | OD | SN | OD | SN | OD | SN |
| 1 × 104 | 4.000 | 43.9 | 4.000 | 34.5 | 4.000 | 32.7 | 4.000 | 52.6 | 4.000 | 32.5 | N/A | N/A | 4.000 | 22.2 |
| 1 × 103 | 4.000 | 42.6 | 4.000 | 38.9 | 4.000 | 14.4 | 4.000 | 36.7 | 4.000 | 29.2 | 4.000 | 34.4 | 2.601 | 22.9 |
| 1 × 102 | 4.000 | 40.1 | 4.000 | 33.1 | 3.0737 | 4.1 | 3.241 | 8.7 | 4.000 | 26.4 | 4.000 | 35.9 | 3.231 | 27.1 |
| 1 × 101 | 4.000 | 34.8 | 3.773 | 33.5 | 0.526 | 2.0 | 2.686 | 2.6 | 4.000 | 27.1 | 4.000 | 48.9 | 1.672 | 24.8 |
| 1 × 100 | 4.000 | 12.8 | 3.662 | 28.1 | 1.189 | 0.9 | 0.888 | 1.2 | 1.652 | 12.6 | 4.000 | 43.4 | 1.187 | 4.0 |
| 1 × 10−1 | 2.782 | 4.4 | 1.496 | 10.8 | 1.849 | 0.8 | 0.047 | 1.3 | 0.072 | 5.8 | 4.000 | 2.6 | 0.793 | 0.8 |
| 1 × 10−2 | 1.428 | 1.4 | 1.928 | 6.2 | 0.046 | 1.0 | 0.052 | 1.1 | 0.069 | 0.9 | 2.099 | 1.2 | 0.506 | 1.1 |
Positives are in italics.
For Hexaplex, an optical density reading of <0.300 was considered negative.
For RVA, a signal-to-background ratio of <2.3 was considered negative.
OD, optical density.
SN, signal-to-noise ratio.
Clinical studies
A total of 424 respiratory specimens (212 patients) were tested, with nasopharyngeal swab specimens (312 samples) accounting for 73%. Other specimens tested included 16 bronchoalveolar lavage, 6 tracheal, 4 sputum, 4 throat, 9 nasal wash, and 17 miscellaneous respiratory samples. There were 169 samples that tested positive by both assays and 20 samples that were positive by only one assay. There were 113 samples originally reported to be positive by Hexaplex that were negative by both Hexaplex and RVA on retesting, and these were called negative. They were presumed to be low-copy-number samples where the RNA had degraded over time. Three samples that were previously negative by Hexaplex and upon retesting were positive by Hexaplex and negative by RVA were defined as negatives (false positives on Hexaplex). Finally, four samples that were previously positive for one virus by Hexaplex were positive on retesting for a different virus by Hexaplex and were negative by RVA. These samples were not evaluated further and were not included in the calculations.
The agreement between the Hexaplex and RVA can be seen in Table 2 . Overall agreement was high (90%; 95% confidence interval [CI], 85–94) and was nearly 100% for the negative samples. The lowest agreement was 69% for HPIV-2-positive samples. This did not correlate with the analytical sensitivity (Table 1). Repeat testing of the amplified PCR product because of positive control failure on one run resulted in three RVA negative samples being determined to be two positives and one indeterminate for HPIV-2. Out of 2,520 analytes tested with the RVA, there were 5 indeterminants (0.2%) and 1 indeterminant Hexaplex out of 2,940 (0.03%) tested. Repeat testing of these samples demonstrated 3 out of 5 RVA indeterminants and the one Hexaplex indeterminant were positive (Table 2, Table 3 ). An influenza virus B RVA indeterminate became discrepant and went from 2.3 to 2.0 signal-to-noise (SN) ratio, and an HPIV-2 sample went from 2.3 to 1.8 SN ratio on retesting, while the Hexaplex results remained positive and went from optical density (OD) readings of 1.2 to 1.6 and 1.6 to 0.9, respectively. The RSV Hexaplex-indeterminate sample was positive on Hexaplex retesting, while the RVA result remained negative. Thus, retesting of the indeterminants resulted in three new discrepants, which were not further tested.
Table 2.
Agreement between multiplex RT-PCR–EHA (Hexaplex) and multiplex RT-PCR–electronic microarray (RVA) in the detection of respiratory viruses in children
| Sample no. | % Agreement on positives | % Agreement on negatives | Ind.a | Dis.b | |
|---|---|---|---|---|---|
| HPIV-1 | 34 | 100 (90–100)c | 100 (98–100) | ||
| HPIV-2 | 15 | 69 (39–100) | 100 (99–100) | 2 | 6 |
| HPIV-3 | 34 | 82 (65–93) | 100 (99–100) | 6 | |
| Influenza virus A | 33 | 88 (72–97) | 100 (99–100) | 1 | 4 |
| Influenza virus B | 29 | 97 (82–100) | 100 (99–100) | 1 | 1 |
| RSV | 44 | 91 (79–98) | 100 (99–100) | 2 | 3 |
| Total | 189 | 90 (85–94) | 100 (99–100) | 6 | 20 |
Table 3.
Agreement after the retesting of 20 discrepant samples
| Sample nos. | % agreement on positives | % agreement on negatives | Ind.a | Dis.b | |
|---|---|---|---|---|---|
| HPIV-1 | 34 | 100 (90–100)c | 100 (99–100) | ||
| HPIV-2 | 16 | 75 (48–93) | 100 (99–100) | 4 | |
| HPIV-3 | 34 | 100 (90–100) | 100 (99–100) | ||
| Influenza A | 34 | 97 (85–100) | 100 (99–100) | 1 | |
| Influenza B | 30 | 93 (78–99) | 100 (99–100) | 2 | |
| RSV | 45 | 98 (88–100) | 100 (99–100) | 1 | 1 |
| Total | 193 | 96 (92–98) | 100 (99–100) | 1 | 8 |
Ind., indeterminates.
Dis., discrepants.
95% confidence interval.
There were 20 samples with originally discrepant results (18 negative RVA; 2 negative Hexaplex). However, on repeat amplification of frozen RNA from each sample using either the standard Hexaplex primer mix or the RVA primer mix, one sample was resolved to be a Hexaplex false positive, 13 of the remaining 19 were now positive, 1 was indeterminate, and 5 were negative. Of the 13 positives, 2 were now positive on Hexaplex and 11 on RVA. Three (9%) out of 33 experimental runs of the RVA assay accounted for 12 of 18 (67%) RVA discrepants, suggesting a specific technical error, not a pervasive one. After repeat testing, 12 of 18 RVA false negatives and 2 of 2 of the Hexaplex false negatives were resolved. No further analysis was done on the remaining five unresolved discrepant results because of their low number. Testing of reagents and machinery did not elucidate the reason for the above-mentioned technical error. The agreement between the two assays is shown in Table 3 after retesting (discrepant analysis). Now the agreement between the two assays on all seven viruses was very high (96% [95% CI, 92–98] and 100% [95% CI, 99–100]).
Cost comparison and work flow
Comparison of the number of hours of technician time and reagents needed to test 6, 14, and 30 clinical samples, including 2 positive controls (i.e., 8, 16, and 32 samples), in the two multiplex RT-PCR assays (Hexaplex [EHA] and RVA) is shown in Table 4 . Costs for RNA isolation were kept the same for the two assays (manual spin columns), and all reagent costs were the same, except for the detection chemistry. The cost of equipment is not included (e.g., NanoChip 400). Although automated extraction would save considerably on technician time, it adds to start-up costs and reagent costs. The costs for automated NA extraction (not including the extractor) was estimated for each assay.
Table 4.
Cost comparison of two “in-house” multiplex RT-PCR assays to detect seven common respiratory viruses
| Manual extraction |
Automated extraction |
||||||
|---|---|---|---|---|---|---|---|
| Sample no. | Parameter | RVA | Hexaplexa | Hexaplexb | RVA | Hexaplexa | Hexaplexb |
| 6 (8)c | Reagent costs | 428c | 358 | 321 | 448 | 378 | 341 |
| Tech time d (h) | 1.8 | 2.4 | 2.4 | 1.0 | 1.45 | 1.45 | |
| Linear time (h) | 5.8 | 6.5 | 6.5 | 4.75 | 4.95 | 4.95 | |
| Total cost/samplee | 59 | 52 | 48 | 59 | 52 | 47 | |
| 14 (16) | Reagent costs | 813 | 715 | 640 | 853 | 755 | 682 |
| Tech time (h) | 2.45 | 4 | 4 | 1.05 | 2.2 | 2.2 | |
| Linear time (h) | 7.05 | 7.75 | 7.75 | 6.65 | 6.2 | 6.2 | |
| Total cost/sample | 55 | 51 | 46 | 55 | 51 | 46 | |
| 30 (32) | Reagent costs | 1,583 | 1,430 | 1,210 | 1,667 | 1,510 | 1,363 |
| Tech time (h) | 4.65 | 8 | 8 | 1.45 | 4.4 | 4.4 | |
| Linear time (h) | 10.65 | 13.5 | 13.5 | 9.45 | 9.4 | 9.4 | |
| Total cost/sample | 53 | 51 | 44 | 53 | 51 | 46 | |
Hexaplex with manufacturer's costs for all reagents and
Hexaplex with manufacturer's costs for the supermix and
The number in parenthesis includes controls.
Tech time at $25/h.
Costs are in U.S. dollars.
The Hexaplex assay was slightly less expensive over all three sample number scenarios. This difference was small, if manufacturer's retail costs were used for all reagents. The costs ranged from $7 to 9 per analyte tested for both the EHA and electronic microarray formats. As the number of samples increased, the cost decreased by a small amount for both detection methods. However, the RVA assay needed significantly less technician time than the Hexaplex. At higher sample numbers, this was almost half as much time (4.65 h versus 8 h). Estimates comparing manual extraction to automated extraction demonstrated significant reductions in technician time without significant decreases in cost to run the assays. The time to run each assay was cut in almost half, with the RVA assay being able to test 32 samples with only 1.45 h of technician time compared to 4.4 h for the Hexaplex. It is possible that at higher sample numbers (e.g., 80), the electronic microarray could demonstrate higher cost savings.
Discussion
Common community-acquired respiratory viruses cause significant morbidity and mortality in all demographic subsets of our population. The groups at highest risk include preschool age children, the elderly, those with chronic diseases, and the immunocompromised (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,36–38). Rapid, accurate, inexpensive point-of-care testing that is able to detect the majority of respiratory viruses would be the ultimate tool to help physicians diagnose this large group of similarly presenting respiratory pathogens. However, this kind of testing is not currently available, nor is it likely to be within the next 5 years. This need is not unique to respiratory viruses. Many clinical disease states are caused by multiple pathogens where clinical diagnosis cannot accurately distinguish the causative agent (e.g., meningoencephalitis and gastroenteritis). The need is greatest in hospitalized patients, where rapid, accurate, pathogen-specific diagnosis can have the greatest positive effect on patient care and health care costs (39–46).
The molecular diagnosis of viral respiratory infections has become commonplace and widely accepted in major medical centers (8, 9, 15, 18, 19, 24, 25, 26, 28, 29, 30). This acceptance has been partly due to significant evidence of dramatic improvements in sensitivity compared to older methods. The majority of this is based on PCR and RT-PCR technology with different endpoint detection strategies (15, 23, 30,47–52). Although other amplification methods have been explored (e.g., nucleic acid sequence-based amplification), none are widely used in clinical diagnostic laboratories for the detection of respiratory viruses at this time (29,50). Medium to large multiplex detection strategies for six or more pathogens have been reported, employing the EHA in 96-well plates using Hexaplex (23), capillary electrophoresis (30), mass spectrometry (47), Luminex beads (48,52), flowthrough microarrays (49), and now, an electronic microarray. There are currently no FDA-cleared molecular amplification methods for detecting the majority of common respiratory viruses, and all methods represent in-house-developed assays or ASRs. This technology only allows fairly expensive testing in medium to high complexity laboratories.
The Hexaplex assay, the first large multiplex clinical assay of its kind, was introduced almost a decade ago (8, 23). This assay, targeting seven common respiratory viruses (influenza virus A and B, RSV A and B, HPIV-1, -2, and -3), has been widely used and has demonstrated excellent sensitivity and specificity, exceeding older methods (8, 15, 20, 21, 22, 23, 24, 25, 26, 27). The Hexaplex uses EHA in 96-well plates for PCR product detection. This becomes more time-consuming and costly as the number of clinical samples increases above 16 samples. One solution to this would be to automate the PCR product detection part of the assay. Nanogen has developed an electronic-microarray system capable of such automated multiplex PCR product detection (53). The microarray can be used to analyze 80 samples across 1 to 10 runs (data not shown). Prodesse and Nanogen co-developed reagents suitable for use on this platform.
This study was undertaken to demonstrate the agreement between multiplex RT-PCR–EHA (Hexaplex) and the RVA ASRs used with the NanoChip 400 system as an assay method in our laboratory in detecting seven common respiratory viruses in children. Other goals included demonstrating LOD using whole virus and comparing costs and work flows of both assays at different levels of utilization (e.g., high-volume laboratories).
The RVA demonstrated good overall LOD using ATCC organisms, but not as good as the standard Hexaplex. Typically, with single-primer-pair PCR assays, the LOD is 1 TCID50/ml or lower, and five of seven viruses demonstrated this on RVA. The overall agreement with first-pass testing (typical clinical laboratory testing) between the two assays was excellent at 90%, and the pattern of discrepant results suggests that it may be much higher (>96%) if the RVA reagents are used on a regular basis. This was best demonstrated by HPIV-3, where the biggest loss of analytical sensitivity was observed. This did not translate into decreased clinical sensitivity after three of the clinical runs were retested. After retesting, the agreement between the two assays went to 100%. The majority of discrepancy between the two assays was in the detection of HPIV-2. Interestingly, the RVA had good agreement in the analytical studies with the Hexaplex and very low LOD. There did appear to be a correlation between low-positive Hexaplex samples (OD ~1.00 or lower) and negative RVA readings (SN < 2.2), but there were too few samples like this and we were unable to do statistical analysis. At this time, it is unclear why for HPIV-2 the RVA ultimately only had 75% agreement with the Hexaplex, but further clinical testing may clarify this issue. More importantly, agreement on the negatives was almost 100%. This lack of RVA false positives and subsequent high probability that any RVA positives are true positives allows high confidence in a clinical setting. It is unclear why the majority of discrepant RVA samples were in three experimental runs (not counting the HPIV-2 discrepants). The same lots of reagents were used in all runs, and no specific technical error in either the NanoChip 400 instrument or any of the other equipment could be identified. The same reagents and equipment were used to retest the samples that now yielded positive results. After the retesting, there were only 8 out of 420 samples still discrepant between the Hexaplex and the RVA, and 4 of them were HPIV-2.
The cost-benefit and time utilization study demonstrated that the Hexaplex was the least expensive assay to run, even with up to 32 samples per day. This did not change with automated extraction. In addition, there are higher start-up costs associated with the electronic-microarray detection. However, the RVA assay (automated detection) with or without automated extraction demonstrated significant technician time savings, and if the reagents become less expensive, this methodology would have a clear advantage in higher-volume laboratories. With the current pricing structure, whether “cost” or “technician time” is more important would help determine which assay would be most useful.
Conclusions
Clearly, until FDA-cleared point-of-care testing devices are available to detect large numbers of pathogens simultaneously with great speed, and accuracy and at low cost (final goal), we must use current technology for the greatest benefit of patients and society. This includes using in-house-developed tests and ASRs. Technological improvements by academia and industry in three major areas are important stepping stones en route to our final goal. These are (i) improved sample preparation methods (e.g., automation of nucleic acid extraction, miniaturization, and use of microfluidics), (ii) improved amplification methods (e.g., multiplex PCR, and microfluidic directed simultaneous multiple single-target amplification with post-PCR pooling of product prior to detection), and (iii) improved detection of amplified targets (e.g., automation of large multiplex detecting and reporting of results). Currently, PCR-based nucleic acid amplification technology is farthest down this road. However, that does not mean that other amplification methods or even proteomic solutions might not be developed in the future. To be most effective in the detection of respiratory viruses, these methods should use a small clinical sample (<500 μl) and be able to detect many pathogens from that same sample at a concentration of ≤1 TCID50/ml, with reporting times in a few hours.
In this paper, we report the first comparison of two “in-house” assays where the major difference was in their PCR product detection strategies (manual versus automated). Our Hexaplex and RVA “in-house” assays for seven common community-acquired respiratory viruses represent older and newer steps in improving laboratory diagnosis for mostly hospitalized or severely ill patients with LRI or illness caused by these viruses. The Hexaplex was less expensive for small to medium sample numbers and had slightly better analytical and clinical sensitivity. The RVA assay offered increasing cost savings as the number of clinical samples increased or in situations with limited technician time. This automated detection assay demonstrated a high level of agreement with the Hexaplex. The RVA assay needs to be further optimized and validated in clinical situations and with fresh specimens to better establish its performance characteristics, especially for the detection of HPIV-2. However, in this initial clinical trial, it performed very well. Further improvement in its analytical LOD for HPIV-3 may be clinically useful in samples with low copy numbers, as in elderly and immunocompromised patients.
Footnotes
NOTE: No responsibility is assumed by the Publisher for any injury and/or damage to persons or property as a matter of products liability, negligence or otherwise, or from any use or operation of any methods, products, instructions or ideas contained in the material herein. No suggested test or procedure should be carried out unless, in the reader's judgment, its risk is justified. Because of rapid advances in the medical sciences, we recommend that the independent verification of diagnoses and drug doses should be made. Discussions, views and recommendations as to medical procedures, choice of drugs and drug dosages are the responsibility of the authors.
SUBSCRIPTION INFORMATION: Access to full text online is included in the price of individual print subscriptions. Please visit www.cmnewsletter.com. Clinical Microbiology Newsletter (ISSN 0196-4399) is issued twice monthly in one indexed volume per year by Elsevier, 360 Park Avenue South, New York, NY 10010. Subscription price per year: Personal: US$79; Institutions: US$479. Periodical postage paid at New York, NY and at additional mailing offices. Postmaster: Send address changes to Clinical Microbiology Newsletter, Elsevier Science Inc., 360 Park Avenue South, New York, NY 10010. For customer service, phone (212) 633-3950; TOLL-FREE for customers in the United States and Canada: 1-888-4ES-INFO (1888-437-4636) or fax: (212) 633-3860.
Reprints: For copies of 100 more of articles in this publication, please contact the Commercial Reprints Department, Elsevier Science, Inc., 360 Park Avenue South, New York, NY 10010-1710. Tel. (212) 633-3813, Fax: (212) 633-3820, e-mail: reprints@elsevier.com
References
- 1.Denny F.W., Clyde W.A., Jr Acute lower respiratory infections in nonhospitallized children. J. Pediatr. 1986;108:635–646. doi: 10.1016/s0022-3476(86)81034-4. [DOI] [PubMed] [Google Scholar]
- 2.Wrights A.L., Taussig L.M., Ray G. The Tucson children's respiratory study. II. Lower respiratory tract illness in the first year of life. Am. J. Epidemiol. 1989;129:1232–1246. doi: 10.1093/oxfordjournals.aje.a115243. [DOI] [PubMed] [Google Scholar]
- 3.Henrickson K.J. Viral pneumonia. Semin. Pediatr. Infect. Dis. 1998;9:217–233. doi: 10.1016/S1045-1870(98)80035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrickson K.J. Parainfluenza viruses. Clin. Microbiol. Rev. 2003;16:242–264. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleming D.M. 1999. Weekly returns service report for 1998. Birmingham Research Unit of the Royal College of General Practitioners, June, 1–21.
- 6.Dowell S.F. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 1996;174:456–462. doi: 10.1093/infdis/174.3.456. [DOI] [PubMed] [Google Scholar]
- 7.Whimbey E., Englund J.A., Ljungman P. Community respiratory viral infections in the immunocompromised host. Proceedings of a symposium. Am. J. Med. 1997;102:1–80. [Google Scholar]
- 8.Henrickson K.J. National disease burden of respiratory viruses detected in children using PCR. Pediatr. Infect. Dis. J. 2004;23:S11–S18. doi: 10.1097/01.inf.0000108188.37237.48. [DOI] [PubMed] [Google Scholar]
- 9.National Institute of Allergy and Infectious Diseases Community-acquired pneumonia in adult and elderly populations. Clinician. 2000;17:1–27. [Google Scholar]
- 10.Osterhaus A., Fouchier R. Human metapneumovirus in the community. Lancet. 2003;361:890–891. doi: 10.1016/S0140-6736(03)12785-7. [DOI] [PubMed] [Google Scholar]
- 11.Falsey A.R. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- 12.Drazen J.M. SARS — looking back over the first 100 days. J. Med. 2003;349:319–320. doi: 10.1056/NEJMp038118. [DOI] [PubMed] [Google Scholar]
- 13.Aberle S.W. Adenovirus DNA in serum of children hospitalized due to an acute respiratory adenovirus infection. J. Infect. Dis. 2003;187:311–314. doi: 10.1086/367808. [DOI] [PubMed] [Google Scholar]
- 14.Fisher R.G. Twenty years of outpatient respiratory syncytial virus infection: a framework for vaccine efficacy trials. Pediatrics. 1997;99:E7. doi: 10.1542/peds.99.2.e7. [DOI] [PubMed] [Google Scholar]
- 15.Weinberg G.A. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J. Infect. Dis. 2004;189:706–710. doi: 10.1086/381456. [DOI] [PubMed] [Google Scholar]
- 16.Hayden F.G. Rhinovirus and the lower respiratory tract. Rev. Med. Virol. 2004;17:17–31. doi: 10.1002/rmv.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marston B.J. Incidence of community-acquired pneumonia requiring hospitalization. Results of a population-based active surveillance study in Ohio. Arch. Intern. Med. 1997;157:1709–1718. [PubMed] [Google Scholar]
- 18.Williams J.V. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Hoek L. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2:e240. doi: 10.1371/journal.pmed.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kehl S.C., Henrickson K.J. Evaluation of the hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol. 2001;39:1696–1701. doi: 10.1128/JCM.39.5.1696-1701.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covalciuc K.A. Comparison of four clinical specimen types for detection of influenza A and B viruses by optical immunoassay (FLU OIA Test) and cell culture methods. J. Clin. Microbiol. 1999;37:3971–3974. doi: 10.1128/jcm.37.12.3971-3974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peiris M., et al 2001. Evaluation of the new Directigen Flu A and B membrane immunoassay for rapid diagnosis of influenza A and B infections. Presented at the Infectious Disease Society of America Symposium on Molecular Diagnostics, Orlando, FL.
- 23.Fan J., Henrickson K.J., Savatski L.L. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex) Clin. Infect. Dis. 1998;26:397–402. doi: 10.1086/516357. [DOI] [PubMed] [Google Scholar]
- 24.Hindiyeh M., Hillyard D., Carroll K. Evaluation of the Prodesse Hexaplex multiplex PCR assay for direct detection of seven respiratory viruses in clinical specimens. Am. J. Clin. Pathol. 2001;116:218–224. doi: 10.1309/F1R7-XD6T-RN09-1U6L. [DOI] [PubMed] [Google Scholar]
- 25.Liolios L. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J. Clin. Microbiol. 2001;39:2779–2783. doi: 10.1128/JCM.39.8.2779-2783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henrickson K.J., H. Werchau, and A. Rohwedder. 2000. Comparison of hexaplex (multiplex-RT-PCR) and tissue culture (TC) to detect common respiratory viruses in children in Germany during 1999–2000. Presented at the 3rd International Symposium for Influenza and Other Respiratory Viruses, St. Lucia.
- 27.Kehl S., and K.J. Henrickson. 1999. Cost effective utilization of a Multiplex RT-PCR assay (Hexaplex®) for respiratory virus detection at a Children's Hospital. Presented at the 2nd International Symposium for Influenza and Other Respiratory Viruses, Cayman Islands.
- 28.Kuroiwa Y. Comparison of an immunochromatography test with multiplex reverse transcription-PCR for rapid diagnosis of respiratory syncytial virus infections. J. Clin. Microbiol. 2004;42:4812–4814. doi: 10.1128/JCM.42.10.4812-4814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox, J.D. et al. 2002. Monoplex and multiplex Nuclisens basic kit NASBA assays for detection of RNA respiratory virus targets, Abstr. T26. In Proceedings of the 18th Annual Clinical Virology Symposium, Clearwater, FL.
- 30.Erdman D.D. GeneScan reverse transcription-PCR assay for detection of six common respiratory viruses in young children hospitalized with acute respiratory illness. J. Clin. Microbiol. 2003;41:4298–4303. doi: 10.1128/JCM.41.9.4298-4303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberg A. 2001. Incidence and diagnosis of viral respiratory tract infections in a lung transplant population. Presented at the Clinical Virology Symposium, Clearwater, FL.
- 32.Monto A.S. Occurrence of respiratory virus: time, place and person. Infect. Dis. J. 2004;23(Suppl):S58–S64. doi: 10.1097/01.inf.0000108193.91607.34. [DOI] [PubMed] [Google Scholar]
- 33.Monto A.S. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 2000;160:3243–3247. doi: 10.1001/archinte.160.21.3243. [DOI] [PubMed] [Google Scholar]
- 34.Henrickson K.J., Kuhn S.M., Savatski L.L. Epidemiology and cost of infection with human parainfluenza virus types 1 and 2 in young children. Clin. Infect. Dis. 1994;18:770–779. doi: 10.1093/clinids/18.5.770. [DOI] [PubMed] [Google Scholar]
Uncited reference
- Additional references cited in the text and original data and material can be found at: http://www.mcw.edu/display/router.asp?docid=18660.


