Abstract
Pteropine orthoreovirus (PRV) is an emerging bat-borne human pathogen causing severe respiratory illness. To date, however, the evaluation of PRV virulence has largely depended on the limited numbers of clinical cases owing to the lack of animal models. To develop an in vivo model of PRV infection, an inbred C3H mouse strain was infected intranasally with pathogenic PRV strain Miyazaki-Bali/2007. C3H mice suffered severe lung infection with significant body weight reduction and died within 7 days after intranasal infection. Infectious viruses were isolated mainly from the lungs and trachea. Histopathological examination revealed interstitial pneumonia with monocytes infiltration. Following repeated intranasal infection, mice developed antibodies to particular structural and non-structural proteins of PRV. The results of these immunological assays will help to develop laboratory protocols for sero-epidemiological studies. Our small rodent model of lethal respiratory infection will further allow investigation of the molecular mechanisms underlying the high pathogenicity of PRV.
Keywords: Reoviridae, Zoonosis, Animal model, Pathogenesis, Respiratory disease
Highlights
-
•
A lethal PRV strain Miyazaki-Bali/2007 murine infection model was established.
-
•
Susceptibility of different mouse strains to PRV infection was investigated.
-
•
Antibody responses to PRV proteins in C3H mice post intranasal infection were studied.
1. Introduction
Pteropine orthoreovirus (PRV) is a member of Family Reoviridae, genus orthoreovirus. A prototype strain of PRV was first isolated from flying fox (Pteropus poliocephalus) in Nelson Bay, Australia in 1968 (Gard and Compans, 1970, Gard and Marshall, 1973). PRV had been a sole member comprising the PRV group for a considerable period until a second case of the PRV strain Pulau was reported from bat species in Malaysia in 2006 (Pritchard et al., 2006). The first human case of PRV infection by the PRV strain Melaka was reported in Malaysia in 2007 from a patient with high fever and respiratory disease (Chua et al., 2007). Since the first case of PRV infection in humans, evidence of PRV infections in humans (Cheng et al., 2009, Chua et al., 2007, Chua et al., 2008, Chua et al., 2011, Voon et al., 2015, Wong et al., 2012, Yamanaka et al., 2014) and bats (Du et al., 2010, Hu et al., 2014, Lorusso et al., 2015) in Association of Southeast Asian Nations (ASEAN) countries and China have been reported. Phylogenetic studies showed there were genetically close relationships between PRV strains of human and bat origin implying that PRVs are bat-borne zoonoses (Chua et al., 2007). In particular, one of the PRV-infected patients had a history of close contact with bats before the onset of illness (Chua et al., 2007). This epidemiological situation supports the hypothesis that patients might obtain infection from bats; however, there has been no direct evidence to show connections between bat and human regarding PRV transmission.
Sero-epidemiological studies reported that the sera from 12 of 272 patients who visited hospitals in Vietnam and 14 of 109 healthy volunteers in Malaysia were sero-positive to PRV (Chua et al., 2007, Singh et al., 2015). Furthermore, a recent epidemiological study based on PCR detected the PRV genome from 34 of 200 patients with acute upper respiratory tract infection indicating that PRV can be considered as a common pathogen in ASEAN countries causing mild respiratory infections (Voon et al., 2015). Taken together, these reports suggest that PRV has evolved to cross the species barrier between bats and humans.
Commercially small animal models including mice and Guinea pigs are commonly utilized as primary disease models for infectious diseases. Previously, studies of the pathogenicity of PRV infection in humans have been evaluated based on the limited numbers of human infection cases owing to the lack of experimental animal models. Experimental infections of PRV were limited to a primary trial using PRV strain Nelson Bay (Gard and Marshall, 1973) inoculated to suckling mice, which were killed by intra-cranial infection. The experiment was, however, obviously dissociated from spontaneous respiratory infection in humans as there were no reports of PRV infection in the central nervous system. However, since the first report of a human case of PRV infection, no animal models for PRV infection reflecting human respiratory infections have been developed.
Here we report a lethal lung infection model of PRV strain Miyazaki-Bali/2007 (MB) isolated from a patient with acute respiratory infection (Yamanaka et al., 2014) in adult, immunocompetent inbred mice. We identified significant differences in the susceptibility to PRV infections between mouse strains. Intranasal infection caused lethal outcome with severe pneumonia whereas oral infections was not established, suggesting that PRV is an air-borne transmissible pathogen. In addition, mice repeatedly infected with PRV developed antibodies to particular viral proteins. Such serologic information will contribute to the development of protocols for sero-epidemiological studies and sero-diagnosis. Furthermore, an animal model for pathogenic PRV infection provides a crucial opportunity to understand virus pathogenesis and to evaluate vaccines and therapeutics to combat this important pathogen.
2. Materials and methods
2.1. Cell and viruses
Mouse fibroblast L929, lung adenocarcinoma A549, and rhesus monkey kidney MA104 cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) (Nacalai Tesque) supplemented with 5% fetal bovine serum (Gibco). The quail fibrosarcoma QT6 cell line was grown in DMEM supplemented with 10% fetal calf serum. PRV strain MB (Yamanaka et al., 2014) was amplified in L929 cells and approximately 10-fold concentrated virus stock was generated using a 10% polyethylene glycol concentration method as described elsewhere (Lewis and Metcalf, 1988). Infectious virus titer was determined by a plaque assay (Kawagishi et al., 2016). Avian reovirus strain 58–132 (Takase et al., 1985), simian rotavirus strain SA11 (Taniguchi et al., 1994), and mammalian orthoreovirus prototype strain T1L were amplified in QT6, MA104, and L929 cells, respectively.
2.2. Fluorescent-focus assay (FFA)
Viruses were inoculated into A549 cells. After absorbance at 37 °C for 1 h, culture medium was replaced with fresh medium and incubated at 37 °C. At 16 h post infection, cells were fixed with 100% ethanol and infected cells were visualized by immunostaining using antiserum against PRV strain MB σC at a dilution of 1:2000, incubated at 37 °C for 60 min, followed by Alexa Flour 488 conjugated anti-mouse IgG second antibody at a dilution of 1:3000 (Invitrogen).
2.3. Antibodies
Polyclonal rabbit anti-σC antiserum was prepared (Sigma Aldrich) against purified His-tagged PRV strain MB σC protein obtained in bacterial culture. In brief, the strain MB σC gene was inserted into the pTrcHis-A plasmid (Thermo Fisher Scientific), which enabled expression of a 6 × His tagged recombinant protein. The plasmid construct was used for transformation of Escherichia coli strain BL21 (Thermo Fisher Scientific). Protein expression was induced by adding 1 mM isopropyl β-D-1-thiogalactopyranoside. The bacterial pellet was lysed by Bugbuster® (Millipore) and target proteins were purified using a His select nickel affinity gel (Sigma Aldrich) according to manufacturer instruction.
2.4. Experimental infection of animals
We purchased four-week old, male C3H/HeNCrl (C3H) mice from Charles River Laboratories and four-week old, male Jcl:ICR (ICR), Balb/cAjcl (Balb/c), and C57BL/6JJcl (C57BL) mice from Japan CLEA. Animals were inoculated intranasally or orally with 20 or 100 μl virus, respectively. To obtain the Kaplan-Meier survival curve, animals were observed up to 22 dpi. Body weight was recorded every 1–2 days. Surviving C3H mice after intranasal infection of PRV strain MB (2 × 105 PFU) were mixed and again infected intranasally with 2 × 106 PFU, up to a total 2 or 4 infections to obtain anti-sera.
To define the extent of viral replication in mouse organs, animals were sacrificed at 2, 4, and 6 days after intranasal infection. Organs were disrupted by repeated freeze-thaw cycles (2 times) followed by homogenization using a bead homogenizer (BeadSmash 12, WAKEN BTECH Co. Ltd.). Serum was separated from the whole blood by clotting at 4 °C and centrifugation at 3000 × g. Virus titers in organs and sera were determined by plaque assay. For pathological study, C3H mice infected intranasally with 2 × 105 PFU of PRV strain MB were sacrificed at day 4. Organs were fixed in 10% buffered formaldehyde and processed for pathological examination including HE staining and IHC analysis using σC-specific antiserum.
The study was approved by the Animal Research Committee of the Research Institute for Microbial Diseases, Osaka University. The experiment was conducted following the guidelines for the Care and Use of Laboratory Animals of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
2.5. Cytokine detection
Inflammatory cytokine IP-10 levels in blood were examined using a Quantikine ELISA kit (R&D Systems) according to manufacturer instruction.
2.6. Viral neutralization (NT) assay
Neutralizing antibody titers of the C3H sera were examined by the neutralization assay. In brief, serially diluted sera (1:20 to 1:5120) in DMEM)) were prepared in a 96-well microplate. An equal volume of virus fluid (50–100 FFU/25 μl) was added and incubated at 37 °C for 60 min in a 5% CO2 incubator. As a positive control, viruses were mixed with an equal volume of DMEM. After incubation, 50 μl neutralized viruses were inoculated onto a monolayer of A549 in a 96-well microplate. After incubation for 16 h, cells were fixed with absolute ethanol and cells expressing viral antigens were detected using murine anti-PRV strain MB σC antibody (Kawagishi et al., 2016) and goat anti-mouse IgG antibody Alexa488 conjugate. The NT titer was indicated by the reciprocal value of the maximum dilution at which the number of foci showed 50% reduction compared with the positive control.
NT assays against MRV, RV, and ARV were performed similarly although virus titration was measured by plaque assay. Briefly, MRV, RV, and ARV incubated with serially diluted sera were inoculated to L929, MA104, or QT6 cells, respectively, and overlaid with agarose gel. Plaque numbers were compared to control samples and the neutralization titer of 50% reduction was calculated. NT titers were expressed as the mean (95% confidence interval (CI)). 95% CI was calculated according to the following formula:
where SD is standard deviation and n is sample number.
2.7. Plasmid construction
For immunological assay, expression plasmid vectors for N-terminal FLAG-tagged PRV proteins were constructed. Genes encoding each viral protein, including λA, λB, λC, µA, µB, µNS, σA, σB, σC, σNS, p17, and p10, were amplified by RT-PCR using specific primers based on sequences deposited in the GenBank (accession numbers: AB908278 to AB908287). Amplified PCR products fused to the FLAG epitope tag at the N-terminus were cloned into the pEF plasmid vector (Mizushima and Nagata, 1990). All plasmids were confirmed by DNA sequencing. Primer sequences used for plasmid construction are available upon request. The native p10 protein is known to be degraded rapidly after synthesis (Shmulevitz et al., 2004). Thus, a V11R mutant with a Val to Arg amino acid substitution at position 11 was generated to achieve increased expression level of p10 (Cheng et al., 2005).
2.8. Immunoblotting
Viral genes for each PRV protein were amplified by PCR using specific primers and inserted into pCAG plasmid expression vectors, which enable the expression of HA-tagged PRV proteins in mammalian cells. The expression plasmids were transfected into 293T cells. After 24 h post infection, cells were lysed by 1% sodium dodecyl sulfate (SDS) or fixed by 100% ethanol for WB or FFA, respectively. For WB, samples were applied to 10% SDS-polyacrylamide gel electrophoresis (PAGE) except for p17 and p10 viral proteins, which were applied to 12% SDS-PAGE. After blotting to a membrane, viral proteins were detected using 1:2000 diluted antisera to PRV developed in C3H by 4-times repeated intranasal infection. For FFA, 293T cells were transfected with HA-tagged PRV proteins and fixed by 10% formalin at 24 h post transfection. Viral proteins were visualized by 1:2000 diluted C3H mouse serum and an anti-HA antibody developed in rabbit followed by anti-mouse IgG-Alexa488 conjugate and anti-rabbit IgG-Alexa594, respectively.
3. Results
3.1. PRV strain MB produces lethal infection by intranasal inoculation in C3H mice
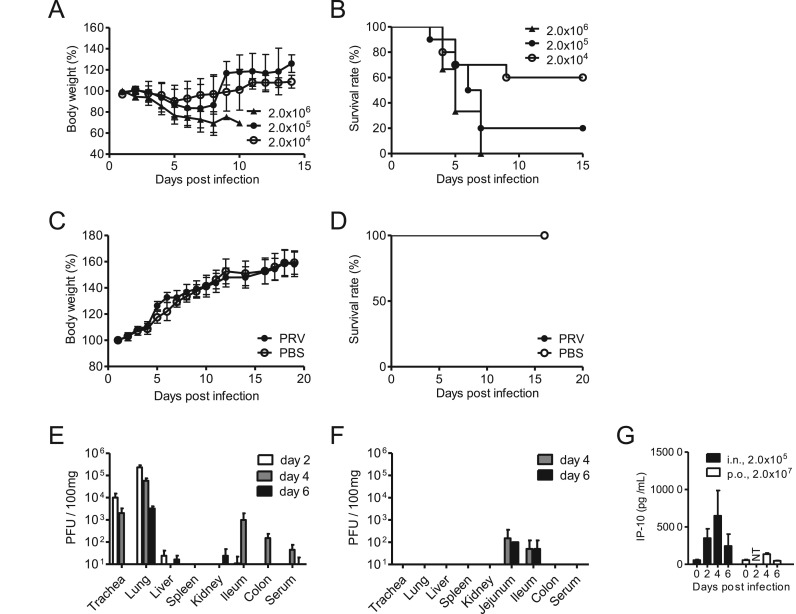
PRV infections are accompanied with severe respiratory symptoms in humans (Tan et al., 2017), suggesting that infection by PRV likely occurs mainly via the intranasal route by air-borne virus particles. To develop a mouse model for respiratory PRV infection, inbred C3H mice (4-week-old, male) were infected with pathogenic PRV strain MB (Yamanaka et al., 2014) via the intranasal route (2.0 × 104, 2.0 × 105, and 2.0 × 106 PFU/head). Mice showed body weight decrease at 3 days post-infection (dpi) and the first deaths occurred at 3 dpi ( Fig. 1A and B). The mouse group receiving the highest infection dose (2.0 × 106 PFU/head) led to 100% mortality followed by 2.0 × 105 (80%) and 2.0 × 104 (40%) (Fig. 1B). In contrast, in the group challenged orally with the virus at the maximum dose (2.0 × 107 PFU), all mice survived with no body weight reduction (Fig. 1C and D).
Fig. 1.
Lethal infection of PRV in C3H mice. (A-D) Four-week old, male C3H mice were infected with PRV strain MB at 2 × 104, 2 × 105, and 2 × 106 PFU via the intranasal route (A, B) or 2 × 107 PFU via the oral route (C, D). Body weight (A, C) and survival rate (B, D) were recorded daily. (E-G) C3H mice were infected with PRV strain MB via the (E) intranasal route (2 × 105 PFU /head) or (F) oral route (2.0 × 107 PFU / head). At day 2, 4, and 6 post infection, animals were sacrificed and infectious viral titers in internal organs were examined. (G) Induction of blood cytokines (IP-10) through the time course of PRV infection. Data are expressed as the means ± SD, n = 6–10. NT: not tested.
Next, the dynamics of virus distribution was investigated following intranasal infection of 80% lethal dose (2.0 × 105 PFU) of PRV strain MB in C3H mice. As expected from the clinical symptoms of PRV infection in humans, respiratory organs constituted the major sites of virus propagation at all time points (Fig. 1E). At 2 dpi, infectious viruses were detected in the lungs (2.3× 105 PFU/100 mg), trachea (1.0 × 104 PFU/100 mg), and liver (2.3 × 101 PFU/100 mg). Especially in the lungs, high titers of PRV were detected throughout the infection (Fig. 1E). In the liver, small numbers of viruses near detection limits were detected on 2 and 6 dpi. Only a limited quantity of infectious viruses (5.0 × 101 PFU/100 mg) were found in the ileum by oral infection (Fig. 1F), suggesting that oral PRV infection leads to inefficient viral spread in the gastrointestinal system.
To assess inflammatory cytokine/chemokine induction after PRV infection as a measure of immune response and symptom generation, we examined the levels of interferon induced protein-10 (IP-10), an inflammatory chemokine induced by various virus infections. The level of blood IP-10 in animals infected intranasally peaked at 4 dpi, followed by subsequent decrease on day 6. In the mice orally infected with PRV, blood IP-10 increased slightly at 4 dpi but soon decreased to the baseline level at 6 dpi (Fig. 1G). The level of blood IP-10 was consistent with viral load in the mice intranasally or orally infected with PRV, suggesting that IP-10 may serve as an index to predict acute infection of PRV.
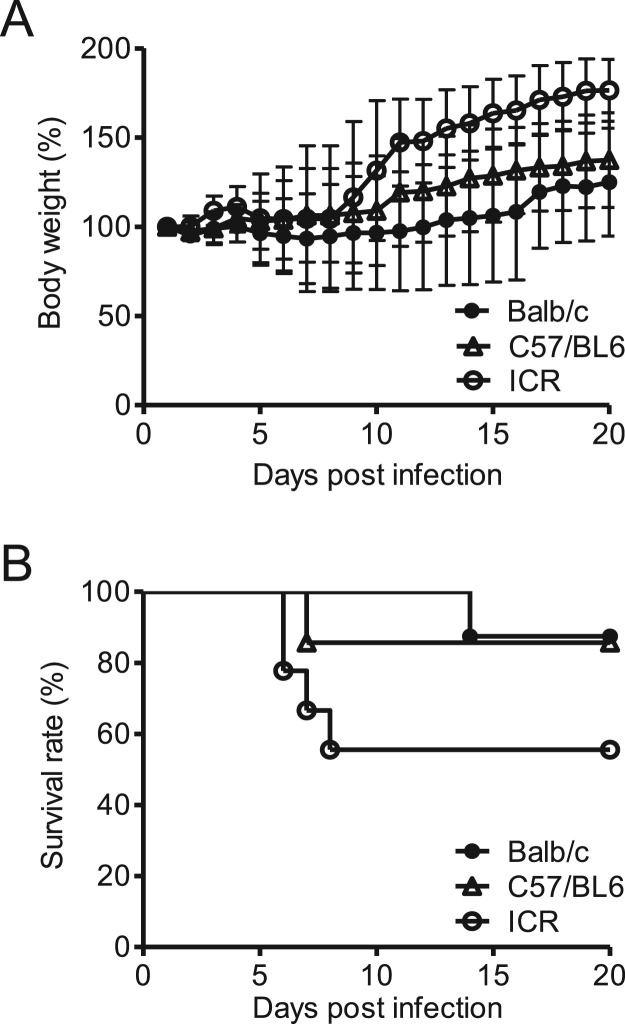
We also investigated the sensitivity of other mouse strains including Balb/c, C57/BL6, and ICR to PRV infection. Balb/c, C57/BL6, and ICR mice were infected with 80% lethal dose for C3H mice (2.0 × 105 PFU) via the intranasal route. Among the three strains, ICR mice were the most sensitive to PRV infection, with relatively high mortality (44.4%) and slight body weight reduction ( Fig. 2A and B). Although Balb/c and C57BL mice exhibited greater reduction in body weight than ICR mice, the mortality of these two mice strains (Balb/c, 12.5% and C57BL, 14.3%) were lower than that of C3H and ICR mice. These data indicated that the C3H mouse strain is the most sensitive to PRV infection among the four mice strains tested and constitutes a suitable animal model to investigate the pathogenicity of respiratory PRV infection.
Fig. 2.
Pathogenicity of PRV in small rodent models. Balb/c, C57/BL6, and ICR mice (4-wk old, male) were infected with 2 × 106 PFU of PRV strain MB intranasally. Body weight (A) and survival rate (B) were recorded daily. Data are expressed as the means ± SD, n = 8.
3.2. Pathological examination in mice infected with PRV
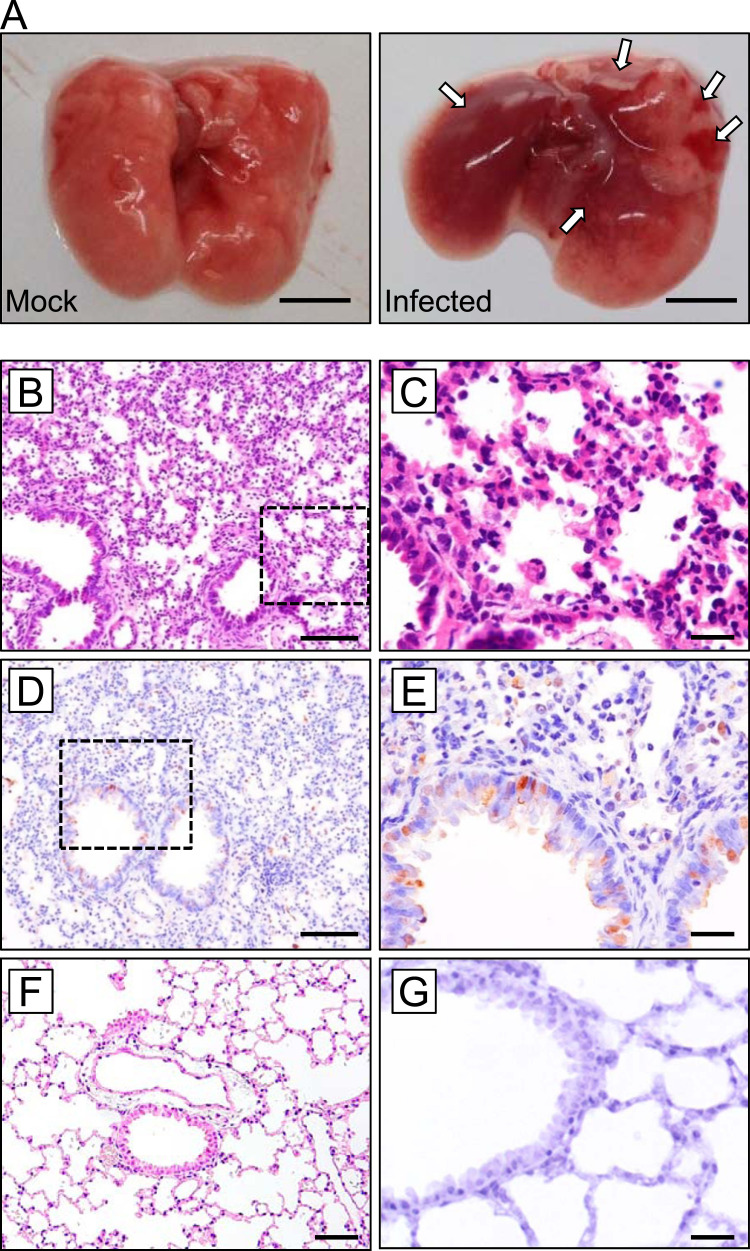
Histo-pathological examination was conducted on the lungs of C3H mice infected intranasally with 2.0 × 105 PFU of PRV strain MB. At 4 dpi, mice were sacrificed and the trachea, lung, small intestine, large intestine, and spleen were sampled. The lungs from infected mice demonstrated focal hemorrhage over the entire organ ( Fig. 3A). Hematoxylin and eosin (HE) staining revealed interstitial pneumonia, with deciduation of bronchial epithelial cells and thickening of the intra-alveolar septum accompanied with infiltration of inflammatory monocytes (Fig. 3B and C). Immunohistochemical (IHC) analyses using σC-specific antiserum revealed the cell tropism of PRV infection. Viral antigens were found in tracheal epithelial cells and alveolar cells, which appeared to comprise type II alveolar epithelial cells (Fig. 3D and E). No positive signals were seen in uninfected control mouse samples, demonstrating the specificity of the antibody used in IHC examinations (Fig. 3F and G). Previous studies demonstrate that PRV strain MB induces syncytium formation in infected cultured cells (Yamanaka et al., 2014, Kawagishi et al., 2016). However, despite extensive examination of HE-stained and IHC-stained samples, no sign of cell-cell fusion was observed in the lung sections.
Fig. 3.
Pathological examination of the C3H mouse infected with PRV (A) The lungs of C3H mice (4 days after infection): Left: mock infected; Right: infected with PRV strain MB. Arrow indicates hemorrhagic region. Scale bars indicate 5 mm. (B, C) Lungs of C3H mouse at 3 dpi were subjected to pathological examination by HE staining and (D, E) IHC using anti-PRV σC antibody. Higher magnification of insets in B and D are shown in C and E, respectively. (F, G) HE and IHC examination of an uninfected control mouse lung section; Original magnification, B, D, and F: 200 ×; C, E, and G: 400 ×. Scale bars indicate 100 µm (B, D, and F) or 20 µm (C, E, and G).
3.3. Humoral immune responses in mice infected with PRV
To assess anti-PRV humoral immune responses, we first investigated neutralizing antibody activity induced in mice infected with PRV strain MB. C3H mice that survived after intranasal infection of 2.0 × 105 PFU were further challenged by intranasal infection of 2.0 × 106 PFU for 1 or 3 times with 2 week intervals (total of 2 or 4 infections). At two weeks after the final infection, infected animals were sacrificed and sera were separated. As our previous study indicated that the PRV cell attachment protein σC is required for entry into A549 cells but not for entry into L929 cells (Kawagishi et al., 2016), we performed NT assay using A549 and L929 cell lines. Mice with intranasal infection of PRV induced specific neutralizing antibodies against PRV when the NT assay was conducted in A549 cells ( Table 1), whereas NT titer was 80 times lower when the experiment was conducted in L929 cells. The results suggest that A549 is a candidate cell line appropriate for NT assays for PRV infection.
Table 1.
Neutralization activity (mean (95% CI)) of anti-PRV serum against Reoviridae viruses.
| PRV MB (in A549) | PRV MB (in L929) | MRV | ARV | RV | |
|---|---|---|---|---|---|
| C3H, MB × 2 (n = 4) | 2880 (1301–3189) | 70.0 (50.4–89.6) | < 20 | < 20 | < 20 |
| C3H, MB × 4 (n = 3) | 8533 (5187–11878) | 113 (0–316) | < 20 | < 20 | < 20 |
MRV: Mammalian orthoreovirus.
ARV: Avian orthoreovirus.
RV: Rotavirus.
We investigated the cross-NT activity between PRV and related viruses of the family Reoviridae. MRV and RV are common human pathogens and the majority of adults possess antibodies to MRV and RV. NT assay using anti-PRV serum against MRV and RV demonstrated that the NT activity of anti-PRV serum was specific to PRV but did not neutralize MRV and RV. Anti-PRV serum also did not neutralize ARV, which is a member of family Reoviridae, indicating that PRV is serologically independent from the common human and animal reoviruses (Table 1). These results of NT tests suggest that antibodies to MRV and RV will not contribute to protection against PRV infection.
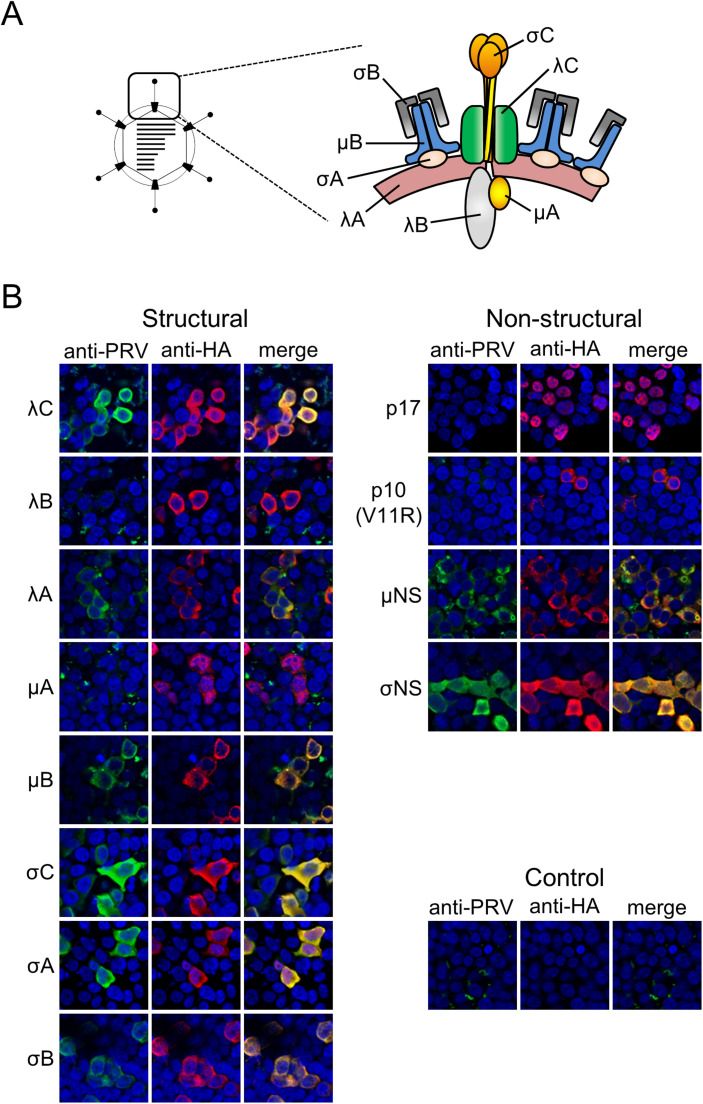
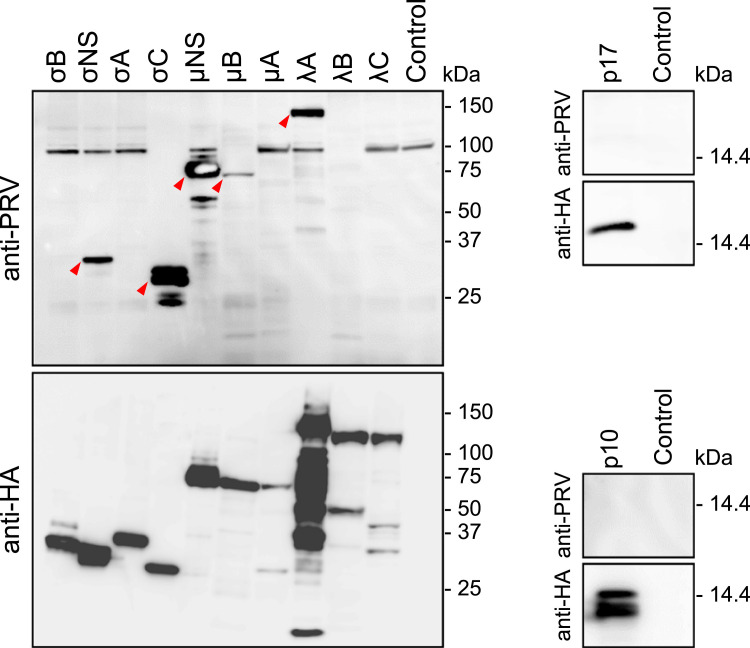
Finally, we examined induced antibodies against PRV proteins in mice generated by intranasal PRV infection. Specific antibody reactions to PRV antigens were firstly investigated by FFA using cells expressing PRV structural and nonstructural proteins. In FFA, there were clear reactions to all the outer capsid proteins (λC, μB, σC, and σB), two inner capsid proteins (λA and σA), and two nonstructural proteins (μNS and σNS), which are components of viral inclusion body ( Fig. 4 and Table 2). Production of antibodies against PRV proteins was further confirmed by western blotting (WB). In WB, mouse serum reacted to fewer numbers of viral proteins compared to in the FFA assay, including μB, σC, λA, μNS, and σNS ( Fig. 5 and Table 2). These results suggest that an immunofluorescence assay using cells infected with PRV or expressing particular PRV proteins may represent an appropriate method for sero-epidemiological studies of PRV.
Fig. 4.
Reactivity of anti-PRV serum to viral proteins. (A) (left) Schematic image of the PRV virion. (right) Predicted structural model of PRV particles depicting the location of structural proteins based on cryoelectron microcopy images (Dryden et al., 1993a, Dryden et al., 1993b, Trask et al., 2012). (B) Anti-PRV serum after 4-time intranasal infections of PRV strain MB was used for FFA against HA-tagged recombinant PRV proteins expressed in 293T cells. Anti-HA antibody was used as a positive control. Alexa-488 or Alexa-594 conjugated secondary antibodies were used to detect anti-PRV or anti-HA antibodies, respectively. Mutant p10 protein (V11R mutant) was used instead of wild-type p10 for efficient detection.
Table 2.
Reactivity of C3H mouse serum to PRV proteinsa.
| Gene segment | Protein | Mass (kDa) | Location in virion | WB | FFAb | |
|---|---|---|---|---|---|---|
| L1 | λC | 141.5 | outer | – | + | 16,000 |
| L2 | λB | 140.1 | inner | – | – | < 250 |
| L3 | λA | 141.7 | inner | + | + | 8000 |
| M1 | μA | 81.6 | inner | – | – | < 250 |
| M2 | μB | 72.5 | outer | + | + | 16,000 |
| M3 | μNS | 69.7 | NS | + | + | 8000 |
| S1 | σC | 35.3 | outer | + | + | 32,000 |
| S1 | p17 | 16.3 | NS | – | – | <250 |
| S1 | p10 | 10.4 | NS | – | – | <250 |
| S2 | σA | 46.7 | inner | – | + | 32,000 |
| S3 | σNS | 39.9 | NS | + | + | 32,000 |
| S4 | σB | 40.1 | outer | – | + | 2000 |
WB: western blotting.
FFA: fluorescent-focus assay.
NS: non-structural.
C3H mouse serum infected intranasally with PRV strain MB for a total of 4 times. 1:2000 diluted serum was used.
Maximum dilution at positive signal.
Fig. 5.
WB analysis of anti-PRV serum against viral proteins. Four-week old, male C3H mice were infected with 2 × 104 PFU of PRV strain MB, followed by 3-time intranasal infection of 2 × 105 PFU of PRV at 2 week intervals. Animals were sacrificed at 2 weeks after the last infection and sera were separated. WB analysis against HA-tagged recombinant viral proteins expressed in 293T cells was performed.
4. Discussion
The development of an animal model would facilitate the study of PRV pathogenicity. Here, we established a small animal model of lethal PRV lung infection using mature, immuno-competent inbred C3H mice (Fig. 1). The phylogenetic analysis implied that genetically related PRV strains are transmitted from bat to human; however, there is no direct evidence of the precise transmission route of PRV. Thus, to understand the transmission of PRV, different infection routes; intranasal and oral, were tested.
Subsequent examination of virus distribution after intranasal infection and pathological examination confirmed that PRV preferentially infects respiratory organs. Severe damage in respiratory organs after intranasal infection observed in mice indicated that PRV infection in mouse is similar to the clinical cases of PRV infection in human (Fig. 1, Fig. 3). Together, the evidence indicates that PRV is an air-borne transmissible disease and the low level of viremia in PRV-infected mice might suggest that the virus is not transmitted by an arthropod vector.
Although human PRV infection cases exclusively exhibited respiratory symptoms, PRV isolates from bats were detected from various samples including saliva, feces (Lorusso et al., 2015), blood (Gard and Marshall, 1973), urine (Pritchard et al., 2006), lung homogenate (Du et al., 2010), and intestinal contents (Hu et al., 2014), indicating that the pathology of PRV infection in bat is dissimilar to human infection. However, information regarding the tissue tropism of PRV in bat is not available. Bats are known to be naturel reservoirs of numerous zoonotic viruses including highly pathogenic viruses, filoviruses, rabies viruses, Nipahviruses, and coronaviruses (Calisher et al., 2006). The importance of bats as reservoirs of zoonotic viruses can be characterized by the feature that bats often permit persistent infection of highly pathogenic viruses without presenting symptoms. As it is not known whether PRV causes pathogenic, respiratory infection in bats, it could not be concluded from this study whether the C3H mouse model reflects PRV infection in bats. Further pathological studies regarding PRV infection in bats would be required to better understand PRV spread and transmission in nature.
Viral infections induce inflammatory cytokines and chemokines that have two opposite roles; host defense and symptom formation, and thus serve as important indicators to predict virus infections (Grebely et al., 2013, Guidotti and Chisari, 2000, Melchjorsen et al., 2003). However, there are no reports regarding cytokine induction after PRV infection. In particular, the inflammatory chemokine IP-10 is induced by various virus infections and recruits activated T-cells to the site of inflammation (Liu et al., 2011). In the current study we found that the levels in blood IP-10 correlated with viral load in the mice intranasally or orally infected with PRV, suggesting that IP-10 serve as an index to predict acute PRV infection.
Among the four mouse strains examined in the current study, the C3H strain was the most sensitive to PRV infection (Fig. 1). In other virus studies, C3H was also reported to be highly sensitive to Sendai virus and measles virus infections (Neighbour et al., 1978, Niewiesk et al., 1993, Parker et al., 1978), whereas this mouse strain is resistant to murine hepatitis virus and murine cytomegalovirus (Allan and Shellam, 1984, Bang and Warwick, 1960). The mechanisms underlying different susceptibilities to these viruses have been considered to be due to the different immune responses or presence of virus binding receptors. Although there was no evidence to explain the highest susceptibility of C3H mice to PRV infection, further analysis regarding the different susceptibilities among the mouse strains (i.e., C3H vs ICR, Balb/c, C57BL/6) would help to understand the molecular mechanisms of PRV pathogenicity, in particular how the genetic background of the host influences the outcome of PRV replication and pathogenesis. It should be noted that only young (4-week-old) male mice were included in this study, although PRV was detected exclusively in adult patients (26–54 years old) (Cheng et al., 2009, Chua et al., 2007, Chua et al., 2008, Chua et al., 2011, Wong et al., 2012, Yamanaka et al., 2014). Further, it should be noted that 4-week-old mice are not immunologically mature (Jackson et al., 2017). To explore the association between the mouse model and natural infection, further studies are required employing female mice or mice of a wider age range. No sign of cell-cell fusion in infected lung tissues was observed. These results suggest that PRV strain MB does not induce syncytium formation in vivo, or that the disruption of damaged cells makes it difficult to identify cell-cell fusion in the targeted tissues.
Using C3H mice sera infected with PRV, serological examination was conducted to investigate whether PRV antibodies contribute to protection against genetically related Reoviridae viruses including ARV, MRV, and RV. The results of NT tests demonstrated that PRV are serologically independent from these viruses. Although ARV-, MRV-, and RV-specific antisera were not tested, investigation regarding whether PRV infection was cross-neutralized by antibodies to other Reoviridae viruses would help to understand the epidemiology of PRV. Furthermore, the significant difference in the NT assay results of PRV between A549 and L929 cells could be explained by the different usage of viral ligands for cell attachment during PRV infection. We have recently demonstrated that PRV utilizes the virus structural protein σC for entry into A549 but not L929 cells (Kawagishi et al., 2016). In the NT assay using A549 cells, antibodies against σC in serum from mice infected with PRV worked effectively as a neutralizing antibody but not in L929 cells. As shown in Fig. 4, Fig. 5, σC comprises one of the major viral proteins that induce specific antibodies. It was thus reasonable that a high amount of neutralizing antibody against σC in infected mice sera caused high NT activity in A549 cells and it could be concluded that A549 cells could therefore be recommended for NT assays in sero-epidemiological studies of PRV.
In comparison, there were significant differences in the reactivity of C3H serum against viral proteins between FFA and WB assays. In general, antibodies that were reactive only in FFA recognize conformational epitopes whereas those reactive only in WB recognize linear epitopes owing to the different methods used in sample preparation. In the present study, mouse serum reacted to λC and σA at quite high titer in the FFA assay but was not reactive in the WB assay, which probably occurred because the antibodies likely recognize only the conformational epitope but not the linear epitope. In FFA and WB assays, a fairly strong reaction was observed to the σC protein. As σC, which is located at the surface of the virion, functions as a cell-attaching viral protein, it is reasonable that antibodies were induced to this protein. σC is known to be highly variable between PRV isolates, probably because neutralizing antibody pressure might drive the rapid mutation of this protein. Owing to the high variation in σC sequences, σC may therefore not represent an appropriate target for sero-epidemiological studies for a broad range of PRV strains but may instead be suitable for detecting specific virus strains.
In contrast, the other viral proteins that induced specific antibodies, including six structural proteins (λC, λA, μB, σC, σA, and σB) and two non-structural proteins (μNS and σNS) are relatively conserved between the PRV isolates reported to date. Notably, although it was anticipated that antibodies were raised preferentially to outer proteins that could readily be exposed to immune cells, the antibodies to the two NS proteins were not expected. μNS and σNS of PRV likely generate viral inclusion bodies (VIBs) during the replication cycle as a common structure in family Reoviridae, although the functions of these proteins are not yet defined. VIBs are a site of virion assembly, which are usually found exclusively in the cytoplasm and not spontaneously released to outside cells. Conversely, our results suggest that VIBs are released outside the cell probably by the disruption of PRV-infected cells. There have been no reports regarding NT activity of antibodies against VIB. Thus, the six structural and two NS proteins constitute candidate viral proteins for sero-epidemiological studies. Especially, λA, μB, μNS, and σNS, which were positive both in FFA and WB assays, are recommended as primary choices for target antigens, although differences in the properties of antibody recognition between host species must be considered (i.e., human patients may develop antibodies to different viral proteins than those recognized in the mouse infection model). In WB analysis, there were discrepancies in the band patterns of λA and σC as revealed by anti-PRV serum and anti-HA antibody. These results indicate that anti-PRV serum recognizes particular sites on each viral protein. Although processing of PRV proteins after synthesis has not been studied, protein degradation or truncated bands revealed by using anti-HA tag antibody may be indicative of specific protein processing important for biological function. Further analysis of the processing of these viral proteins could enable the characterization of each PRV protein.
The high pathogenicity of PRV in human has not been fully investigated owing to the limited number of clinical cases and lack of a suitable animal model. Our established animal model of lethal lung infection of PRV could thus contribute to primary examination of the pathogenicity of PRV strains isolated from humans and bats. Furthermore, we have recently developed a plasmid-based reverse-genetics (RG) system of PRV (Kawagishi et al., 2016) that enabled us to generate mutant viruses affecting virus virulence. Combined studies incorporating the PRV RG system and the lethal mouse model established in the current study will likely facilitate disclosure of the mechanisms underlying the high virulence of PRV and promote the development and evaluation of vaccines and treatments against this pathogenic reovirus.
Acknowledgments
The authors would like to thank Naoko Nagasawa and Misa Onishi for their technical assistance. We also thank Toru Okamoto for technical advice, Kozo Takase for providing avian orthoreovirus, Koki Taniguchi for providing rotavirus, and Terence S. Dermody for providing mammalian orthoreovirus. This work was supported in part by grants-in-aid from the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED) and JSPS KAKENHI Grant Numbers JP17H05814, JP16K19138, and JP26292149.
References
- Allan J.E., Shellam G.R. Genetic control of murine cytomegalovirus infection: virus titres in resistant and susceptible strains of mice. Arch. Virol. 1984;81:139–150. doi: 10.1007/BF01309303. [DOI] [PubMed] [Google Scholar]
- Bang F.B., Warwick A. Mouse macrophages as host cells for the mouse hepatitis virus and the genetic basis of their susceptibility. Proc. Natl. Acad. Sci. USA. 1960;46:1065–1075. doi: 10.1073/pnas.46.8.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Lau C.S., Lai A., Ho E., Leung P., Chan F., Wong A., Lim W. A novel reovirus isolated from a patient with acute respiratory disease. J. Clin. Virol. 2009;45:79–80. doi: 10.1016/j.jcv.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Cheng L.T., Plemper R.K., Compans R.W. Atypical fusion peptide of Nelson Bay virus fusion-associated small transmembrane protein. J. Virol. 2005;79:1853–1860. doi: 10.1128/JVI.79.3.1853-1860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Crameri G., Hyatt A., Yu M., Tompang M.R., Rosli J., McEachern J., Crameri S., Kumarasamy V., Eaton B.T., Wang L.F. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. USA. 2007;104:11424–11429. doi: 10.1073/pnas.0701372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Voon K., Crameri G., Tan H.S., Rosli J., McEachern J.A., Suluraju S., Yu M., Wang L.F. Identification and characterization of a new orthoreovirus from patients with acute respiratory infections. PLoS One. 2008;3:e3803. doi: 10.1371/journal.pone.0003803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Voon K., Yu M., Keniscope C., Abdul Rasid K., Wang L.F. Investigation of a potential zoonotic transmission of orthoreovirus associated with acute influenza-like illness in an adult patient. PLoS One. 2011;6:e25434. doi: 10.1371/journal.pone.0025434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden K.A., Wang G., Yeager M., Nibert M.L., Coombs K.M., Furlong D.B., Fields B.N., Baker T.S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden K.A., Wang G., Yeager M., Nibert M.L., Coombs K.M., Furlong D.B., Fields B.N., Baker T.S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Lu Z., Fan Y., Meng K., Jiang Y., Zhu Y., Wang S., Gu W., Zou X., Tu C. Xi River virus, a new bat reovirus isolated in southern China. Arch. Virol. 2010;155:1295–1299. doi: 10.1007/s00705-010-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard G., Compans R.W. Structure and cytopathic effects of Nelson Bay virus. J. Virol. 1970;6:100–106. doi: 10.1128/jvi.6.1.100-106.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard G.P., Marshall I.D. Nelson Bay virus. A novel reovirus. Arch. Gesamt. Virusforsch. 1973;43:34–42. doi: 10.1007/BF01249346. [DOI] [PubMed] [Google Scholar]
- Grebely J., Feld J.J., Applegate T., Matthews G.V., Hellard M., Sherker A., Petoumenos K., Zang G., Shaw I., Yeung B., George J., Teutsch S., Kaldor J.M., Cherepanov V., Bruneau J., Shoukry N.H., Lloyd A.R., Dore G.J. Plasma interferon-gamma-inducible protein-10 (IP-10) levels during acute hepatitis C virus infection. Hepatology. 2013;57:2124–2134. doi: 10.1002/hep.26263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti L.G., Chisari F.V. Cytokine-mediated control of viral infections. Virology. 2000;273:221–227. doi: 10.1006/viro.2000.0442. [DOI] [PubMed] [Google Scholar]
- Hu T., Qiu W., He B., Zhang Y., Yu J., Liang X., Zhang W., Chen G., Wang Y., Zheng Y., Feng Z., Hu Y., Zhou W., Tu C., Fan Q., Zhang F. Characterization of a novel orthoreovirus isolated from fruit bat, China. BMC Microbiol. 2014;14:293. doi: 10.1186/s12866-014-0293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.J., Andrews N., Ball D., Bellantuono I., Gray J., Hachoumi L., Holmes A., Latcham J., Petrie A., Potter P., Rice A., Ritchie A., Stewart M., Strepka C., Yeoman M., Chapman K. Does age matter? The impact of rodent age on study outcomes. Lab. Anim. 2017;51:160–169. doi: 10.1177/0023677216653984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi T., Kanai Y., Tani H., Shimojima M., Saijo M., Matsuura Y., Kobayashi T. Reverse genetics for fusogenic bat-borne orthoreovirus associated with acute respiratory tract infections in humans: role of outer capsid protein sigmaC in viral replication and pathogenesis. PLoS Pathog. 2016;12:e1005455. doi: 10.1371/journal.ppat.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Guo S., Hibbert J.M., Jain V., Singh N., Wilson N.O., Stiles J.K. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22:121–130. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Teodori L., Leone A., Marcacci M., Mangone I., Orsini M., Capobianco-Dondona A., Camma C., Monaco F., Savini G. A new member of the Pteropine Orthoreovirus species isolated from fruit bats imported to Italy. Infect. Genet. Evol. 2015;30:55–58. doi: 10.1016/j.meegid.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Melchjorsen J., Sorensen L.N., Paludan S.R. Expression and function of chemokines during viral infections: from molecular mechanisms to in vivo function. J. Leukoc. Biol. 2003;74:331–343. doi: 10.1189/jlb.1102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S., Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neighbour P.A., Rager-Zisman B., Bloom B.R. Susceptibility of mice to acute and persistent measles infection. Infect. Immun. 1978;21:764–770. doi: 10.1128/iai.21.3.764-770.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiesk S., Brinckmann U., Bankamp B., Sirak S., Liebert U.G., ter Meulen V. Susceptibility to measles virus-induced encephalitis in mice correlates with impaired antigen presentation to cytotoxic T lymphocytes. J. Virol. 1993;67:75–81. doi: 10.1128/jvi.67.1.75-81.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.C., Whiteman M.D., Richter C.B. Susceptibility of inbred and outbred mouse strains to Sendai virus and prevalence of infection in laboratory rodents. Infect. Immun. 1978;19:123–130. doi: 10.1128/iai.19.1.123-130.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L.I., Chua K.B., Cummins D., Hyatt A., Crameri G., Eaton B.T., Wang L.F. Pulau virus; a new member of the Nelson Bay orthoreovirus species isolated from fruit bats in Malaysia. Arch. Virol. 2006;151:229–239. doi: 10.1007/s00705-005-0644-4. [DOI] [PubMed] [Google Scholar]
- Shmulevitz M., Corcoran J., Salsman J., Duncan R. Cell-cell fusion induced by the avian reovirus membrane fusion protein is regulated by protein degradation. J. Virol. 2004;78:5996–6004. doi: 10.1128/JVI.78.11.5996-6004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Shimojima M., Ngoc T.C., Quoc Huy N.V., Chuong T.X., Le Van A., Saijo M., Yang M., Sugamata M. Serological evidence of human infection with Pteropine orthoreovirus in Central Vietnam. J. Med. Virol. 2015;87:2145–2148. doi: 10.1002/jmv.24274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase K., Nishikawa H., Nonaka F., Yamada S. Pathogenic characteristics of highly virulent avian reovirus, strain 58-132, isolated from a chicken with tenosynovitis. Nihon juigaku zasshi. Jpn. J. Vet. Sci. 1985;47:567–574. doi: 10.1292/jvms1939.47.567. [DOI] [PubMed] [Google Scholar]
- Tan Y.F., Teng C.L., Chua K.B., Voon K. Pteropine orthoreovirus: an important emerging virus causing infectious disease in the tropics? J. Infect. Dev. Ctries. 2017;11:215–219. doi: 10.3855/jidc.9112. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Nishikawa K., Kobayashi N., Urasawa T., Wu H., Gorziglia M., Urasawa S. Differences in plaque size and VP4 sequence found in SA11 virus clones having simian authentic VP4. Virology. 1994;198:325–330. doi: 10.1006/viro.1994.1035. [DOI] [PubMed] [Google Scholar]
- Trask S.D., McDonald S.M., Patton J.T. Structural insights into the coupling of virion assembly and rotavirus replication. Nat. Rev. Microbiol. 2012;10:165–177. doi: 10.1038/nrmicro2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon K., Tan Y.F., Leong P.P., Teng C.L., Gunnasekaran R., Ujang K., Chua K.B., Wang L.F. Pteropine orthoreovirus infection among out-patients with acute upper respiratory tract infection in Malaysia. J. Med. Virol. 2015;87:2149–2153. doi: 10.1002/jmv.24304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.H., Cheng P.K., Lai M.Y., Leung P.C., Wong K.K., Lee W.Y., Lim W.W. Virulence potential of fusogenic orthoreoviruses. Emerg. Infect. Dis. 2012;18:944–948. doi: 10.3201/eid1806.111688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A., Iwakiri A., Yoshikawa T., Sakai K., Singh H., Himeji D., Kikuchi I., Ueda A., Yamamoto S., Miura M., Shioyama Y., Kawano K., Nagaishi T., Saito M., Minomo M., Iwamoto N., Hidaka Y., Sohma H., Kobayashi T., Kanai Y., Kawagishi T., Nagata N., Fukushi S., Mizutani T., Tani H., Taniguchi S., Fukuma A., Shimojima M., Kurane I., Kageyama T., Odagiri T., Saijo M., Morikawa S. Imported case of acute respiratory tract infection associated with a member of species nelson bay orthoreovirus. PLoS One. 2014;9:e92777. doi: 10.1371/journal.pone.0092777. [DOI] [PMC free article] [PubMed] [Google Scholar]